Abstract
Studies in mouse, human, and chicken suggest that activation-induced deaminase (AID) is involved in three known processes leading to antibody diversification: somatic hypermutation, gene conversion, and class-switch recombination. Developing rabbit appendix provides a particularly good site for studying all three of these B cell maturation events. We report here successful cloning of rabbit AID and isolation of AID protein from rabbit appendix-cell nuclear and cytoplasmic extracts. We succeeded in identifying and locating AID protein in cells by immunohistochemical and immunofluorescent staining techniques and examined colocalization of AID and other molecules important for Ab diversification. This report extends our knowledge about AID to a mammalian species that uses gene conversion to diversify rearranged Ig genes. Although much work remains to understand fully the mechanism of action of AID and its association with other cellular components, the rabbit system now offers a particularly useful model for future studies of these dynamics.
Keywords: mass spectrometry, gene conversion, somatic hypermutation
The rabbit is an excellent animal model for study of somatic hypermutation (SHM) and gene conversion (GC) of rearranged variable regions (V) of Ab heavy (H) and light (L) chain genes. The appendix of neonatal rabbits is a critical site for development of the primary Ab repertoire (1, 2). Rearrangements of H and L chain genes occur in sites including fetal liver, omentum, and bone marrow. After birth, B cells with rearranged genes migrate into gut-associated lymphoid tissue such as the appendix. Gut microflora are required for B cells to proliferate and develop in appendix follicles (3). At ≈3 wk of age, diversification of rearranged gene sequences by GC and SHM starts within the appendix-follicle microenvironment (2). GC diversifies sequences encoding V regions of H and L chains through introduction of sequences from upstream donor V gene segments. B cells that have undergone class-switch recombination (CSR), particularly to IgA, are also found in rabbit appendix at ≈3 wk of age (2). Activation-induced deaminase (AID) is involved in three processes of Ab diversification and maturation, SHM, CSR (4–7), and GC (8, 9), that all coexist in young rabbit appendix. Thus far, the only published evidence for the role of AID in GC comes from two studies showing that in the chicken DT40 bursal tumor cell line that carries out GC in vitro, no GC occurs when AID is knocked out on both chromosomes (8, 9). Although there are several hypotheses about how AID functions, data are accumulating that suggest that AID deaminates cytosine to uracil on the non-template strand of DNA during transcription (10). It is also possible that AID deaminates cytidine to uridine on an unknown mRNA (11, 12). Data are still emerging that suggest AID has different roles and cofactor interactions when participating in CSR and SHM (13, 14).
We used 1- to 6-wk-old ali/ali VH mutant rabbits to study rabbit AID. These mutants have a small (≈10 kb) deletion encompassing the VH1a2 allotype-encoding VH1 gene at the 3′ end of the VH cluster that rearranges in most normal B cells in WT rabbits of the a2 allotype (15–17). As ali/ali rabbits mature, they produce increasing proportions (10–40%) of their total serum Ig with serological properties indistinguishable from normal a2 allotype despite the absence of a VH1a2 gene. These a2+ immunoglobulins result from rearrangement of upstream genes such as the first functional gene, VH4; GC then alters the framework region sequences so that they resemble the deleted VH1a2 gene sequence (18–20).
In follicles of the developing young appendix, a dark zone (DZ) and light zone (LZ) can be defined histologically. B cells in the DZ are characterized by intense proliferative activity (centroblasts) (2, 21, 22). Those in the LZ express higher amounts of surface Ig (centrocytes). Cells also undergo positive and negative selection within the developing appendix; cells that do not survive undergo apoptosis and are removed by macrophages (21, 22). We cloned and sequenced cDNA encoding rabbit AID, generated Abs to two different AID peptide segments, and used purified Abs for immunohistochemical and immunofluorescent staining to localize the AID molecule in appendix follicles and single B cells. In addition, we prepared extracts of appendix cells and recovered AID using anti-AID peptide Abs for affinity purification.
Materials and Methods
Rabbits. Rabbits were obtained from the allotype-defined pedigreed colony maintained at the National Institute of Allergy and Infectious Diseases. Normal rabbits of known Ig allotypes from our breeding colony, ≈1 yr of age, were immunized. Homozygous mutant ali/ali rabbits, 1–6 wk of age, provided appendix for immunohistochemistry, immunofluorescence, and extraction of cytoplasmic and nuclear proteins.
Rabbit AID cDNA Cloning and Sequencing. Total RNA was obtained from immunized rabbit spleen that was immediately placed in TRIzol reagent (Life Technologies), tissue was homogenized, and nucleoprotein complexes were removed by centrifugation at 2,500 × g for 10 min at 4°C. RNA was then separated from supernatant with 1-bromo-3-chloropropane (BCP) (Molecular Research Center, Cincinnati) and precipitated with isopropyl-alcohol. First-strand cDNA was synthesized by using the Super-Script first-strand synthesis kit (Invitrogen), using synthesized primers corresponding to the known human (NM_020661) and mouse (NM_009645) AID sequence: upstream 5′-ATGGACAGCCTCTTGATGAA-3′ and downstream 5′-TCAAA(A/G)TCCCAAAGTACGAAATGC-3′ at the ends of the ORF. Touchdown PCR conditions were melting at 94°C for 1 min, annealing at 66°C for 30 sec, and elongation at 72°C for 2 min. The annealing temperature was dropped at a rate of 2°C per cycle for five cycles to 56°C for the remaining 25 cycles. A final extension was at 72°C for 5 min. The PCR products were cloned into pCR4-TOPO (Invitrogen), for sequencing from the T3 and T7 sites in the vector by using T3 and T7 primers. Nucleotide sequencing was carried out by using the BigDye terminator v3.0 cycle sequencing kit (Applied Biosystems) and an automated sequencer (Model 377, Applied Biosystems). Three additional independent PCR cloning and sequencing experiments were conducted to rule out errors introduced by PCR. To obtain the 3′ sequence, oligo(dT) and upstream primers were used, and cloning and sequencing were repeated as described above.
Production of Abs to AID Peptides. Two peptides were synthesized on a MAP-4 carrier backbone (Multiple Peptide Systems, San Diego): RAID (ISDWDLDPGR), which is near the proposed catalytic center of rabbit AID; and HAID (LRDAFRTLGL), which is from the human C terminus and differs from the rabbit C-terminal sequence by one amino acid. Rabbits were immunized s.c. with 0.5 mg of MAP-peptide in 0.5 ml of PBS (pH 7.4) emulsified with 0.5 ml of complete Freund's adjuvant on day 0 and boosted s.c. with the same doses in incomplete Freund's adjuvant on days 21, 42, 63, and 84. Sera, taken 1 wk after each boost, were assayed by ELISA for reactivity with MAP-peptide bound to the plate. Sera taken after the last boost were used for purification of peptide-specific Abs with affinity columns of RAID-MAP-4 and HAID-MAP-4, each conjugated to a bio-support medium by using the UltraLink EDC/DADPA immobilization kit (Pierce). Specific Abs were eluted according to the manufacturer's protocol at low pH and exchanged into PBS.
Abs Used in Assays. Affinity-purified anti-RAID, affinity-purified anti-HAID, and control normal rabbit IgG (Jackson ImmunoResearch) were conjugated to biotin (EZ-Link Sulfo-NHS-LC-Biotinylation, Pierce) or to Alexa Fluor 568 or 647 with a labeling kit (Molecular Probes). Also used were rat anti-phospho-Histone 3 (Sigma), mouse anti-rabbit macrophage (Dako), mouse anti-human Ki67 cross-reactive with rabbit (Roche Molecular Biochemicals), polyclonal rabbit anti-UNG (uracil DNA glycosylase) conjugated to biotin or Alexa Fluor 488 (Imgenex, Sorrento Valley, CA), anti-single-stranded DNA (Apostain reagent, Apotech, San Diego), FITC-goat anti-rabbit IgM (Southern Biotechnology Associates) or IgA (Bethyl Laboratories, Montgomery, TX), and FITC-species specific secondary Abs (Jackson ImmunoResearch).
Preparation of Appendix Cytoplasmic and Nuclear Extracts. Appendixes were collected from 4.5-wk-old ali/ali rabbits, washed with cold PBS, and gently teased to produce single-cell suspensions. We used the NE-PER Nuclear and Cytoplasmic Extraction kit (Pierce) to extract nuclear and cytoplasmic contents. Halt Proteinase inhibitors (Pierce) were added to the extracts. Samples in Slide-A-Lyzer dialysis cassettes (Pierce) were concentrated with PEG 8000, and protein concentrations were estimated by using a Micro BCA kit (Pierce).
Western Blots. For Western blots, 25, 50, 100, and 200 μg of total protein were loaded on 4–20% SDS/PAGE gels and proteins transferred to PVDF membranes (Invitrogen). Blots were probed with biotinylated anti-RAID and anti-HAID Abs at a final concentration of 20 ng/ml, followed by avidin-HRP. Signals were developed and detected by using a SuperSignal West Femto kit (Pierce) and exposed for 5–30 min. A positive control for AID detection was recombinant mouse AID expressed in Escherichia coli (a gift from D. Garboczi, National Institute of Allergy and Infectious Diseases). In peptide blocking tests, 0.1 μg/ml RAID or HAID peptide was added to the incubation solution containing anti-RAID Ab.
Immunoaffinity Purification. Nuclear extracts from two 4.5-wk-old appendixes (total of 3–5 × 109 cells) were first dialyzed against 1× PBS for 4–5 h and then preincubated for 4 h at room temperature with normal rabbit IgG conjugated to agarose (CarboxyLink coupling gel, Pierce) to preclear nonspecific binding. The precleared lysates were then incubated at 4°C for 10–16 h with anti-RAID-peptide Ab similarly conjugated to agarose, washed, and eluted to recover purified proteins. In some experiments, proteins were further purified by using ≈108 Dynabeads M-280 (Dynal, Oslo) conjugated with 10 μg of anti-RAID Ab according to the manufacturer's protocol. After concentration and dialysis against incubation buffer (20 mM Hepes, pH 7.6/25% glycerol/1.5 mM MgCl2/0.2 mM EDTA/0.2 M KCl/1 mM DTT/1 mM PMSF/1× Halt Proteinase inhibitors) for 4 h, proteins were incubated with anti-RAID-coupled beads overnight at 4°C. Beads were sequentially washed with incubation buffer containing 0.4, 0.6, and 0.8 M KCl. The beads were boiled in 2× SDS loading buffer for 5 min, and AID was detected by Western blots with the protocol described above and silver staining with the SilverQuest kit (Invitrogen). Bands were excised from gels, destained, and individually digested for mass spectrometry analyses.
Mass Spectrometry. Protein gel bands were digested with trypsin, and the peptides were extracted as described in ref. 23. Samples were desalted with C18 Zip Tips (Millipore) according to the manufacturer's protocols and stored at -20°C until mass spectrometry analysis. Samples (0.5 μl) were cocrystallized with 0.5 μl of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/1% trifluoroacetic acid and spotted directly on a stainless steel matrix-assisted laser desorption ionization (MALDI) target plate. Mass spectra were acquired by using an Applied Biosystems 4700 MALDI-TOF/TOF (tandem time-of-flight) mass spectrometer (Applied Biosystems). For all mass spectra the laser frequency was 200 Hz, and for collision-induced dissociation the collision energy was 1 keV (1 eV = 1.602 × 10-19 J) (air was used as collision gas). The MALDI-TOF/TOF spectra were internally calibrated (<20 ppm) by using trypsin autolysis products. Postacquisition baseline correction and smoothing was carried out by using software provided with the instrument. Bioinformatic analysis was carried out by using global proteome surveyor (gps) software (Applied Biosystems).
Immunohistochemistry. Serial sections (7 μm) of 4.5-wk-old ali/ali rabbit appendix were cut in a cryostat microtome at -20°C, kept overnight in a desiccator, fixed with acetone for 10 min and cold methanol for 15 min at -20°C, and blocked with 10% rabbit serum. For biotinylated Abs, 10 μg/ml biotinylated RAID or UNG Ab was incubated for 1 h at room temperature and then visualized with an avidin-biotin complex conjugated to alkaline phosphatase and Vector Blue kit (Vector Laboratories); sections were counterstained with nuclear fast red (Vector Laboratories). For unconjugated Abs, primary Abs were incubated for 1 h at room temperature with 10 μg/ml final concentration. Secondary biotinylated Abs were incubated at room temperature for 30 min, and the same protocol was used to visualize positive staining.
Immunoflourescence. Appendixes of 4.5-wk-old ali/ali rabbits were removed and cleaned in chilled PBS. The appendix was chopped in small pieces in PBS and pressed through a stainless steel mesh screen with a rubber-tipped syringe plunger. Debris and cell clumps were removed by passing the suspension through 70 μM nylon mesh. Dead cells were excluded by incubation with ethidium monoazide (EMA) (final concentration 5 μg/ml; Molecular Probes) in the dark on ice for 15 min, followed by a 10-min exposure to a 40-W fluorescent light at an 18-cm distance. Cells were washed twice with PBS and fixed with 2% paraformaldehyde for 30 min on ice. EMA-negative cells were sorted on a FACStar Plus-Violet (Becton Dickinson), and sorted cells were centrifuged onto glass slides with a Cytospin cytocentrifuge (Shandon). The cytospin slides and serial tissue sections of 4.5-wk-old ali/ali appendixes were fixed with 4% paraformaldehyde for 10 min and then 8% paraformaldehyde for 50 min at room temperature, rinsed with 50 mM NH4Cl for 5 min and 0.1% Triton X-100 for 10 min, washed with PBS, and blocked with 10% rabbit serum for 15 min. Sections were incubated with 10 μg/ml Alexa Fluor 568-conjugated anti-RAID-peptide Abs in 3× PBS for 1 h and washed with 1× PBS containing 0.1% Triton X-100 three times for 10 min. For double or triple staining, additional staining steps were done, and finally, 0.05% DAPI was used to stain nuclei. We did double staining with anti-RAID and anti-phospho-Histone 3, -IgM, -IgA, -Ki67, or -single-strand DNA Ab. We did triple staining with anti-RAID anti-IgM and anti-macrophage Abs. The slides were mounted with Prolong anti-fade (Molecular Probes). Images were obtained with a Leica confocal microscope and edited by using imaris software (Bit-plane). To count AID+ cells recovered in sorted EMA-negative cell populations, cytospin slides were stained with anti-IgM, anti-AID, and DAPI. The confocal microscope was used to capture 30 images at ×63 magnification (30–50 cells per field; ≈1,200 total cells). AID+ IgM high and low B cells were counted three times to estimate the percentage of detectable AID+ cells.
Flow Cytometry. Appendix cells (20 × 106) were suspended in 1 ml of PBS, EMA was added, and the suspension was incubated and exposed to light as described above. Samples (100 μl; ≈2 × 106 cells) were incubated with 1 μg of anti-rabbit IgM-FITC on ice for 40 min and then washed twice with PBS and 2% heat-inactivated FCS solution (PBS-FCS). The cells were then fixed in 70% ethanol for 1 h, washed twice with PBS-FCS, and resuspended in 100 μl of PBS-FCS containing 10% normal rabbit serum and 0.1% Triton X-100. The cells were further incubated for 40 min with anti-rabbit-AID-Alexa 647. The cells were washed twice with PBS-FCS, resuspended in PBS with 0.0001% DAPI, and analyzed immediately on an LSR-II cell analyzer (Becton Dickinson). Appropriate isotype controls were included.
Results
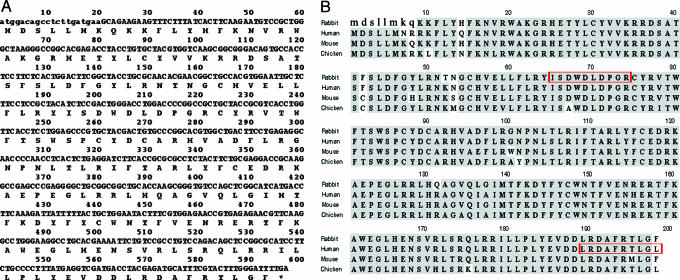
Rabbit AID Cloning and Sequence Comparison. The rabbit AID cDNA sequence obtained by reverse transcriptase PCR and the amino acid sequence deduced from the cDNA sequence are shown in Fig. 1A. Fig. 1B shows a comparison of the rabbit AID sequence with those of human, mouse, and chicken. Oligo(dT) priming and amplification were used to determine the encoded rabbit C-terminal sequence shown. The DNA sequence is 89%, 87%, and 79% identical to human, mouse, and chicken AID sequences, respectively. Lowercase letters at the 5′ end indicate the location of the primer sequence used for initial PCR amplification. Although we did not sequence the 5′ end, rabbit (Oryctolagus cuniculus) whole-genome shotgun (WGS) sequence generated by the Broad Institute of the Massachusetts Institute of Technology and Harvard [National Center for Biotechnology Information (NCBI) trace archive number TI 626458610] contains rabbit AID exon 2 consisting of all but the first four bases of rabbit AID coding sequence at the 5′ end (base numbers 5–156). This sequence has one silent difference from that shown. At the 3′ end, NCBI trace archive number TI 651771662 contains sequence corresponding to our bases 543–597; it has one silent change and one change that leads to replacement of Phe-198 seen in mice with Leu seen in human. These may reflect polymorphisms in rabbits. At the protein level, the amino acid sequences shown are 93%, 94%, and 88% identical to human, mouse, and chicken AID.
Fig. 1.
Rabbit AID sequence and comparisons with other species. (A) DNA sequence and deduced encoded protein sequence of rabbit AID. Lowercase letters at the 5′ end indicate the location of the primer sequence used for initial PCR amplification. (B) Rabbit AID amino acid sequence compared with human, mouse, and chicken AID sequences. The red rectangles represent the locations of peptides synthesized for use as immunogens.
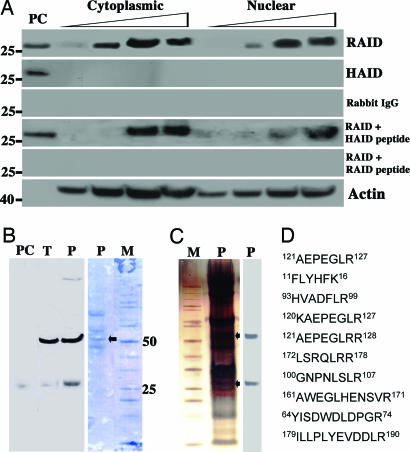
Western Blotting and AID Purification. Affinity-purified anti-RAID Abs detected a control recombinant mouse AID band of ≈26 kDa and a band of the same size in both nuclear and cytoplasmic extracts (Fig. 2A). This binding can be inhibited specifically by RAID peptide and not by HAID peptide (Fig. 2 A). Positive ≈52-kDa bands were observed on Western blots after partial purification of nuclear extracts (Fig. 2B). In later experiments with an additional purification step with Dynabeads, Western blotting detected two bands in purified AID from nuclear extracts, one at ≈52 kDa and the other at ≈26 kDa (Fig. 2C). The partially purified ≈52-kDa band observed with Coomassie blue staining, and later, the two positive bands from silver-stained gels, were excised, in-gel trypsin was digested, and eluted peptides were identified by MALDI-TOF/TOF MS. The purifications were repeated twice more with the first protocol. Rabbit AID peptides were identified in each of the three experiments from the ≈52-kDa band and in both bands in the two experiments where the additional purification step was included (Fig. 2D). It is likely these represent AID monomer and homodimer. The latter can be partially dissociated by boiling in loading buffer containing 8 M urea (Fig. 6 and Supporting Methods, which are published as supporting information on the PNAS web site).
Fig. 2.
Results of Western blotting of nuclear and cytoplasmic extracts from 4.5-wk-old ali/ali appendix. (A) Twenty-five, 50, 100, and 200 μg of total protein from cytoplasmic and nuclear extracts were loaded per well. Recombinant mouse AID expressed in E. coli dissolved in 8 M urea was the positive control (PC). Blots were probed with anti-RAID, anti-HAID, or normal rabbit IgG (exposed for 30 min) and anti-β-actin (exposed for 5 min). Results of blocking with HAID or RAID peptide followed by probing with anti-RAID Ab are also shown. (B) Partially purified protein from two 4.5-wk-old ali/ali appendixes was loaded on the gel stained with Coomassie blue (B) or silver (C). The blots were probed with anti-RAID Ab, and corresponding bands (indicated with arrows) were cut from the gels and analyzed by MS to identify the AID sequence. (D) Peptide sequences of rabbit AID detected by MS. PC, positive control; T, total extract; P, purified; M, molecular weight marker.
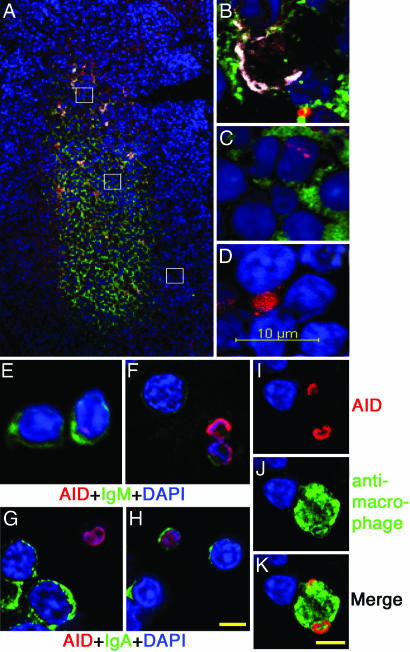
Immunohistochemistry and Immunofluorescence. Appendix tissue sections from ali/ali rabbits 1–6 wk of age were stained with anti-RAID Ab (Fig. 7 and Supporting Methods, which are published as supporting information on the PNAS web site). AID+ B cells were detected in both DZ and LZ of appendix follicles at ≈3–6 wk of age but not before the onset of GC, SHM, and CSR at 1 and 2 wk. AID was also found in Peyer's patches of WT immunized mice but not in AID-/- mice (Fig. 8 and Supporting Methods, which are published as supporting information on the PNAS web site). More AID+ cells were detected in 4.5-wk-old ali/ali appendix compared with other time points and also to 4.5-wk-old WT rabbits. Immunohistochemical staining revealed UNG and RAD51C, a protein involved in the GC process in yeast (24), mainly in light zones of appendix follicles; very few positive cells were found outside this area (Fig. 7). Immunofluorescent staining of tissue sections detected two different patterns of AID staining (Fig. 3). Fig. 3A shows a typical germinal center with green staining of IgM high LZ B cells and DAPI-stained nuclei of DZ cells (mainly IgM low B cells, blue). Stained RAID-positive cells are red and macrophages are shown in white pseudocolor. The RAID staining was inhibitable by RAID peptide (Fig. 9 and Supporting Methods, which are published as supporting information on the PNAS web site). Fig. 3B shows a macrophage that has RAID-positive remnants of engulfed lymphocytes. The first type of AID+ cells, found mainly in the LZ, contained AID foci inside the nuclei (Fig. 3C). The second type of AID+ cells that were more frequently found in the DZ and the area between DZ and LZ had smaller nuclei compared with B lymphocytes in the LZ, and frequently occurred as doublets (Fig. 3D). To see more details, we made cytospin slides from appendix cell suspensions. Anti-RAID stained IgM high and IgM low B cells (Fig. 3 E and F) and also stained some IgA+ cells (Fig. 3 G and H). AID is readily detected in a macrophage that presumably engulfed dying B cells (Fig. 3 I–K). We also found AID+ apoptotic cells using the Apostain anti-single-stranded DNA reagent (data not shown). Laser scanning cytometry was used to estimate numbers of RAID-positive cells (Fig. 10, Table 1, and Supporting Methods, which are published as supporting information on the PNAS web site). Approximately half of the cells identified as macrophages had detectable AID (Fig. 10A). The percentages of B cells that were scored as AID+ ranged from ≈0.5–1.0% of total cells in nine follicles (Fig. 10B).
Fig. 3.
Seven-micrometer 4.5-wk-old ali/ali appendix sections were stained with Alexa Fluor 568-anti-RAID (red), FITC-anti-IgM (green), Alexa Fluor 647-anti-macrophage (white), and DAPI (blue). (A) Whole follicle. (B–D) High-magnification images captured from A in the order from top to bottom. Double staining with anti-RAID in red, anti-IgM (E and F), anti-IgA (G and H), or anti-macrophage (I–K) in green, and DAPI in blue on cytospin slides from 4.5-wk-old ali/ali appendix cell suspension. (Scale bar: 5 μm.)
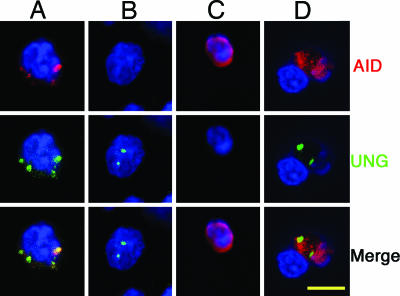
AID Is Colocalized with UNG in Some but Not All Cells. AID can also be found colocalized with UNG in some but not all AID-positive B cells (Fig. 4 A and B). We did not find colocalization of AID and UNG in B cells occurring in doublets (Fig. 4C). AID and UNG are detected as if colocalized most clearly within macrophages (Fig. 4D).
Fig. 4.
Double staining with anti-RAID in red, anti-UNG (A–D) in green, and DAPI in blue on cytospin slides of sorted EMA-negative cells from 4.5-wk-old ali/ali appendix cell suspension. (Scale bar: 5 μm.)
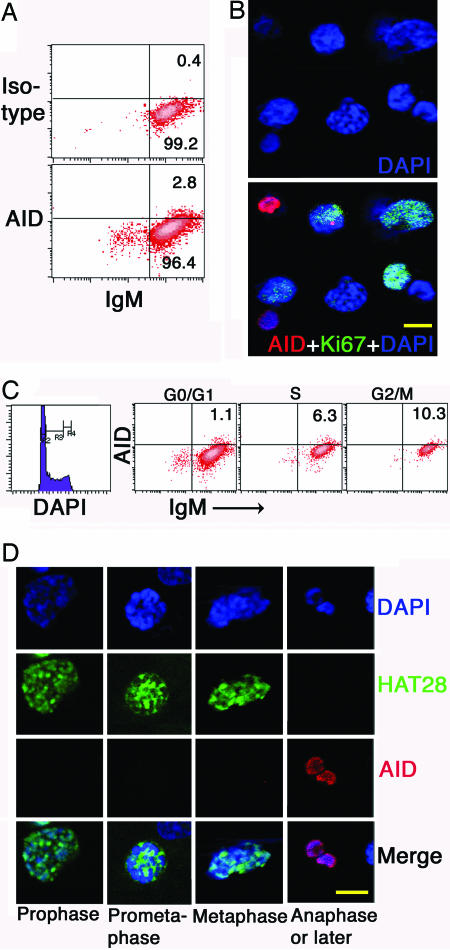
Detectable AID May Correlate with Cell-Cycle Division Stage. Cells of 4.5-wk-old ali/ali appendix were sorted after gating out dead cells by using EMA followed by fixation. Immunofluorescent staining was done on sorted cytocentrifuged cells. Low percentages of detectable AID+ cells were found on the cytospin slides (≈3–5%) as well as by flow cytometry (Fig. 5A). Possible explanations for the low percentages of AID+ cells are addressed in Discussion. A few proliferating (Ki67-positive) cells contained AID foci (Fig. 5B); however, the small AID+ cells do not stain for Ki67 (Fig. 5B). We used DAPI to gate appendix cells at different stages of the cell cycle for flow cytometric analysis. In the experiment shown in Fig. 5C, there were higher proportions of AID-positive cells in the G2/M phase than other phases. The absolute numbers of AID+ cells among 20,904 cells scored in this experiment were 174 of 15,791 in G0/G1 phase, 233 of 3,697 in S phase, and 146 of 1,416 in G2/M. Percentages of total cells that were AID+ were similar in each fraction (0.8%, 1.1%, and 0.7%). In contrast, in three experiments, proportions of AID+ cells within each stage of the cell cycle were 0.5–1.1% in G0/G1 phase, 1.6–6.3% in S phase, and 4.7–10.3% in G2/M phase. Double staining with anti-RAID and anti-phospho-Histone 3, a marker of early stages of mitosis (25), showed that the small AID+ cells were not stained by it (Fig. 5D).
Fig. 5.
Cell cycle and detection of AID. (A) Flow cytometry of 4.5-wk-old ali/ali appendix cells stained with anti-IgM and anti-AID or rabbit IgG isotype control. (B) Cytospin slides of sorted EMA-negative cells from 4.5-wk-old ali/ali appendix stained by anti-Ki-67 and anti-RAID. (C) FACS analysis of AID and cell division cycle. Data shown represent one of three analyses. (D) Double staining on cytospin slides of sorted EMA-negative cells from 4.5-wk-old ali/ali appendix with anti-RAID and anti-phospho-Histone 3 (HAT28).
Discussion
VH mutant ali/ali rabbits face an additional challenge compared with WT rabbits when they diversify their rearranged Ig genes in the appendix after birth, probably because the mutants lack the predominantly rearranged VH1a2 gene (15–17). Earlier suggestions that B cells expressing surface Ig receptors with VH framework region sequences characteristic of the VH1a2 sequence are preferentially selected for survival (22) via interaction of framework region sequences with endogenous superantigens, as well as those present on gut microflora (26–28), have recently received additional support (27, 28). Compared with WT rabbits, there are more apoptotic B cells in ali/ali rabbit appendix follicles during the stage when they are diversifying rearranged heavy and light chain genes (22, 26). Between 3 and 6 wk of age, the mutants have smaller appendixes. Although they lack the VH1a2 gene, they start to develop a2+ B cells in the appendix by GC-like alteration of other rearranged upstream genes especially VH4 (19, 27, 29). It is not surprising that peak expression of AID is detected in appendix cells of 4.5-wk-old ali/ali mutants when selection processes are driving cells undergoing GC toward survival and proliferation.
By 10 wk, ali/ali rabbits have appendix sizes comparable with those of the WT a2 animals (22, 26), and they have also developed a diversified repertoire resembling normal animals. The GC mechanism plays a critical role in producing B cells with a2 framework region sequence. Both SHM and GC also contribute to the repertoire diversification process. The AID that we detect in appendix of these animals during this period is likely to be functioning in these processes and probably also for CSR to IgA. Despite this, our anti-RAID reagent only detected AID protein in at most 5% of total appendix cells at 4.5 wk. One possible explanation is that our Ab cannot detect all AID protein molecules present in the cells. AID-binding proteins may block accessibility to the epitope detected by our anti-RAID Ab. Apoptosis appears to render the RAID epitope more available for detection than in living B cells. Another explanation may be that AID is only transiently present and is then degraded because AID is dangerous for the stability of genomic DNA (6, 30). Apobec3G, another member of same deaminase family, is known to be ubiquitinylated and degraded during HIV infection (31, 32). Finally, the reported transport of AID out of the nucleus suggests that the retention time of AID in the nucleus may be very short (33–35). The LZ of the rabbit appendix appears to be an important area for AID-related DNA repair. We found that repair molecules such as RAD51C and H2AX can only be found in LZ (data not shown). These results suggest that repair of the DNA breaks that may result from GC, SHM, and CSR may occur in the appendix LZ even though AID can be found in both LZ and DZ. If this scenario is correct, migration of AID+ B cells from DZ to LZ would not only be important for positive selection processes thought to occur in the LZ, but also for completion of the diversification processes themselves.
B cells probably use different repair systems at different stages of the cell cycle to repair the DNA breaks in the Ig locus that may be introduced directly or indirectly by AID. Nonhomologous end joining can occur at all cell cycle stages but preferentially occurs during G1/early S phase (36, 37), and homologous recombination occurs preferentially in late S/G2 phase (38, 39). We speculate that the higher percentage of AID+ cells detected by flow cytometry among those at the G2/M phase may reflect GC or SHM events, and those AID+ cells in G0/G1 phase may be undergoing nonhomologous end joining during CSR. Although lower percentages of AID+ cells were found at earlier stages of the cell cycle, 73–75% of the cells were in G0/G1 phase. In view of the high absolute number of G0/G1 phase cells, comparable numbers were recovered in both the CSR and GC/SHM categories. However, it should be recognized that if epitope-masking restricts detection of AID protein in some cells, this could affect quantitation of AID+ cells during some stages of the cell cycle.
In summary, the results reported here document that the mutant ali/ali rabbit is a valuable model to investigate cells and mechanisms of AID function during Ab diversification processes in developing lymphoid tissues. Further studies will now be possible to investigate AID protein and its interactions with other repair molecules and to compare and contrast developing appendix and germinal centers in spleens from immunized rabbits where GC, SHM, and CSR also occur.
Supplementary Material
Acknowledgments
We thank G. Rai, M. Mage, and R. Pospisil for critical comments about this paper; D. Garboczi for his generous gift of recombinant mouse AID used as a positive control; O. Schwartz for his help with confocal imaging; D. Stephany for help with flow cytometry; C. Eigsti for help with cell sorting; C. B. Alexander for outstanding technical support; and W. Bonner, P. Henkart, and M. Catalfamo (National Institutes of Health) for helpful discussions and advice about this work. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases/National Institutes of Health. B.L.H., T.P.C., and T.D.V. are supported by National Cancer Institute Contract NO1-CO-12400.
Author contributions: R.G.M. designed research; G.Y., H.O., R.K.S., B.A.N., B.L.H., T.P.C., and T.D.V. performed research; R.K.S. and B.A.N. contributed new reagents/analytic tools; G.Y., H.O., R.K.S., B.L.H., T.P.C., T.D.V., and R.G.M. analyzed data; and G.Y., R.K.S., B.A.N., and R.G.M. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AID, activation-induced deaminase; CSR, class-switch recombination; DZ, dark zone; EMA, ethidium monoazide; GC, gene conversion; H, Ab heavy chain; L, Ab light chain; LZ, light zone; SHM, somatic hypermutation; UNG, uracil DNA glycosylase; V, Ab variable region.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY928183).
References
- 1.Weinstein, P. Z., Anderson, P. O. & Mage, R. G. (1994) Immunity 1, 647-659. [DOI] [PubMed] [Google Scholar]
- 2.Sehgal, D., Obiakor, H. & Mage, R. G. (2002) J. Immunol. 168, 5424-5433. [DOI] [PubMed] [Google Scholar]
- 3.Lanning, D., Sethupathi, P., Rhee, K. J., Zhai, S. K. & Knight, K. L. (2000) J. Immunol. 165, 2012-2019. [DOI] [PubMed] [Google Scholar]
- 4.Okazaki, I. M., Kinoshita, K., Muramatsu, M., Yoshikawa, K. & Honjo, T. (2002) Nature 416, 340-345. [DOI] [PubMed] [Google Scholar]
- 5.Yoshikawa, K., Okazaki, I. M., Eto, T., Kinoshita, K., Muramatsu, M., Nagaoka, H. & Honjo, T. (2002) Science 296, 2033-2036. [DOI] [PubMed] [Google Scholar]
- 6.Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature 418, 99-103. [DOI] [PubMed] [Google Scholar]
- 7.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553-563. [DOI] [PubMed] [Google Scholar]
- 8.Harris, R. S., Sale, J. E., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Curr. Biol. 12, 435-438. [DOI] [PubMed] [Google Scholar]
- 9.Aarkawa, H., Hauschild, J. & Buerstedde, J. M. (2002) Science 295, 1301-1306. [DOI] [PubMed] [Google Scholar]
- 10.Ramiro, A. R., Stavropoulos, P., Jankovic, M. & Nussenzweig, M. C. (2003) Nat. Immunol. 4, 452-456. [DOI] [PubMed] [Google Scholar]
- 11.Begum, N. A., Kinoshita, K., Kakazu, N., Muramatsu, M., Nagaoka, H., Shinkura, R., Biniszkiewicz, D., Boyer, L. A., Jaenisch, R. & Honjo, T. (2004) Science 305, 1160-1163. [DOI] [PubMed] [Google Scholar]
- 12.Begum, N. A., Kinoshita, K., Muramatsu, M., Nagaoka, H., Shinkura, R. & Honjo, T. (2004) Proc. Natl. Acad. Sci. USA 101, 13003-13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu, X., Geraldes, P., Platt, J. L. & Cascalho, M. (2005) J. Immunol. 174, 934-941. [DOI] [PubMed] [Google Scholar]
- 14.Nagaoka, H., Ito, S., Muramatsu, M., Nakata, M. & Honjo, T. (2005) Proc. Natl. Acad. Sci. USA 102, 2022-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight, K. L. & Becker, R. S. (1990) Cell 60, 963-970. [DOI] [PubMed] [Google Scholar]
- 16.Allegrucci, M., Newman, B. A., Young-cooper, G. O., Alexander, C. B., Meier, D., Kelus, A. D. & Mage, R. G. (1990) Proc. Natl. Acad. Sci. USA 87, 5444-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ros, F., Puels, J., Reichenberger, N., van Schooten, W., Buelow, R. & Platzer, J. (2004) Gene 330, 49-59. [DOI] [PubMed] [Google Scholar]
- 18.Chen, H. T., Alexander, C. B., Young-Cooper, G. O. & Mage, R. G. (1993) J. Immunol. 150, 2783-2793. [PubMed] [Google Scholar]
- 19.Sehgal, D., Mage, R. G. & Schiaffella, E. (1998) J. Immunol. 160, 1246-1255. [PubMed] [Google Scholar]
- 20.Zhu, X., Boonthum, A., Zhai, S. K. & Knight, K. L. (1999) J. Immunol. 163, 3313-3320. [PubMed] [Google Scholar]
- 21.MacLennan, I. C. (1994) Annu. Rev. Immunol. 12, 117-139. [DOI] [PubMed] [Google Scholar]
- 22.Pospisil, R., Young-Cooper, G. O. & Mage, R. G. (1995) Proc. Natl. Acad. Sci. USA 92, 6961-6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilm, M., Shevchenko, A., Houthaeve, T., Breit, S., Schweigerer, L., Fotsis, T. & Mann, M. (1996) Nature 379, 466-469. [DOI] [PubMed] [Google Scholar]
- 24.French, C. A., Tambini, C. E. & Thacker, J. (2003) J. Biol. Chem. 278, 45445-45450. [DOI] [PubMed] [Google Scholar]
- 25.Goto, H., Tomono, Y., Ajiro, K., Kosako, H., Fujita, M., Sakurai, M., Okawa, K., Iwamatsu, A., Okigaki, T., Takahashi, T., et al. (1999) J. Biol. Chem. 274, 25543-25549. [DOI] [PubMed] [Google Scholar]
- 26.Pospisil, R. & Mage, R. G. (1998) Cell. Immunol. 185, 93-100. [DOI] [PubMed] [Google Scholar]
- 27.Rhee, K. J., Jasper, P. J., Sethupathi, P., Shanmugam, M., Lanning, D. & Knight, K. L. (2005) J. Exp. Med. 201, 55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weill, J. C. & Reynaud, C. A. (2005) J. Exp. Med. 201, 7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pospisil, R., Alexander, C. B., Obiakor, H., Sinha, R. K. & Mage, R. G. (2006) Dev. Comp. Immunol., in press. [DOI] [PubMed]
- 30.Martin, A. & Scharff, M. D. (2002) Proc. Natl. Acad. Sci. USA 99, 12304-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P. & Yu, X. F. (2003) Science 302, 1056-1060. [DOI] [PubMed] [Google Scholar]
- 32.Sheehy, A. M., Gaddis, N. C. & Malim, M. H. (2003) Nat. Med. 9, 1404-1407. [DOI] [PubMed] [Google Scholar]
- 33.Brar, S. S., Watson, M. & Diaz, M. (2004) J. Biol. Chem. 279, 26395-26401. [DOI] [PubMed] [Google Scholar]
- 34.Ito, S., Nagaoka, H., Shinkura, R., Begum, N., Muramatsu, M., Nakata, M. & Honjo, T. (2004) Proc. Natl. Acad. Sci. USA 101, 1975-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride, K. M., Barreto, V., Ramiro, A. R., Stavropoulos, P. & Nussenzweig, M. C. (2004) J. Exp. Med. 199, 1235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valerie, K. & Povirk, L. F. (2003) Oncogene 22, 5792-5812. [DOI] [PubMed] [Google Scholar]
- 37.Lieber, M. R., Ma, Y., Pannicke, U. & Schwarz, K. (2003) Nat. Rev. Mol. Cell Biol. 4, 712-720. [DOI] [PubMed] [Google Scholar]
- 38.Rothkamm, K., Krüger, I., Thompson, L. H. & Löbrich, M. (2003) Mol. Cell. Biol. 23, 5706-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin, H. L., Suzuki, Y., Matsumoto, Y., Tomita, M., Furusawa, Y., Enomoto, A., Morita, A., Aoki, M., Yatagai, F., Suzuki, T., et al. (2004) Radiat. Res. 162, 433-441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.