Abstract
Nontypeable Haemophilus influenzae (NTHI) initiates infection by colonizing the upper respiratory tract mucosa. NTHI disease frequently occurs in the context of respiratory tract inflammation, where organisms encounter damaged epithelium and exposed basement membrane. In this study, we examined interactions between the H. influenzae Hap adhesin and selected extracellular matrix proteins. Hap is an autotransporter protein that undergoes autoproteolytic cleavage, with release of the adhesive passenger domain, Haps, from the bacterial cell surface. We found that Hap promotes bacterial adherence to purified fibronectin, laminin, and collagen IV and that Hap-mediated adherence is enhanced by inhibition of autoproteolysis. Adherence is inhibited by pretreatment of bacteria with a polyclonal antiserum recognizing Haps. Purified Haps binds with high affinity to fibronectin, laminin, and collagen IV but not to collagen II. Binding of Haps to fibronectin involves interaction with the 45-kDa gelatin-binding domain but not the 30-kDa heparin-binding domain of fibronectin. Taken together, these observations suggest that interactions between Hap and extracellular matrix proteins may play an important role in NTHI colonization of the respiratory tract.
Extracellular matrix (ECM) consists of a diverse group of proteins that form the scaffolding responsible for the development, growth, and maintenance of mammalian tissues. In epithelial tissues, certain ECM proteins function together with cells to form barriers intended to prevent penetration of these tissues by microorganisms. Both pathogenic and commensal bacteria have evolved mechanisms designed to subvert epithelial barriers (3, 5, 14, 23, 28). These mechanisms include bacterial surface proteins called adhesins that bind to ECM and bacterial proteases that degrade ECM components and permit migration of bacteria to deeper tissue spaces where they may gain easier access to nutrients and safe harbor from the host immune response.
Haemophilus influenzae is a gram-negative bacterium that is often found as a commensal inhabitant of the respiratory tract in healthy adults but also represents a common cause of both localized respiratory tract and invasive systemic disease (25). In studies examining interactions between H. influenzae clinical isolates and human respiratory tract tissue, bacteria were often associated with damaged epithelium and exposed ECM (16, 17). Furthermore, examination of bronchial biopsies from patients with persistent H. influenzae bronchitis revealed organisms in the subepithelial compartment, suggesting that this pathogen is capable of penetrating the basement membrane (11). Additional analysis of interactions between clinical isolates and purified ECM components demonstrated that many H. influenzae strains were capable of binding to fibronectin, laminin, and various collagens (2, 27). In recent work, Virkola and coworkers found that H. influenzae hemagglutinating pili mediated attachment to both fibronectin and heparin-binding growth-associated molecule, although nonpiliated strains were also capable of binding to these proteins (26).
In addition to producing pili, H. influenzae elaborates numerous nonpilus adhesins. One of these adhesins, called Hap, is ubiquitous among both nontypeable (nonencapsulated) and type b encapsulated clinical isolates and promotes binding to a variety of cultured human epithelial cells (20). Hap is a member of the autotransporter family of gram-negative bacterial virulence factors (7). In recent work, we demonstrated that the Hap autotransporter consists of an amino-terminal signal sequence that directs transport across the bacterial inner membrane, a 110-kDa adhesive passenger domain called Haps, and a 45-kDa translocator domain called Hapβ (8). As with other autotransporters, the translocator domain is thought to insert itself into the outer membrane as a pore-forming beta-barrel through which the adhesive passenger domain is extruded to reach the bacterial cell surface. In addition to its adhesive activity, the Haps passenger domain also harbors serine protease activity that directs autoproteolytic cleavage of Haps from Hapβ under certain circumstances. Mutation of the Haps active-site serine residue at position 243 to an alanine (HapS243A) inhibits release of Haps from the bacterial surface and results in increased adherence to epithelial cells.
In the present study, we sought to determine whether Hap is capable of interacting with ECM proteins. In initial experiments, we found that Hap promotes attachment of bacteria to purified fibronectin, laminin, and collagen IV. In addition, we observed that purified Haps passenger domain binds with high affinity to these ECM proteins and that Haps interacts specifically with the 45-kDa gelatin-binding domain of fibronectin. These results provide additional insight into the mechanisms by which H. influenzae colonizes the human respiratory tract.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. H. influenzae strains were grown in brain heart infusion broth supplemented with NAD and hemin (BHIs) or on BHIs agar or chocolate agar as described previously (1) and were stored at −80°C in brain heart infusion broth with 20% glycerol. Tetracycline was used at a concentration of 5 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Reference |
|---|---|---|
| H. influenzae DB117 | Laboratory strain, rec-1, capsule-deficient serotype d | 19 |
| Plasmids | ||
| pGJB103 | E. coli-H. influenzae shuttle vector, Tcr | 24 |
| pJS106 | pGJB103 with a 6.7-kb PstI fragment containing hap | 20 |
| pHapS243A | pJS106 with S243A mutation | 8 |
SEM of bacterial interactions with purified ECM proteins.
Glass coverslips were spotted with 30-μl aliquots of plasma fibronectin, laminin, or collagen IV at 1 mg/ml and were incubated for 15 min at room temperature and then were washed once with phosphate-buffered saline (PBS). Subsequently, the coverslips were added to petri dishes containing suspensions of bacteria that had been incubated in BHIs to late exponential phase (optical density at 600 nm = 0.9) and then chilled to stop growth. Coverslips were incubated while shaking in the bacterial suspensions for 2 h at room temperature, washed once with PBS, and then fixed with glutaraldehyde and prepared for scanning electron microscopy (SEM). Bacterial association was scored by an independent observer.
Quantitative adherence assays.
Adherence assays were performed as described previously (21). Twenty-four-well tissue culture plates coated with human plasma fibronectin, murine laminin, or murine collagen IV (BD Biosciences) were rehydrated in 0.5 ml of minimum essential medium (Sigma), and plates were warmed to 37°C for 1 h in a tissue culture incubator infused with 5% CO2. Percent adherence was calculated by dividing the number of adherent CFU per well by the number of inoculated CFU.
Far-Western dot immunoblots.
Purified human plasma fibronectin, murine laminin, and murine collagen IV were obtained from BD Biosciences. Fibronectin fragments and bovine serum albumin (BSA) were obtained from Sigma. Haps was purified as described previously (8). ECM proteins were spotted in 50-μg quantities into wells of a 96-well dot blot manifold apparatus holding a nitrocellulose membrane. Samples were incubated for 30 min and then pulled through the membrane by vacuum suction. Subsequently, the membrane was blocked in Tris-buffered saline plus 5% skim milk for 1 h. The membrane was then incubated with purified Haps diluted to a final concentration of 10 μg/ml in Tris-buffered saline plus 5% skim milk. Next, the membrane was incubated with anti-Haps antiserum RabK2 (9) diluted 1:1,000 and then a secondary anti-rabbit immunoglobulin G antibody conjugated to horseradish peroxidase (Sigma) diluted 1:10,000. Detection of Haps binding was accomplished by incubating the membrane in a chemiluminescent horseradish peroxidase substrate solution (Pierce, Rockford, Ill.) and exposing the membrane to film.
Enzyme-linked immunosorbent assays (ELISAs).
Collagen II-coated 96-well plates were prepared using purified bovine tracheal collagen II (Sigma) and a protocol provided by the manufacturer. In particular, collagen II was dissolved in ice-cold 0.5 M acetic acid, pH 2.0, to a final concentration of 1 mg/ml. Wells of tissue culture-treated 96-well plates (Costar) were incubated with 40 μl of dissolved collagen at room temperature for 1 h with gentle shaking and were then washed once with PBS. In control plates, coating was confirmed using mouse ascites fluid against collagen II (Sigma), a secondary anti-mouse antibody conjugated to horseradish peroxidase, and a chromogenic horseradish peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.), followed by measurement of absorbance at 650 nm. Ninety-six-well plates coated with fibronectin, laminin, and collagen IV were obtained from BD Biosciences.
Wells were blocked with 200 μl of PBS-1% BSA for 1 h at room temperature and then washed once with PBS. One hundred microliters of purified Haps or purified HifB (22) diluted in PBS plus 1% BSA to final concentrations of 0.5, 1, 5, 10, 50, or 100 nM was added to triplicate wells, and plates were incubated for 1 h at room temperature. Wells were then incubated successively for 1 h with anti-Haps antiserum RabK2 or anti-HifB antiserum Rab452 (22), each diluted 1:1,000 in PBS-1% BSA, and then a secondary anti-rabbit immunoglobulin G antibody conjugated to horseradish peroxidase diluted 1:10,000 in PBS-1% BSA. Wells were washed three times with PBS after each incubation step. After the final wash, binding by Haps or HifB binding was detected by incubating wells with 200 μl of a chromogenic horseradish peroxidase substrate (Kirkegaard & Perry Laboratories) and measurement of absorbance at 650 nm. Data were analyzed using PadPrism software (version 3.0a).
RESULTS
Hap promotes adherence of bacteria to fibronectin, laminin, and collagen IV.
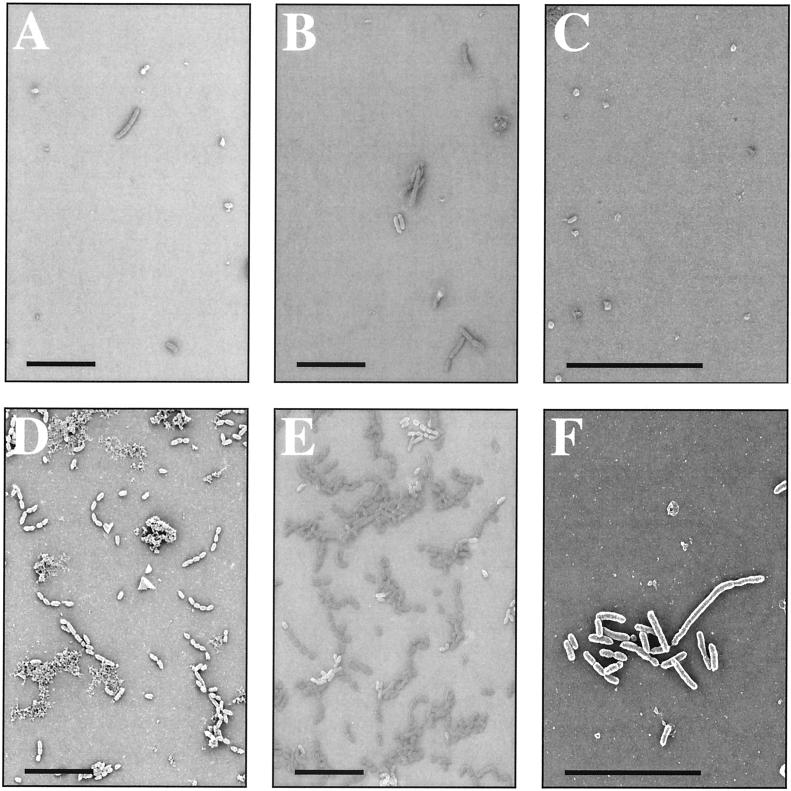
In considering how Hap might contribute to colonization of the human respiratory tract by H. influenzae, we first sought to determine whether expression of Hap could promote binding of live bacteria to purified ECM proteins. In initial experiments we used SEM to examine binding of H. influenzae strains DB117/pHapS243A, which expresses a derivative of Hap that does not undergo autoproteolysis and therefore retains all of the Haps adhesive passenger domain on the bacterial cell surface, and H. influenzae strain DB117/pGJB103 (empty vector). Glass coverslips were spotted in duplicate with aqueous solutions of purified plasma fibronectin, laminin, or collagen IV and then incubated for 2 h with late-exponential-phase bacterial cultures. Assessment of bacterial adherence by DB117/pHapS243A revealed numerous organisms associated in clusters with all ECM proteins (Fig. 1). In contrast, examination of adherence by DB117/pGJB103 revealed very few organisms on any of the coverslips (Fig. 1).
FIG. 1.
SEM of H. influenzae strains DB117/pGJB103 and DB117/pHapS243A after incubation with ECM proteins. Shown are representative fields of SEM samples with DB117 expressing empty vector (top panels) or HapS243A (bottom panels). Suspensions of bacteria were incubated with glass slides previously coated with purified fibronectin (A and D), laminin (B and E), or collagen IV (C and F). Bars correspond to 10 μm.
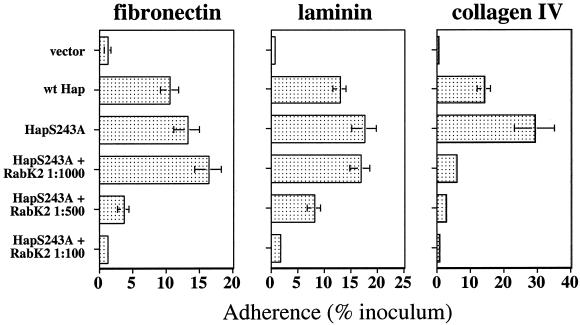
To extend these results, we quantitated Hap-mediated adherence to ECM proteins in standard 30-min adherence assays using tissue culture plates coated with preparations of purified plasma fibronectin, laminin, or collagen IV. As shown in Fig. 2, DB117 expressing wild-type Hap adhered significantly to each of the ECM proteins at levels ranging from 10 to 15% of the inoculum, while DB117 harboring vector alone exhibited minimal adherence. Since processing of the wild-type Hap protein culminates in autoproteolytic cleavage of the Haps adhesive domain from a portion of the precursor population, we next evaluated bacteria expressing HapS243A. As expected, DB117 expressing HapS243A adhered at higher levels to each of the ECM proteins than did bacteria expressing wild-type Hap (Fig. 2), consistent with earlier studies of Hap-mediated adherence to epithelial cells (9).
FIG. 2.
Adherence to ECM proteins by H. influenzae strain DB117 expressing wild-type Hap or HapS243A and inhibition of adherence by anti-Haps antiserum. Adherence to fibronectin, laminin, and collagen IV was calculated by dividing the number of adherent bacteria by the number of inoculated bacteria. Error bars represent the mean ± standard error of the mean of measurements made in triplicate from representative experiments. Adherence was inhibited by increasing concentrations of antiserum RabK2, which reacts with Haps. A 1:100 dilution of preimmune rabbit serum had no effect on adherence. The x axis varies from one panel to the next.
In additional experiments, we examined the ability of RabK2, a polyclonal antiserum raised against purified Haps, to inhibit Hap-mediated adherence to ECM proteins. As shown in Fig. 2, preincubation of DB117/pHapS243A with increasing concentrations of RabK2 resulted in dose-dependent decreases in adherence to each of the ECM proteins. Inhibition by RabK2 was most striking in experiments with collagen IV, in which even a 1:1,000 dilution of antiserum caused marked decreases in adherence relative to the untreated control. In assays with fibronectin and laminin, significant decreases in adherence were observed with 1:500 and 1:100 dilutions of RabK2. In contrast, preincubation of DB117/pHapS243A with a 1:100 dilution of preimmune rabbit serum had no significant effect on adherence (97.0% ± 12.8% of untreated control with fibronectin, 103.2% ± 7.9% of untreated control with laminin, and 104.1% ± 3.6% of untreated control with collagen IV [means ± standard errors of the means]). Taken together, these results strongly suggest that Haps present on the surface of bacterial cells is capable of interacting with fibronectin, laminin, and collagen IV to promote adherence of live organisms to each of these ECM proteins.
Beyond being present in plasma and ECM, fibronectin is associated with the surface of epithelial cells. Accordingly, we wondered whether fibronectin might serve as a receptor in interactions between bacteria expressing Hap and cultured epithelial cells. To explore this possibility, we preincubated confluent monolayers of Chang or A549 epithelial cells with various dilutions of a polyclonal antiserum directed against fibronectin (Sigma) and then inoculated DB117 expressing HapS243A. In uninfected control samples, fluorescence microscopy confirmed antibody binding to cells as large aggregates, suggesting clumps of fibronectin (data not shown). In samples inoculated with DB117/pHapS243A, even with the highest concentration of antifibronectin antiserum, we observed no reduction in adherence (data not shown), suggesting that fibronectin is probably not the receptor for Hap on epithelial cells. Alternatively, cellular fibronectin may be one of several receptors present in quantities large enough that blockage of fibronectin does not interfere with overall adherence levels.
Purified Haps binds to purified fibronectin, laminin, and collagen IV.
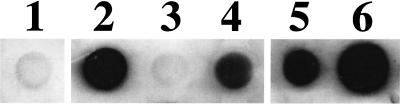
To extend our observations with bacteria expressing Hap, we examined whether purified Haps (the Hap adhesive domain) could interact with fibronectin, laminin, and collagen IV in protein binding assays. Initially, 50-μg samples of these ECM proteins were immobilized on a nitrocellulose membrane, which was then overlaid with purified Haps diluted to a final concentration of 10 μg/ml. As shown in Fig. 3, far-Western immunoblot assays demonstrated binding of Haps to fibronectin, laminin, and collagen IV. No binding of Haps was detected to a control spot containing 50 μg of BSA. Since many fibronectin-binding proteins are known to interact with specific domains of fibronectin, we prepared two additional spots containing purified fibronectin proteolytic fragments (Sigma), one representing the 30-kDa heparin-binding domain and the other representing the 45-kDa gelatin-binding domain. In this experiment, Haps bound to the 45-kDa fragment but not the 30-kDa fragment, suggesting that Hap interacts specifically with the fibronectin gelatin-binding region (Fig. 3).
FIG. 3.
Far-Western dot immunoblot of purified Haps binding to ECM proteins. Samples were loaded with 50 μg of purified protein as follows: lane 1, BSA; lane 2, fibronectin; lane 3, fibronectin 30-kDa heparin-binding fragment; lane 4, fibronectin 45-kDa gelatin-binding fragment; lane 5, laminin; lane 6, collagen IV. Immunoblotting was performed with antiserum RabK2 after incubation of the membrane with purified Haps diluted to 10 μg/ml.
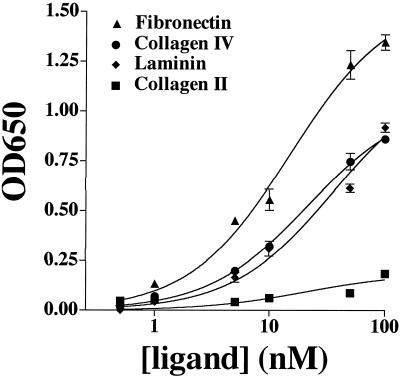
Finally, we attempted to quantitate the strength of Haps binding interactions with ECM proteins by ELISA. Samples of purified Haps at concentrations ranging from 0.5 to 100 nM were added to wells of 96-well plates coated with plasma fibronectin, laminin, collagen IV, or collagen II, and binding was detected with a polyclonal antiserum directed against Haps. Dose-dependent binding of Haps to wells coated with plasma fibronectin, laminin, and collagen IV was observed (Fig. 4). Conversely, Haps did not bind appreciably to wells coated with collagen II (Fig. 4) or to wells coated with BSA alone (data not shown). As a negative control in all of the ELISAs, we examined binding of purified HifB, a periplasmic chaperone involved in the assembly of H. influenzae pili and lacking adhesive activity. No appreciable binding of HifB to any of the ECM proteins was detected (data not shown). Nonlinear regression analysis of the ELISA data was used to estimate dissociation constants for the binding interactions between purified Haps and each of the ECM proteins. The binding constants predicted from this analysis were approximately 15 nM for plasma fibronectin, 20 nM for collagen IV, and 35 nM for laminin, suggesting that Haps binding to ECM ligands is a strong interaction and roughly equivalent among the three proteins examined. The R2 values for all of the predicted binding curves ranged from 0.97 to 0.99, indicating excellent fit of the curves to the ELISA data. Differences in the absolute signal intensity between individual experiments likely reflect differences in incubation time with the detection substrate and not differences in binding affinities.
FIG. 4.
Dose-dependent binding of purified Haps to ECM proteins. Shown are results of binding of increasing concentrations of purified Haps to fibronectin, collagen IV, laminin, and collagen II by purified Haps. Binding was quantitated by ELISA at an absorbance of 650 nm. Points represent the means (error bars, standard errors of the means) of measurements made in triplicate. Binding curves were calculated by nonlinear regression analysis using GraphPad Prism software (version 3.0).
DISCUSSION
In previous work, we demonstrated that Hap promotes bacterial adherence to cultured epithelial cells (20). Inhibition of autoproteolysis by mutation of the Hap active-site serine residue results in retention of the Haps passenger domain on the bacterial cell surface and increased adherence to epithelial cells (9). In the present study, we found that Hap also promotes bacterial adherence to the ECM proteins fibronectin, laminin, and collagen IV. Similar to the situation with epithelial cells, bacteria expressing HapS243A adhere more efficiently to ECM proteins than bacteria expressing wild-type Hap, suggesting that Haps possesses the binding domains responsible for interactions with ECM proteins.
Inhibition of Hap-mediated adherence to fibronectin, laminin, and collagen IV by antiserum RabK2 provides further evidence for the presence of ECM binding domains within Haps. RabK2 reacts specifically with epitopes in Haps and does not react with the Hapβ translocator domain. It is possible that some antibodies in RabK2 react with the C terminus of Haps and obscure a binding domain in the N terminus of Hapβ. However, a binding domain in Hapβ would presumably be equally abundant on bacteria expressing either wild-type Hap or HapS243A. The difference in adherence efficiencies between DB117/pJS106 and DB117/pHapS243A argues against this possibility. Studies with Hap in-frame deletion mutants have focused our search for ECM binding domains on the C-terminal half of Haps (D. L. Fink and J. W. St. Geme III, unpublished observations), and scanning mutagenesis within this region may eventually pinpoint individual residues involved in binding domains.
Examination of interactions between Haps and fibronectin fragments suggests that Hap may interact specifically with the 45-kDa gelatin-binding domain of fibronectin. These data do not exclude the possibility of interactions between Hap and other fibronectin domains, which remain to be evaluated. Successful mapping of fibronectin binding domains has been achieved for certain bacterial adhesins by far-Western immunoblot of fibronectin fragments produced by limited proteolysis with thermolysin (Magnus Höök, personal communication). This strategy has so far been unsuccessful with purified Haps, further suggesting that Hap may recognize a conformational epitope on natively folded fibronectin. Future studies will evaluate interactions between Haps and purified thermolysin-digested fibronectin fragments.
Our results demonstrate that Hap promotes adherence to three distinct ECM proteins. Similar broad-spectrum binding profiles have been reported for several other bacterial adhesins, including FnbpA and Emp of Staphylococcus aureus, Ace of Enterococcus faecalis, and YadA of Yersinia spp. (4, 10, 13). Purified Haps binds to fibronectin, laminin, and collagen IV with comparably high affinities in ELISAs, suggesting that Hap may interact with a particular structural epitope present on all three proteins. Candidates for this type of receptor structure include complex sugars of glycosylated proteins, as is the case with binding of H. influenzae pili and the HMW1 adhesin to their respective receptors. However, preliminary studies with chemical or enzymatic disruption of carbohydrate structures on fibronectin and laminin indicate that sugars may not be involved in interactions between Hap and ECM proteins. Hap may instead recognize a protein fold common among fibronectin, laminin, and collagen IV. Curiously, the binding constants for interactions between purified Haps and ECM proteins are also similar to those calculated for binding of Haps to Chang and A549 epithelial cells (Fink et al., submitted for publication), raising the possibility that an ECM protein may be the cellular receptor for Hap. Although pretreatment of cells with antifibronectin antibodies does not affect Hap-mediated bacterial adherence, any of a number of cell-associated ECM proteins could potentially serve as a receptor. Alternatively, the cellular Hap receptor may possess a conformational epitope similar to those present in ECM proteins.
In considering the role of Hap in NTHI colonization of the respiratory tract, it is noteworthy that fibronectin, laminin, and collagen IV are all components of basement membrane underlying respiratory epithelium. Additionally, fibronectin is present on the surface of epithelial cells. In contrast to the situation with collagen IV, Haps does not bind to collagen II, which is found mainly in cartilage and would not typically be encountered by H. influenzae during natural infection. NTHI disease is often associated with underlying chronic obstructive pulmonary disease or recent respiratory viral infection (6, 12, 15). Given that respiratory inflammation involves increased production of fibronectin and collagen and deposition of these proteins on epithelial cells (18), Hap-mediated bacterial attachment to ECM may be an important factor in initial colonization of the inflamed respiratory tract as well as contiguous spread of organisms to new sites of infection. Furthermore, damaged epithelium and exposed basement membrane are often found in the context of respiratory tract inflammation. NTHI associates preferentially with damaged epithelium in studies with nasopharyngeal tissue and explants from persistently infected patients (11, 16), and Hap may contribute to these interactions.
In summary, the H. influenzae Hap adhesin promotes bacterial adherence to fibronectin, laminin, and collagen IV. Retention of the Haps passenger domain on the bacterial cell surface enhances adherence to these ECM proteins, while treatment with an antiserum reactive against Haps inhibits adherence. In assays with purified proteins, Haps binds with high affinity to fibronectin, laminin, and collagen IV. Purified Haps interacts specifically with the 45-kDa gelatin-binding domain of fibronectin and not with the 30-kDa heparin-binding domain. These data suggest a possible mechanism for previously observed interactions between nontypeable H. influenzae and ECM and further advocate that these interactions may be important to colonization of the respiratory tract by NTHI. Future studies will seek to identify the Haps binding domains responsible for adherence to ECM proteins and their cognate receptor sites on fibronectin, laminin, and collagen IV.
Acknowledgments
This work was supported by an Established Investigator Award from the American Heart Association (to J.W.S.), a research grant from the March of Dimes (to J.W.S.), and funds from Wyeth Vaccines.
We thank Jerry Pinkner for valuable assistance in the purification of Haps; Rob Smith for his assistance with electron microscopy; and Dave Cutter, Sue Grass, and Sven Laarmann for technical help.
Editor: D. L. Burns
REFERENCES
- 1.Anderson, P., R. B. Johnston, Jr., and D. H. Smith. 1972. Human serum activity against Haemophilus influenzae type b. J. Clin. Investig. 51:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bresser, P., R. Virkola, M. Jonsson-Vihanne, H. M. Jansen, T. K. Korhonen, and L. van Alphen. 2000. Interaction of clinical isolates of nonencapsulated Haemophilus influenzae with mammalian extracellular matrix proteins. FEMS Immunol. Med. Microbiol. 28:129-132. [DOI] [PubMed] [Google Scholar]
- 3.de Bentzmann, S., C. Plotkowski, and E. Puchelle. 1996. Receptors in the Pseudomonas aeruginosa adherence to injured and repairing airway epithelium. Am. J. Respir. Crit. Care Med. 154:S155-S162. [DOI] [PubMed] [Google Scholar]
- 4.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291:209-218. [DOI] [PubMed] [Google Scholar]
- 5.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 6.Foxwell, A. R., J. M. Kyd, and A. W. Cripps. 1998. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol. Mol. Biol. Rev. 62:294-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrixson, D. R., M. L. de la Morena, C. Stathopoulos, and J. W. St. Geme III. 1997. Structural determinants of processing and secretion of the Haemophilus influenzae Hap protein. Mol. Microbiol. 26:505-518. [DOI] [PubMed] [Google Scholar]
- 9.Hendrixson, D. R., and J. W. St Geme III. 1998. The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol. Cell 2:841-850. [DOI] [PubMed] [Google Scholar]
- 10.Hussain, M., K. Becker, C. von Eiff, J. Schrenzel, G. Peters, and M. Herrmann. 2001. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 183:6778-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moller, L. V., W. Timens, W. van der Bij, K. Kooi, B. de Wever, J. Dankert, and L. van Alphen. 1998. Haemophilus influenzae in lung explants of patients with end-stage pulmonary disease. Am. J. Respir. Crit. Care Med. 157:950-956. [DOI] [PubMed] [Google Scholar]
- 12.Musher, D. M., K. R. Kubitschek, J. Crennan, and R. E. Baughn. 1983. Pneumonia and acute febrile tracheobronchitis due to Haemophilus influenzae. Ann. Intern. Med. 99:444-450. [DOI] [PubMed] [Google Scholar]
- 13.Nallapareddy, S. R., X. Qin, G. M. Weinstock, M. Hook, and B. E. Murray. 2000. Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect. Immun. 68:5218-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 15.Rayner, R. J., E. J. Hiller, P. Ispahani, and M. Baker. 1990. Haemophilus infection in cystic fibrosis. Arch. Dis. Child. 65:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Read, R., R. Wilson, A. Rutman, V. Lund, H. Todd, A. Brian, P. Jeffery, and P. Cole. 1991. Interaction of nontypable Haemophilus influenzae with human respiratory mucosa in vitro. J. Infect. Dis. 163:549-558. [DOI] [PubMed] [Google Scholar]
- 17.Read, R. C., A. A. Rutman, P. K. Jeffery, V. J. Lund, A. P. R. Brian, E. R. Moxon, P. J. Cole, and R. Wilson. 1992. Interaction of capsulate Haemophilus influenzae with human airway mucosa in vitro. Infect. Immun. 60:3244-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roman, J. 1996. Extracellular matrix and lung inflammation. Immunol. Res. 15:163-178. [DOI] [PubMed] [Google Scholar]
- 19.Setlow, J. K., D. C. Brown, M. E. Boling, A. Mattingly, and M. P. Gordon. 1968. Repair of deoxyribonucleic acid in Haemophilus influenzae. I. X-ray sensitivity of ultraviolet-sensitive mutants and their behavior as hosts to ultraviolet-irradiated bacteriophage and transforming deoxyribonucleic acid. J. Bacteriol. 95:546-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.St. Geme, J. W., III, M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 21.St. Geme, J. W., III, S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypeable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 90:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.St. Geme, J. W., III, J. S. Pinkner, G. P. Krasan, J. Heuser, E. Bullitt, A. L. Smith, and S. J. Hultgren. 1996. Haemophilus influenzae pili are composite structures assembled via the HifB chaperone. Proc. Natl. Acad. Sci. USA 93:11913-11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas, D. D. 1996. Aspects of adherence of oral spirochetes. Crit. Rev. Oral Biol. Med. 7:4-11. [DOI] [PubMed] [Google Scholar]
- 24.Tomb, J. F., G. J. Barcak, M. S. Chandler, R. J. Redfield, and H. O. Smith. 1989. Transposon mutagenesis, characterization, and cloning of transformation genes of Haemophilus influenzae Rd. J. Bacteriol. 171:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turk, D. C. 1984. The pathogenicity of Haemophilus influenzae. J. Med. Microbiol. 18:1-16. [DOI] [PubMed] [Google Scholar]
- 26.Virkola, R., M. Brummer, H. Rauvala, L. van Alphen, and T. K. Korhonen. 2000. Interaction of fimbriae of Haemophilus influenzae type b with heparin-binding extracellular matrix proteins. Infect. Immun. 68:5696-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Virkola, R., K. Lahteenmaki, T. Eberhard, P. Kuusela, L. van Alphen, M. Ullberg, and T. K. Korhonen. 1996. Interaction of Haemophilus influenzae with the mammalian extracellular matrix. J. Infect. Dis. 173:1137-1147. [DOI] [PubMed] [Google Scholar]
- 28.Westerlund, B., and T. K. Korhonen. 1993. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9:687-694. [DOI] [PubMed] [Google Scholar]