Abstract
An understanding of the nature of the chromosomes of the filariae is expected to greatly assist the future interpretation of genome data. Filarial development is not eutelic, and there does not seem to be a fixed number of cell divisions in the way that there is in Caenorhabditis. It is not clear whether the chromosomes of the filariae have localized centromeres or whether they are holocentric. Sex determination is by a chromosomal "balance" X0 system in most filariae, but in some Onchocercidae there has been a chromosomal fusion to create a neo-XY system. It is presumed that the molecular basis of sex determination in filariae is similar to Caenorhabditis. The ancestral karyotype of the filariae is probably 5A+X0, but in some Onchocercidae this has been reduced to 4A+XY, and in O. volvulus and O. gibsoni it has been further reduced to 3A+XY. Onchocerca volvulus and O. gibsoni both have supernumary (B-) chromosomes and in O. volvulus there is a single active nucleolus organising region near the middle of the long autosome.
Background
Filariae and other nematodes have small genomes relative to other multicellular eukaryotes. Onchocerca volvulus and Wuchereria bancrofti have estimated haploid genomes of 1.5 and 0.81 × 108 nucleotide pairs respectively [1], and this corresponds, for example, to genome sizes of 30 and 2.78 × 108 in humans and Anopheles gambiae respectively [2]. Because of the small genome size, the chromosomes of the filariae are correspondingly small. This makes it difficult to see their gross morphological features, which are at the limits of resolution of the light microscope, and consequently the cytogenetics of the Filariae have not been well studied. However, the physical structure of the chromosomes is a reflection of the organisation of the DNA and its genetics. For example, the chromosomes determine the number of linkage groups and the pattern of sex linkage, and how this can vary between species. The interpretation of the available nuclear genome sequence of Brugia malayi and the expressed sequence tag (EST) libraries for Onchocerca volvulus, Onchocerca ochengi, Wuchereria bancrofti, Brugia malayi, Dirofilaria immitis and Litomosoides sigmodontis [3] as well as any possible O. volvulus genome sequencing project will be helped by an understanding of their chromosomes.
Eutely, centromeres and sex determination
The observations of Goldschmidt [4] established the idea that postembryonic growth in nematodes occurred without further cell division, and this became known as eutely. For more than 70 years this idea became established in the general literature (for example, [5]), but it is now clear that there are many species where it is not true. For example in Caenorhabditis elegans there is a 1.47-fold increase in the number of somatic nucleii [6] and in Romanomermis culicivorax there is an 8-fold increase during the parasitic phase [7]. However, these two species contrast another feature of nematode development. In C. elegans it is clear that there is a fixed number of cell divisions (including post-embryonic cell divisions) during development leading to a constant number of somatic nucleii in the adult [6]. This is not true of R. culicivorax where the number of somatic nucleii varies and is correlated with adult size [7]. There have been no specific studies of these patterns in Filariae, but Bain [8] has reported somatic cell divisions in developing larvae of O. volvulus in the vector. Furthermore, counts of lacto-acetic orcein stained nucleii [9] of intrauterine microfilariae and infective L3 larvae of O. volvulus have indicated a mean of 280 nucleii in microfilariae and approximately 900 in L3s [10]. It is clear that the concept of eutely can not be applied to filariae, because there is obviously an increase in cell number between these two post-embryonic stages. It is also unlikely that filariae have a fixed number of cell divisions because larvae at the same stage of development were found to show variation in numbers of somatic nuclei [9].
It seems that most nematodes probably have holocentric chromosomes (the chromosomes attach to the meiotic and mitotic spindle microtubules along their whole length, instead of this function being concentrated into a single centromere) [11]. A consequence of this is that broken fragments of chromosomes can still assort regularly at cell division. However, it is clear that the trichurids (at least) have normal (localised) centromeres, but the situation is not well understood for the filariae. Procunier and Hirai [12] interpreted their mitotic and meiotic metaphases from O. volvulus and O. gutturosa in terms of localised centromeres, but it is not obvious that their drawn figures are correct interpretations of their photographs, and they explained that "the position of the centromere is not always obvious" and in the longest chromosome its position "appears to vary between individuals". The chromosomes of the filariae are very small, difficult to interpret and no other authors have directly addressed this issue. However, the orientation of the chromosomes at metaphase has been remarked upon. Delves et al. [13] showed that synaptonemal complexes were present in early female meiosis of Dirofilaria immitis, but at metaphase I the chromosomes appeared to be pairing end to end. This was particularly obvious for the X chromosome pair (which is the longest chromosome in D. immitis) because a more normal side by side pairing was observed in only 10% of ova. This same end to end pairing is apparent in the photographs of other authors for other filariae (for example [14]). In most animals meiotic metaphase chromosomes show evidence of crossing over (which is the visible manifestation of recombination), but this is not obvious for filariae. Procunier and Hirai [12] interpreted their figures to show crossing over, but no other authors have done so, and it is not clear whether this is due to the small size of the chromosomes or some more fundamental biological reason. In any case, recombination is expected to occur in filariae because they have synaptonemal complexes [13] and recombination has been proven in some other nematodes such as Caenorhabditis [15], and Globodera [16].
Some nematodes such as Strongyloides exhibit forms of parthenogenesis, and a few such as Mermis subnigrescens have environmental sex determination (dependant upon the number of mermithids parasitising a particular insect host). However, amongst the vertebrate parasites chromosomal sex determination seems to be the rule. And there seems to be an X0 system in all species where it is known except the Oxyuridae (which have a system of haplo-diploidy), Ascaridae (which have multiple sex chromosomes) and Filarioidea (where some species of Onchocercidae have an XY system). It is clear that the X0 system is fundamental to nematodes (including the filariae), and the few XY systems which occur amongst the filariae are secondary derivatives. In other organisms with X0 sex chromosomes, such as Orthoptera, the fusion of an autosome to the X chromosome to create a neo-XY is very well documented [17]. Genetically, an X0 sex determining system has to be a 'balance' system, and there is a good understanding of the molecular basis of sex determination in Caenorhabditis (which has an X0 system, with X0 males and XX hermaphrodites) [18], and it is probable that filariae are basically similar.
Chromosome numbers and karyotype evolution
Table 1 summarises all published records of filarial karyotypes, and there are apparently three basic types, 5A+X0, 4A+XY and 3A+XY. There are a few exceptions in the literature, and it is not always clear the extent to which these might be errors resulting from the difficult nature of filarial chromosomes, or indicative of natural variation. Litomosoides sigmodontis has been reported to have either 5A+X0 (new unpublished data, see Figure 1) or 4A+X0 [19,20]. In view of the karyotypes of other X0 species it is likely that 5A+X0 is correct, and 4A+X0 is either the result of natural variation for an autosome-autosome translocation or a mistake (due to small difficult chromosomes). Indeed, there are clearly six chromosomes present in some of McLaren's [20] illustrations. Similarly Taylor [19] reported Dirofilaria immitis to have 4A+X0, whilst other authors reported 5A+X0 [20] or 4A+XY [21,13]. It is possible that this reflects natural variation within Dirofilaria, but Taylor [19] did not have access to a past body of published work that later cytogeneticists have been able to build upon, and it seems most likely that 4A+X0 was a mistake. Post et al. [22] reported that Onchocerca tarsicola from Germany was 4A+XY (i.e. n = 5), but the same species from Sweden was n = 3. This difference was attributed to possible intraspecific geographic variation. Most authors agree that O. volvulus has 3A+XY (i.e. n = 4). However, Basáñez et al. [23] reported n = 4 or 5, and Miller [24] reported n = 5. It is likely that these reports are the result of B-chromosome variation (see below), but this can not explain the old report by Salazar Mallen et al. [25] of n = 2. In the absence of corroborating evidence this report might be thought of as an error resulting from the experimental use of tissue sections, which is an older cytogenetic technique that is more problematic than the newer squash techniques [26].
Table 1.
List of karyotypes recorded from the Filarioidea
| Species | Karyotype | Authors |
| With X0 System: | ||
| Acanthoceilonema viteae | 5A+X0 | [34] |
| 5A+X0 | [20] | |
| Dipetalonema setariosum | 5A+X0 | [20] |
| Setaria equina | 5A+X0 | [35] |
| Setaria digitata | 5A+X0 | [21] |
| Litomosoides sigmodontis | 4A+X0 | [19] |
| 4A+X0 | [20] | |
| 5A+X0 | Jolley & Post, unpublished | |
| Litomosoides galizai | 5A+X0 | Wade & Post, unpublished |
| Mononema martini | 5A+X0 | Wade & Post, unpublished |
| Loa loa | 5A+X0 | [36] |
| With XY System: | ||
| Dirofilaria immitis | 4A+X0 | [19] |
| 5A+X0 | [20] | |
| 4A+XY | [21] | |
| 4A+XY | [13] | |
| Brugia pahangi | 4A+XY | [37] |
| Brugia malayi | 4A+XY | [37] |
| 4A+XY | [38] | |
| Wuchereria bancrofti | 4A+XY | [24] |
| Onchocerca gutturosa | 4A+XY | [39] |
| 4A+XY | [38] | |
| 4A+XY | [14] | |
| Onchocerca lienalis | 4A+XY | [14] |
| Onchocerca armilatta | 4A+XY | [14] |
| Onchocerca tarsicola | Germany 4A+XY (n = 5) | [22] |
| Sweden n = 3 | [22] | |
| Onchocerca dukei | 4A+XY | [22] |
| Onchocerca ochengi | 4A+XY | [14] |
| Onchocerca gibsoni | 3A+XY | [14] |
| Onchocerca volvulus | n = 2 | [25] |
| n = 4 or 5 | [23] | |
| n = 5 | [24] | |
| 3A+XY (n = 4) | [39] | |
| 3A+XY | [12] | |
| 3A+XY | [40] | |
| 3A+XY | [41] | |
| 3A+XY | [14] |
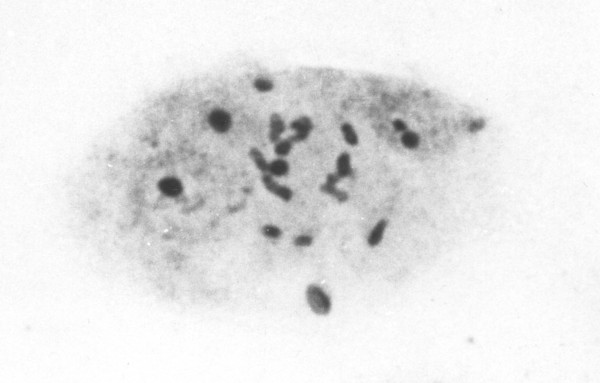
Figure 1.
Litomosoides sigmodontis karyotype. Aceto-orcein stained spermatozoa from the seminal receptacle of an adult female worm, showing five or six condensed chromosomes (arrowed).
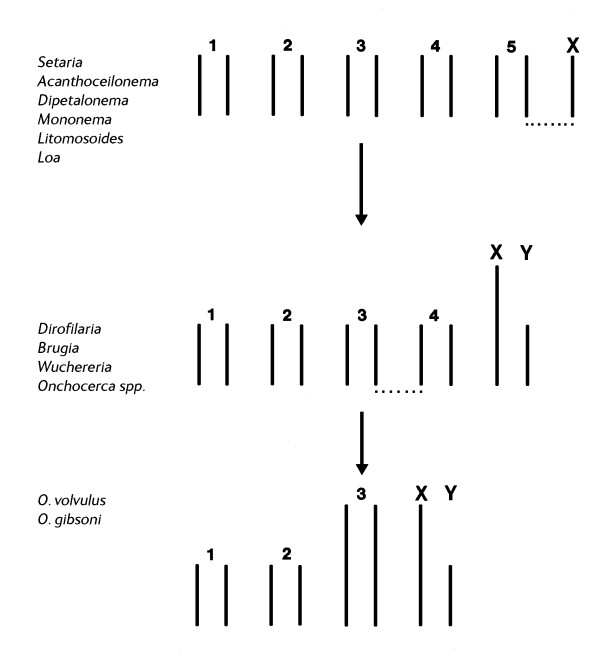
Figure 2 shows the likely karyotype evolution in the filariae. 5A+X0 is likely to be ancestral because the X0 sex chromosome system is the almost universal sex chromosome system amongst nematodes (see above), and hence the XY system is almost certainly a derived (neo-XY) system, resulting from the fusion of an autosome to the old X-chromosome. There are a number of chromosomal mechanisms for such a fusion [27]. If the chromosomes have localized centromeres a Robertsonian translocation is most likely (i.e. one that occurs at the centromere so that the resultant fragments still have a single centromere and hence can disjoin regularly at meiosis and mitosis). If the chromosomes are holocentric almost any sort of fusion can occur, because all fragments will show centromeric activity and can disjoin regularly. In support of the postulated fusion (by whatever chromosomal mechanism), the five autosomes and the X-chromosome of the 5A+X0 species are all approximately the same size as each other, whereas in the 4A+XY species the four autosomes and the Y-chromosome are of comparable size, whereas the X-chromosome (which would be an X-autosome fusion product) is visibly larger. The 3A+XY karyotype observed in O. volvulus and O. gibsoni is likely to have been derived from the 4A+XY karyotype by a fusion between two autosomes [12]. The relative sizes of the different chromosomes are consistent with this hypothesis in that one of the autosomes is clearly the largest chromosome, and presumably the autosome-autosome fusion product.
Figure 2.
Karyotype evolution in the Filariae.
The pattern of karyotype evolution (Figure 2) is likely to reflect organismal evolution and indicate taxonomic relationships. All species of filarioids which have been examined for their chromosomes are from the family Onchocercidae, and from three subfamilies, Setariinae (Seteria spp), Dirofilariinae (Dirofilaria and Loa) and Onchocercinae (all other species in Table 1). The presumed ancestral state (5A+X0) occurs in the Setariinae, most of the Onchocercinae and Loa loa (but not Dirofilaria). Current phylogenetic opinion has been unable to resolve the major clades of the Onchocercinae, which reduce to a star polytomy [28]. Loa forms a clade from this unresolved polytomy, but Dirofilaria is consistently grouped along the Onchocerca clade. The reduction to 4A+XY has occurred in both Dirofilaria and Onchocerca, and hence it supports the molecular data, but it also indicates that the Onchocerca/Dirofilaria clade is most closely related to the Wuchereria/Brugia clade because it is found in both.
The observation that O. volvulus apparently shares the same karyotype as O. gibsoni, but not O. ochengi or O. dukei, might be taken as evidence for a close phylogenetic relationship, and Muller [29] also considered that O. volvulus was most closely related to O. gibsoni on the basis of the structure of the cuticle. However, Bain [30] held that O. volvulus was taxonomically more closely related to O. ochengi, and recent molecular phylogeny reconstruction using the 12s, 16s and ND5 mitochondrial genes very strongly supports this view (Morales Hojas, Cheke and Post, unpublished data). If this is true, it would indicate that the similar karyotypes of O. volvulus and O. gibsoni have been produced by two different fusion events (which may not even have involved the same pairs of autosomes), and this is likely to be reflected in the precise position of the translocation breakpoints at the molecular level.
B-chromosomes and nucleolus organising region
Both O. volvulus and O. gibsoni have B-chromosomes [12,14]. These are supernumery chromosomes which are not present in every individual, and when they are present they can be haploid, diploid, triploid, tetraploid, etc, in different individuals. The origin and subsequent evolution of B-chromosomes is a long-standing problem in genetics [31]. B-chromosomes can have definite phenotypic affects, but it has always been unclear whether this was the result of specific genes or a mere consequence of the presence of an extra chromosome. It is largely unknown whether B-chromosomes carry genes and whether those genes are repeated elsewhere in the more constant part of the genome. In any case, if the origin of the 3A+XY karyotype was the result of some sort of autosome-autosome fusion this would result in one new large autosome (see Figure 2), and possibly a small chromosomal fragment [27]. The exact nature of the fragment will depend upon the exact nature of the fusion and there are a number of potential chromosomal mechanisms, which partly depend upon the exact nature of the chromosomes. Procunier and Hirai [12] interpreted the B-chromosomes of O. volvulus as chromosomal fragments which resulted from a Robertsonian translocation (i.e. a translocation at the site of the centromere in a species with localised centromeres) which resulted in a new large autosome and a small chromosomal fragment. In such cases, both products would be able to assort regularly because both would have a centromere. If Onchocerca have holocentric chromosomes, any chromosome fragment will show centromeric activity and disjoin regularly.
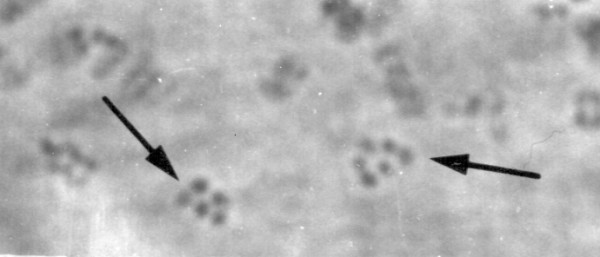
There have been no attempts to characterize the different chromosomes of any filariae using chromosome banding or molecular cytogenetics. The only differences that have been noted are size differences and sex chromosome differences (see above). However, There has been one unpublished study of silver staining of O. volvulus (Post & Bella, unpublished data). Silver staining is a technique that stains the nucleolus and hence reveals the active nucleolus organising regions, which is where the active ribosomal genes (rDNA) are found [32]. In O. volvulus there was a single nucleolus organizing region, situated at the centre of the long autosome (Figure 3). There may be other inactive clusters of rDNA elsewhere in the genome, and it is interesting to note that rDNA has sometimes been implicated at the sites of chromosome mutations (such as translocations) in other organisms such as humans [33].
Figure 3.
Onchocerca volvulus NOR. Silver stained seven-cell morula embryo showing six interphase nuclei and the central cell in mitotic metaphase with four chromosome pairs including the longest pair each with a central Nucleolus Organising Region darkly stained.
Conclusion
1. Eutely is certainly not a characteristic of filarial development, and there does not seem to be a constant number of somatic cells at different developmental stages. However, it is still possible that the rather constant pattern of cell lineages seen in Caenorhabditis is present in filariae in some more flexible form.
2. Although some authors have interpreted their metaphase chromosome figures as indicating localised centromeres this is unusual in nematodes, which usually have holocentric chromosomes. Meiotic metaphase chromosomes exhibit an unusual appearance of end to end pairing in filariae, which is not consistent with having a localised centromere. The nature of the centromere in filariae remains to be resolved.
3. Sex determination in filariae is chromosomal and fundamentally of the X0 type, although in some species of the Onchocercidae this has been secondarily modified into a neo-XY system by the fusion of an autosome onto the old X chromosome. X0 sex determination is genetically a balance system, and likely to be similar to that described at the molecular level in Caenorhabditis.
4. The primitive karyotype of the Filarioidea is presumed to be 5A+X0, because the X0 chromosomal sex determining system seems to be almost universal amongst nematodes in general. However, in some Onchocercidae there has been an X-autosome fusion to produce a neo-XY system with four autosomal pairs (4A+XY). In O. volvulus and O. gibsoni there has been a further fusion of two autosomes to yield a karyotype of 3A+XY. This second step might have occurred independently in the two species.
5. Both O. volvulus and O. gibsoni have B-chromosomes, probably resulting from the autosome-autosome translocation which reduced the karyotype to n = 4. However, nothing is known about these chromosomes in terms of their genetics or potential phenotypic effects.
6. In O. volvulus there is a single active nucleolus organizing region (where the active rDNA occurs) near the centre of the long autosome, near the site of the chromosomal fusion event.
Competing interests
The author(s) declare that they have no competing interests.
Acknowledgments
Acknowledgements
Philip McCall (Liverpool School of Tropical Medicine) provided the cryopreserved larval stages of Onchocerca volvulus. Litomosoides galizai and Monanema martini were kindly supplied by Dr Odile Bain (Museum National d'Histoire Naturelle, Paris), and specimens of Litomosoides sigmodontis were provided by Dr A Beg (University of Salford, UK). The silver staining of the nucleolus organising region was carried out in collaboration with Dr Pepe Bella (Universidad Autonoma de Madrid). The specimen illustrated in Figure 1 was prepared by Mr S Jolley (University of Salford, UK).
References
- Hammond MP, Bianco AE. Genes and genomes of parasitic nematodes. Parasitol Today. 1992;8:299–305. doi: 10.1016/0169-4758(92)90100-G. [DOI] [PubMed] [Google Scholar]
- GOLD Genomes OnLine Database http://www.genomesonline.org/
- Nematode.net Genome Sequencing Center http://www.nematode.net/
- Goldschmidt RB. Histologische untersuchungen an nematoden. Zool Jahrb Abt Anat Ontog Tiere. 1903;18:1–57. [Google Scholar]
- Barnes RD. Invertebrate Zoology. second . London: WB Saunders company; 1968. [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Curran J, Webster JM. Postembryonic growth of Romanomermis culicivorax Ross and Smith, 1976: an example of accretionary growth in the Nematoda. Can J Zool. 1983;61:1793–1796. [Google Scholar]
- Bain O. Morphologie des stades larvaires d'Onchocerca volvulus chez Simulium damnosum et redescription de la microfilaire. Ann Parasitol. 1969;44:69–82. [PubMed] [Google Scholar]
- Mimori T, Tada I, Shiwaku K, Ufomadu GO, Nwoke BEB. A biometric study of Onchocerca volvulus microfilariae from Nigeria using the nuclear counting method. Z Parasitenkd. 1986;72:835–836. doi: 10.1007/BF00925105. [DOI] [PubMed] [Google Scholar]
- Post RJ, McCall PJ, Trees AJ, Delves CJ, Kouyate B. The cytotaxonomy of Onchocerca [abstract] British Simuliid Group Bulletin. 1993;2:3. [Google Scholar]
- Triantaphyllou AC. Genetics and Cytology. In: Zuckerman BM, Mai WF, Rohde RA, editor. Plant Parasitic Nematodes, Cytogenetics, Host-Parasite Interactions, and Physiology. Vol. 2. London: Academic Press; 1971. pp. 1–34. [Google Scholar]
- Procunier WS, Hirai H. The Chromosomes of Onchocerca volvulus. Parasitol Today. 1986;2:307–309. doi: 10.1016/0169-4758(86)90125-0. [DOI] [PubMed] [Google Scholar]
- Delves CJ, Howells RE, Post RJ. Gametogenesis and fertilization in Dirofilaria immitis (Nematoda: Filarioidea) Parasitology. 1986;92:181–197. doi: 10.1017/s003118200006354x. [DOI] [PubMed] [Google Scholar]
- Post RJ, McCall PJ, Trees AJ, Delves CJ, Kouyate B. Chromosomes of six species of Onchocerca (Nematoda: Filarioidea) Trop Med Parasit. 1989;40:292–294. [PubMed] [Google Scholar]
- Barnes TM, Kohara Y, Coulson A, Hekimi S. Meiotic recombination, noncoding DNA and genome organization in Caenorhabditis elegans. Genetics. 1995;141:159–179. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voort R, van Eck HJ, van Zandvoort PM, Overmars H, Helder J, Bakker J. Linkage analysis by genotyping of sibling populations: a genetic map for the potato cyst nematode constructed using a "pseudo-F2" mapping strategy. Mol Gen Genet. 1999;261:1021–1031. doi: 10.1007/s004380051051. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Orthoptera Grasshoppers and Crickets. Berlin: Gebrüder Borntraeger; 1999. [John B (Series Editor): Animal Cytogenetics vol 3.1] [Google Scholar]
- Stothard P, Pilgrim D. Sex determination gene and pathway evolution in nematodes. Bioessays. 2003;25:221–231. doi: 10.1002/bies.10239. [DOI] [PubMed] [Google Scholar]
- Taylor AER. The spermatogenesis and embryology of Litomosoides carinii and Dirofilaria immitis. J Helminthol. 1960;34:3–12. doi: 10.1017/s0022149x00020289. [DOI] [PubMed] [Google Scholar]
- McLaren DJ. The structure and development of the spermatozoon of Dipetalonema vitae (Nematoda: Filarioidea) Parasitology. 1973;66:447–463. doi: 10.1017/s0031182000046011. [DOI] [PubMed] [Google Scholar]
- Sakaguchi Y, Kihara S, Tada I. The chromosomes and gametogenesis of Dirofilaria. Jap J Parasit. 1980;29:53–57. [Google Scholar]
- Post RJ, Bain O, Kläger S. Chromosome numbers in Onchocerca dukei and O. tarsicola. J Helminthol. 1991;65:208–210. doi: 10.1017/s0022149x00010725. [DOI] [PubMed] [Google Scholar]
- Basáñez MG, Botto C, Yarzábal L. Caracteristicas de un deme de Onchocerca volvulus s.l. del alto Orinoco (Territorio Federal Amazonas, Venezuela). Estudio citogenetico preliminary. Caracas Publ Cient. 1983;2:79–82. [Google Scholar]
- Miller MJ. Observations on spermatogenesis in Onchocerca volvulus and Wuchereria bancrofti. Can J Zool. 1966;44:1003–1006. doi: 10.1139/z66-104. [DOI] [PubMed] [Google Scholar]
- Salazar Mallén M, González Barranco QFBD, Sámano A. Chromosomas de Onchocerca volvulus. Salud Pùbl Méx Epoca V. 1962;4:983–984. [Google Scholar]
- Darlington CD, La Cour LF. The Handling of Chromosomes. London: George Allen & Unwin; 1976. [Google Scholar]
- White MJD. Animal Cytology and Evolution. Third. Cambridge: University Press; 1973. [Google Scholar]
- Casiraghi M, Bain O, Guerrero R, Martin C, Pocacqua V, Gardner SL, Franceschi A, Bandi C. Mapping the presence of Wolbachia pipientis on the phylogeny of filarial nematodes: evidence for symbiont loss during evolution. Int J Parasit. 2004;34:191–204. doi: 10.1016/j.ijpara.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Muller R. Identification of Onchocerca. In: Taylor AER, Muller R, editor. Problems in the Identification of Parasites and their Vectors. Oxford: Blackwell; 1979. pp. 175–206. 17th Symposium of the British Society for Parasitology. [Google Scholar]
- Bain O. Evolutionary relationships among filarial nematodes. In: Klei TR, Rajan TV, editor. The Filaria. Boston: Kluwer Academic; 2002. pp. 21–29. Black SJ, Seed JR (Series Editors): World Class Parasites, vol 5. [Google Scholar]
- Jones RN, Rees H. B Chromosomes. London: Academic Press; 1982. [Google Scholar]
- De La Torre J, Bella JL, López-Fernandez C, Gosálvez J. New approaches to the role of sulfhydryl groups in silver stainability of protein in grasshopper chromosomes. Genome. 1988;30:133–137. [Google Scholar]
- Grabowski M, Fauth C, Wirtz A, Speicher MR. Breakpoint within the nucleolus organizer region resulting in a reciprocal translocation t(4;14)(q21;p12) Am J Med Gen. 2000;92:264–268. doi: 10.1002/(SICI)1096-8628(20000605)92:4<264::AID-AJMG8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Terry A, Terry RJ, Worms MJ. Dipetalonema witei, filarial parasite of the Jird, Meriones libycus. II. The reproductive system, gametogenesis and development of the microfilaria. J Parasitol. 1961;47:703–711. [PubMed] [Google Scholar]
- Walton AC. Some parasites and their chromosomes. J Parasitol. 1959;45:1–20. [PubMed] [Google Scholar]
- Post RJ, Pinder M. Oogenesis and embryogenesis in Loa loa. J Helminthol. 1995;69:351–356. [Google Scholar]
- Sakaguchi Y, Tada I, Ash LR, Aoki Y. Karyotypes of Brugia pahangi and Brugia malayi (Nematoda: Filarioidea) J Parasitol. 1983;69:1090–1093. [PubMed] [Google Scholar]
- Delves CJ. PhD thesis. University of Liverpool, Liverpool School of Tropical Medicine; 1986. Developmental Processes in Filarial Worms. [Google Scholar]
- Hirai H, Sakaguchi Y, Tada I. Chromosomes of Onchocerca volvulus and O. gutturosa. Z Parasitenkd. 1985;71:135–139. doi: 10.1007/BF00932927. [DOI] [PubMed] [Google Scholar]
- Hirai H, Tada I, Takahashi H, Nwoke BEB, Ufomadu GO. Chromosomes of Onchocerca volvulus (Spirurida: Onchocercidae): A comparative study between Nigeria and Guatemala. J Helminthol. 1987;61:43–46. doi: 10.1017/s0022149x0000969x. [DOI] [PubMed] [Google Scholar]
- Akoh JI, Tada I, Uchida A, Sato Y, Hirai H. Cytogenetic analysis of chromosomal polymorphism in southern and northern guinea savanna grassland strains of O. volvulus from Nigeria. Proceedings of the Nigeria/Japan Joint Conference, Jos. 1987. pp. 198–199.