Abstract
Most cases of severe Staphylococcus aureus disease cannot be explained by the action of a single virulence determinant, and it is likely that a number of factors act in combination during the infective process. This study examined the relationship between disease in humans and a large number of putative virulence determinants, both individually and in combination. S. aureus isolates (n = 334) from healthy blood donors and from patients with invasive disease were compared for variation in the presence of 33 putative virulence determinants. After adjusting for the effect of clonality, seven determinants (fnbA, cna, sdrE, sej, eta, hlg, and ica) were significantly more common in invasive isolates. All seven factors contributed independently to virulence. No single factor predominated as the major predictor of virulence, their effects appearing to be cumulative. No combinations of the seven genes were either more or less likely to cause disease than others with the same number of virulence-associated genes. There was evidence of considerable horizontal transfer of genes on a background of clonality. Our findings also suggested that allelic variants of a polymorphic locus can make different contributions to the disease process, further study of which is likely to expand our understanding of staphylococcal disease pathogenesis.
Staphylococcus aureus is a major cause of severe community-acquired and nosocomial sepsis (12, 36). The relative importance of host factors versus bacterial virulence determinants in disease pathogenesis is unknown, but it is widely held that bacterial factors including toxins, cell wall-associated adhesins, and secreted exoproteins are involved in the process (34). There is clear evidence for this in the case of toxin-mediated S. aureus diseases such as toxic shock syndrome caused by toxic shock syndrome toxin 1, scalded skin syndrome resulting from the action of epidermolytic toxins, and food poisoning following the ingestion of preformed enterotoxins (6). However, the vast majority of cases of severe S. aureus disease cannot be explained by the action of a single virulence determinant and it is likely that a number of factors act in combination during the infective process. Support for this comes from animal models where outcome has been compared following infection with isogenic mutants deficient in a single putative virulence determinant, in which infection was attenuated but not prevented (17, 27). In addition, clinical isolates associated with human infection may be naturally deficient in a range of putative virulence determinants. It seems reasonable to postulate, therefore, that S. aureus strains associated with human infection have variable combinations of pathogenic determinants and that either the presence or the expression of given combinations varies depending on the type of infection and genetic susceptibility of the affected host.
The aim of this study was to examine a large number of putative virulence determinants in a natural population of S. aureus isolates and explore their relationship, both individually and in combination, to invasive S. aureus disease in humans. A panel of isolates was assembled during a prospective case control study in which strains were collected from healthy blood donors and from patients with invasive disease, the quality of clinical information being ensured by the prospective application of defined case definitions. The presence of virulence determinants was compared between the isolate groups associated with carriage and disease, analysis of which included adjustment for the potentially confounding effects of clonality and the hitchhiker effect. This incorporated a framework provided by multilocus sequence typing (MLST), a technique which also gave us insights into the horizontal gene transfer of virulence determinants. Finally, we explored the possibility that allelic variants at a given polymorphic locus (namely, sdrE/bbp) do not make the same contribution to the pathogenic process.
MATERIALS AND METHODS
Bacterial isolates.
We examined 33 bacterial determinants in 155 isolates recovered from patients with invasive S. aureus disease (94 hospital acquired and 61 community acquired) and 179 isolates recovered from healthy individuals. These isolates were collected within Oxfordshire, United Kingdom, between 1997 and 1998 by using a prospective case control design. Cases of invasive infection were identified prospectively through the microbiology laboratory serving the main hospitals in the Oxford area. Clinical details were recorded, and the bacterial isolate was stored. Community-acquired disease was defined as admission to a hospital with an illness consistent with invasive S. aureus disease, with isolation of S. aureus from a normally sterile site within 24 h of admission. Hospital-acquired disease was defined as an illness consistent with invasive S. aureus disease and isolation of the organism from a normally sterile site 48 h or more after admission for another condition. Control bacteria were obtained from nasal swabs of blood donors drawn from the same population catchment area as Oxford Radcliffe Hospitals.
Evaluation of bacterial determinants.
Twenty-nine determinants were examined for the presence of the gene by PCR, and four determinants were evaluated by phenotypic tests. The bacterial determinants examined incorporated the major putative virulence factors reported in the published literature at the design stages of this study (Table 1).
TABLE 1.
Bacterial determinants examined in this study
| Bacterial determinant | Putative function | Reference | Method of detection |
|---|---|---|---|
| Adhesins | |||
| FnBPA | Adhesin for fibronectin | 14 | PCR for fnbA |
| ClfA and ClfB | Adhesins for fibrinogen | 24, 28 | PCR for clfA and clfB |
| Cna | Adhesin for collagen | 32 | PCR for cna |
| Protein A | Binds Fc domain of immunoglobulin and von Willibrand factor | 13, 15 | PCR for spa |
| SdrC, SdrD, and SdrE | Unknown; putative adhesins | 21 | PCR for sdrC, sdrD, and sdrE |
| Bbp | Adhesin for bone sialoprotein | 38 | PCR for bbp |
| EbpS | Adhesin for elastin | 10, 30 | PCR for ebpS |
| Map/Eap | Major histocompatibility complex class II analogue protein | 20, 25, 29 | PCR for map/eap |
| Toxins | |||
| TSST-1a | Exotoxin with superantigen activity | Reviewed in reference 7 | PCR for tst |
| Enterotoxins A, B, C, D, E, G, H, I, and J | Exotoxins with superantigen activity | Reviewed in reference 7 | PCR for sea, seb, etc. |
| Exfoliative toxins A and B | Exotoxins with superantigen activity | Reviewed in reference 7 | PCR for eta and etb |
| Alpha-toxin | Cytolytic pore-forming toxin | Reviewed in reference 2 | Lysis of rabbit erythrocytes |
| Beta-toxin | Sphingomyelinase | Reviewed in reference 1 | Hot-cold lysis of sheep erythrocytes |
| Delta-toxin | Cytolytic toxin | Reviewed in reference 1 | Syngergy of lysis between beta-toxin producer and test isolate on washed sheep erythrocytes |
| Panton-Valentine leukocidin | Bicomponent leukocidin | 33, 37 | PCR for pvl |
| Gamma-toxin | Bicomponent leukocidin | 33, 37 | PCR for hlg |
| Others | |||
| ica locus | Polysaccharide intercellular adhesin | 9 | PCR for icaA |
| Coagulase | Binds prothrombin, activating conversion of fibrinogen to fibrin | 16, 22 | Coagulase test |
| Efb | Binds to fibrinogen | 3 | PCR for efb |
| V8 protease | Serine protease | 11 | PCR for V8 |
| agr subgroup | Global regulator | 18, 19 | PCR for agr subgroups I, II, III, and IV |
TSST-1, toxic shock syndrome toxin 1.
PCR analysis.
Genomic DNA from S. aureus was extracted by using the Wizard Genomic DNA purification kit (Promega), with the modification that 30 μg of lysostaphin/ml (Ambi) was added at the cell lysis step. sea-i and tst were amplified by multiplex PCR with the primers and conditions described elsewhere (26); sej, eta, and etb were amplified with the primers and conditions provided by G. O'Neill (personal communication). Primers and conditions used to amplify efb, pvl, and hlg were as described in references 5 (efb) and 23 (pvl and hlg). Primer design for the remaining determinants was based on gene sequences available from GenBank (Table 2). This was straightforward with the exception of map/eap, the agr subgroup, and sdrE and bbp. There were three GenBank sequences available for map (AJ243790, clinical isolate 7; AJ245439, strain Wood 46; AJ223806, strain Newman) and one for a gene termed map/eap (AJ290973, strain Newman). These sequences were aligned, and common forward and reverse primers were designed. To distinguish between agr groups I, II, III, and IV, use was made of sequence differences in or around agrD, the region encoding the autoinducing peptide. The forward primers for agr subgroups I and IV started 9 bases upstream of the region encoding the peptide; primers for agr subgroups II and III started at the ninth and first bases, respectively, of the region encoding the peptide. The reverse primers for all agr subgroups were within agrC.
TABLE 2.
PCR primers and conditions
| PCR product | Primer descriptions and sequences | Positive control (product length [bp]) | PCR conditions cycling | Mg concn (mM) |
|---|---|---|---|---|
| fnbA | Forward, position 431 (accession no. J04151); 5′-CACAACCAGCAAATATAG-3′ | 8325-4 (1,362) | 1 min, 94°C; 1 min, 50°C; 2 min, 72°C | 1.5 |
| Reverse, position 1792; 5′-CTGTGTGGTAATCAATGTC-3′ | ||||
| clfA | Forward, position 368 (accession no. Z18852); 5′-GTAGGTACGTTAATCGGTT-3′ | Newman (1,584) | 1 min, 94°C; 1 min, 45°C; 2 min, 72°C | 3 |
| Reverse, position 1951; 5′-CTCATCAGGTTGTTCAGG-3′ | ||||
| clfB | Forward, position 425 (accession no. AJ224764); 5′-TGCAAGATCAAACTGTTCCT-3′ | Newman (596) | 1 min, 94°C; 1 min, 45°C; 1 min, 72°C | 3 |
| Reverse, position 1020; 5′-TCGGTCTGTAAATAAAGGTA-3′ | ||||
| cna | Forward, position 1719 (accession no. M81736); 5′-AGTGGTTACTAATACTG-3′ | Phillips (variable)a | 1 min, 94°C; 1 min, 55°C; 2 min, 72°C | 1.5 |
| Reverse, position 3457; 5′-CAGGATAGATTGGTTTA-3′ | ||||
| spa | Forward, position 1 (accession no. J01786); 5′-TCGAAATAGCGTGATTTTGC-3′ | 8325-4 (1,892) | 1 min, 94°C; 1 min, 65°C; 2 min, 72°C | 1.5 |
| Reverse, position 1892; 5′-GCACTGAGCAACAAAAGATG-3′ | ||||
| sdrC | Forward, position 481 (accession no. AJ005645); 5′-ACGACTATTAAACCAAGAAC-3′ | Newman (560) | 1 min, 94°C; 1 min, 45°C; 1 min, 72°C | 1.5 |
| Reverse, position 1040; 5′-GTACTTGAAATAAGCGGTTG-3′ | ||||
| sdrD | Forward, position 361 (accession no. AJ005646); 5′-GGAAATAAAGTTGAAGTTTC-3′ | Newman (500) | 1 min, 94°C; 1 min, 45°C; 1 min, 72°C | 1.5 |
| Reverse, position 860; 5′-ACTTTGTCATCAACTGTAAT-3′ | ||||
| sdrE | Forward, position 650 (accession no. AJ005647); 5′-CAGTAAATGTGTCAAAAGA-3′ | Isolate 476 (767) | 1 min, 94°C; 1 min, 45°C; 1.5 min, 72°C | 3 |
| Reverse, position 1416; 5′-TTGACTACCAGCTATATC-3′ | ||||
| bbp | Forward, position 836 (accession no. Y18653); 5′-CAGTAAATGTGTCAAAAGA-3′ | Isolate 252 (1,055) | 1 min, 94°C; 1 min, 45°C; 1.5 min, 72°C | 3 |
| Reverse, position 1890; 5′-TACACCCTGTTGAACTG-3′ | ||||
| ebpS | Forward, position 175 (accession no. U48826); 5′-CAATCGATAGACACAAATTC-3′ | Isolate 252 (526) | 1 min, 94°C; 1 min, 50°C, 1 min, 72°C | 4 |
| Reverse, position 700; 5′-CAGTTACATCATCATGTTTA-3′ | ||||
| map/eap | Forward, positions 203 (accession no. AJ243790), 255 (AJ290973), 257 (AJ245439), and 128 (AJ223806); 5′-TAACATTTAATAAGAATCAA-3′ | Newman (943-949) | 1 min, 94°C; 1 min, 45°C; 1.5 min, 72°C | 3 |
| Reverse, positions 1151, 1203, 1199, and 1076, respectively; 5′-CCATTTACTGCAATTGT-3′ | ||||
| icaA | Forward, position 2711 (accession no. AF086783); 5′-GATTATGTAATGTGCTTGGA-3′ | Isolate 252 (770) | 1 min, 94°C; 1 min, 50°C; 1 min, 72°C | 4 |
| Reverse, position 3480; 5′-ACTACTGCTGCGTTAATAAT-3′ | ||||
| V8 | Forward, position 4 (accession no. Y00356); 5′-TTGTTCTTCGAAACTT-3′ | V8 (1,550) | 1 min, 94°C; 1 min, 50°C; 2 min, 72°C | 3 |
| Reverse, position 1553; 5′-GGCTTTGGCTTTATTG-3′ | ||||
| agr subgroup I | Forward, position 2405 (accession no. X52543); 5′-ATCGCAGCTTATAGTACTTGT-3′ | Isolate 2 (739) | 1 min, 94°C; 1 min, 50°C; 1 min, 72°C | 3 |
| Reverse, position 3143; 5′-CTTGATTACGTTTATATTTCATC-3′ | ||||
| agr subgroup II | Forward, position 642 (accession no. AF001782); 5′-AACGCTTGCAGCAGTTTATTT-3′ | Isolate 19 (691) | 1 min, 94°C; 1 min, 50°C; 1 min, 72°C | 3 |
| Reverse, position 1332; 5′-CGACATTATAAGTATTACAACA-3′ | ||||
| agr subgroup III | Forward, position 642 (accession no. AF001783); 5′-TATATAAATTGTGATTTTTTATTG-3′ | Isolate 252 (712) | 1 min, 94°C; 1 min, 50°C; 1 min, 72°C | 3 |
| Reverse, position 1353; 5′-TTCTTTAAGAGTAAATTGAGAA-3′ | ||||
| agr subgroup IV | Forward, position 1346 (accession no. AF88215); 5′-GTTGCTTCTTATAGTACATGTT-3′ | Isolate 3049 (683) | 1 min, 94°C; 1 min, 50°C; 1 min, 72°C | 3 |
| Reverse, position 2028; 5′-CTTAAAAATATAGTGATTCCAATA-3′ |
Variable product size depending on the number of B repeats. (multiples of ∼560 nucleotides).
Primer design for amplification of sdrE and bbp was complicated by the fact that these independently described genes are known to share significant homology (38). We considered whether these genes were alleles of the same locus. The sequences for sdrE (GenBank accession number AJ005647) and bbp (accession number Y18653) were blasted against the available genome sequence for two isolates undergoing sequencing at the Sanger Centre, Cambridge, United Kingdom (http://www.sanger.ac.uk/Projects/S_aureus/blast_server.shtml). These isolates were provided to the Sanger Centre by us and are called isolates 252 (methicillin-resistant isolate) and 476 (methicillin-sensitive isolate) in this paper. Blast analysis demonstrated that sdrE and bbp colocalized to identical positions in the respective isolates. Isolate 252 shared 94% homology at the amino acid level with bbp and 79% homology with sdrE. In contrast, isolate 476 shared 80% homology with bbp and 93% homology with sdrE. The sequences for sdrE and bbp were aligned using Gene Jockey II (Biosoft, Cambridge, United Kingdom), and primers were designed to distinguish between what we assume to be two alleles of a single gene by using a common forward primer but unique reverse primer. These primers were both piloted by using 50 randomly selected isolates. A clear distinction was seen between the primer pairs with the amplification of a single band of the appropriate size for SdrE (766 bp), bbp (1,054 bp), or neither; these primers were used to evaluate the remaining isolates.
PCR amplifications were performed in a PTC-200 DNA engine (MJ Research, Waltham, Mass.) with Taq polymerase (Bioline). The final concentrations of the PCR mixtures were 1× reaction buffer, variable MgCl2 concentrations depending on optimization results (Table 2), 100 pmol of forward and reverse primers, 1 μl of 1:5 dilution template DNA, 200 μM deoxynucleoside triphosphate mix, and 2.5 U of Taq polymerase. The PCR primers and cycling conditions not referred to elsewhere are shown in Table 2. Aliquots of the reaction mixtures were analyzed by 1% agarose gel electrophoresis. A positive control (Table 2) and a negative control (reaction mixture minus DNA) were included in each PCR run.
Phenotypic tests.
Production of coagulase was evaluated using standard methodology (8). Production of hemolysins was determined by streaking isolates onto a range of blood agar plates and incubating them in air at 37°C for 18 h. Isolates were considered positive for alpha-toxin if hemolytic on 5% rabbit blood agar, positive for beta-toxin if hemolytic on 5% sheep blood agar following incubation at 37°C for 18 h and then 4°C for 30 min (hot-cold lysis), and positive for delta-toxin if synergy was observed between the test isolate and a beta-toxin producer on 3% washed sheep erythrocytes. Positive controls were NCTC 5655 (alpha-toxin positive), NCTC 7428 (beta-toxin positive), and NCTC 9715 (delta-toxin positive).
Analysis.
The isolates used in this study were chosen for two reasons. First, the case control study design helps to avoid the potential confounding effect from comparing isolates from different geographical locations or time periods. Second, having defined the population structure of these isolates, we were able to adjust for the effects of clonality on associations between bacterial determinants and invasive disease. For example, a given determinant could become overrepresented in the invasive group if it is linked to a true virulence gene within a given lineage. We recognize that analysis linked to interpretation of MLST results becomes model dependent. In view of this, in the primary analysis, isolates were evaluated according to origin (nasal carriage isolate or invasive isolate) and further subdivided depending on whether the disease was hospital or community acquired. The second round of analysis used isolates grouped by lineage as defined by MLST results and analysis repeated for associations within each lineage. A lineage (or clonal complex) was defined for the purposes of this analysis as a group of organisms that had sequences at five or more of the seven MLST loci examined that were identical to those from at least one other organism in the group. Further detail of the genetic structure of S. aureus can be found on the MLST website (http://www.mlst.net).
Contingency tables were used to compare proportions between groups. To adjust for the hitchhiker effect, analyses were stratified by MLST-defined lineage by either the Mantel-Haenszel method or conditional logistic regression. In this way, the presence of particular determinant(s) was only compared between disease and carriage isolates within lineages, and these separate effects summated to give an overall odds ratio. Strains that were unrelated to any other at five of seven loci were arbitrarily grouped as a lineage for the purpose of this stratification. No corrections were made for multiple comparisons, though in an attempt to recognize this, statistical significance was set at P = 0.01 for the purpose of interpretation and discussion. For the same reason, conservative 99% confidence intervals are used throughout.
RESULTS
Univariate analysis.
Eight of the 33 determinants (24%) were significantly more common in invasive isolates than in carriage isolates (Table 3). The eight determinants were as follows: three genes encoding cell wall-associated adhesins (fnbA, encoding fibronectin binding protein A; cna, encoding collagen binding protein; and sdrE, encoding a protein of unknown function), four genes encoding toxins (sea and sej, encoding staphylococcal enterotoxins A and J, respectively; eta, encoding exfoliative toxin A; and hlg, encoding gamma-toxin), and icaA (used here as a marker for the entire ica operon), which is involved in biofilm production.
TABLE 3.
Presence of putative virulence determinants in carriage and disease isolates as determined by univariate analysis and analysis stratified by clonal complex
| Virulence determinant | No. of isolates
|
No. of genesa
|
||||
|---|---|---|---|---|---|---|
| Unstratified analysis
|
Stratified analysis
|
|||||
| Carriage (n = 178) (%) | Invasive (n = 155) (%) | Odds ratio (99% CI) | P | Odds ratiob (99% CI) | P | |
| Adhesins | ||||||
| fnbA | 154 (87) | 152 (98) | 7.9 (1.7-77) | 0.0001c | 12.0 (1.6-89) | 0.0001 |
| clfA | 174 (98) | 155 (100) | 0.13 | 0.13 | ||
| clfB | 178 (100) | 155 (100) | 1 | |||
| cna | 57 (32) | 81 (52) | 2.3 (1.3-4.3) | 0.0002 | 2.0 (1.1-3.7) | 0.003 |
| spa | 160 (90) | 145 (94) | 1.6 (0.54-5.4) | 0.24 | 1.7 (0.55-5.5) | 0.20 |
| sdrC | 178 (100) | 156 (100) | 1 | |||
| sdrD | 73 (41) | 74 (48) | 1.3 (0.71-2.3) | 0.27 | 1.9 (0.8-4.4) | 0.05 |
| sdrE | 72 (40) | 87 (56) | 1.9 (1.0-3.4) | 0.006 | 3.1 (1.4-6.7) | 0.0001 |
| bbp | 76 (43) | 59 (38) | 0.82 (0.45-1.5) | 0.43 | 0.74 (0.37-1.5) | 0.27 |
| ebpS | 110 (62) | 105 (68) | 1.3 (0.70-2.4) | 0.30 | 1.5 (0.65-3.4) | 0.20 |
| map/eap | 165 (93) | 149 (96) | 2.0 (0.50-9.4) | 0.24 | 1.7 (0.42-6.5) | 0.34 |
| Toxins | ||||||
| tst | 44 (25) | 46 (30) | 1.3 (0.68-2.6) | 0.27 | 1.2 (0.46-2.9) | 0.68 |
| sea | 30 (17) | 48 (32) | 2.3 (1.1-4.7) | 0.003 | 2.6 (1.0-6.6) | 0.007 |
| seb | 13 (7) | 14 (9) | 1.3 (0.41-3.9) | 0.72 | 0.79 (0.22-2.8) | 0.33 |
| sec | 20 (11) | 15 (10) | 0.84 (0.30-2.2) | 0.72 | 0.85 (0.32-2.3) | 0.66 |
| sed | 9 (5) | 8 (5) | 1.0 (0.24-4.2) | 1 | 1.2 (0.34-4.4) | 0.68 |
| see | 0 | 0 | 1 | |||
| seg | 113 (64) | 85 (55) | 0.69 (0.38-1.3) | 0.12 | 0.42 (0.2-0.9) | 0.003 |
| seh | 18 (10) | 24 (15) | 1.6 (0.65-4.1) | 0.19 | 1.4 (0.63-3.3) | 0.25 |
| sei | 106 (60) | 81 (52) | 0.73 (0.40-1.3) | 0.18 | 0.50 (0.24-1.0) | 0.01 |
| sej | 12 (7) | 38 (25) | 4.5 (1.8-13) | <0.001 | 4.4 (1.7-11) | <0.0001 |
| eta | 11 (6) | 34 (22) | 4.2 (1.6-13) | <0.001 | 6.3 (2.0-20) | <0.0001 |
| etb | 6 (3) | 0 (0) | 0.03 | 0.06 | ||
| pvl | 3 (2) | 6 (4) | 2.3 (0.31-27) | 0.31 | 2.7 (0.38-19) | 0.18 |
| hlg | 157 (89) | 150 (97) | 3.6 (0.97-19) | 0.01 | 4.5 (1.0-19) | 0.004 |
| Alpha-toxin | 178 (100) | 153 (99) | 0.21 | 0.15 | ||
| Beta-toxin | 160 (89) | 122 (79) | 0.45 (0.18-1.0) | 0.02 | 0.43 (0.17-1.1) | 0.01 |
| Delta-toxin | 144 (80) | 106 (69) | 0.54 (0.27-1.1) | 0.02 | 0.55 (0.27-1.1) | 0.03 |
| Others | ||||||
| ica | 136 (77) | 143 (92) | 3.5 (1.4-9.8) | <0.0001 | 3.5 (1.4-9.1) | 0.0002 |
| V8 | 168 (94) | 149 (96) | 1.5 (0.34-7.5) | 0.61 | 1.8 (0.42-7.9) | 0.28 |
| efb | 105 (60) | 105 (68) | 1.5 (0.79-2.7) | 0.11 | 1.7 (0.86-3.4) | 0.03 |
| Coagulase | 175 (99) | 155 (100) | 1 | 0.30 | ||
| agr | 0.64 | 0.21 | ||||
| Subgroup I | 65 (37) | 53 (34) | 0.91 (0.57-1.5) | 0.73 | 0.43 (0.15-1.2) | 0.10 |
| Subgroup II | 43 (24) | 34 (22) | 0.89 (0.51-1.5) | 0.70 | 3.3 (1.0-11) | 0.04 |
| Subgroup III | 65 (37) | 65 (42) | 1.3 (0.80-2.0) | 0.31 | 0.78 (0.22-2.8) | 0.70 |
| Subgroup IV | 5 (3) | 2 (1) | 0.46 (0.04-2.8) | 0.46 | 0.79 (0.13-14) | 0.79 |
CI, confidence interval.
Odds ratio and P value after Mantel-Haenszel stratification for MLST-defined lineage. This was carried out to account for any influence of underlying clonality on the results.
Bold indicates statistically significant values (P ≤ 0.01).
Adjusting for the effect of clonality.
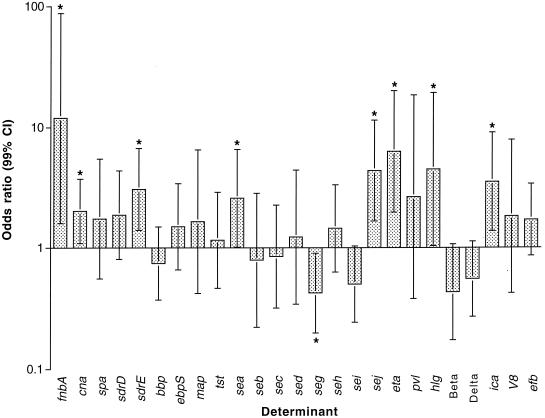
That such a large proportion of the genes examined were associated with disease (albeit all positively) raises the question of whether the result can be explained by linkage disequilibrium between these genes and one or more true virulence determinants elsewhere in the genome (the hitchhiker effect). In an attempt to adjust for this, we repeated the analysis, this time stratifying for clonality. The clonal structure of this collection of isolates has been defined by MLST (http://www.mlst.net); using this information and the Mantel-Haenszel method, we compared the prevalence of each gene in the disease and carriage groups within but not between clonal complexes (Table 3 and Fig. 1). Following this stratification, all eight genes remained positively associated with disease. seg was associated with disease on stratified analysis alone. We also repeated the analysis, omitting epidemic methicillin-resistant S. aureus clone 16 (EMRSA-16) (sequence type ST36), by far the single largest clone, containing 22 disease strains from a single group of hospitals but no carriage strains, and found that without this clone sea was not associated with disease. In view of this, sea was not included as a virulence-associated gene in the remaining analysis. The other results were not affected, leaving seven putative virulence determinants positively associated with disease across multiple S. aureus lineages.
FIG. 1.
Presence or absence of putative virulence determinants. Odds ratios for disease, adjusted for the effects of clonality by Mantel-Haenszel stratification of MLST-defined lineage. Error bars denote 99% confidence intervals (CIs), and an asterisk indicates a significant association with disease (P < 0.01). Genes that were either ubiquitous (such as clfA and clfB) or very rare or absent (such as see and etb) are not shown.
Disease origin: community or hospital.
The relative importance of the 33 bacterial factors in hospital-acquired versus community-acquired isolates was assessed by a comparison of carriage isolates and community-acquired disease alone. The presence of genes encoding six of the seven virulence-associated factors from the primary analysis (fnbA, cna, sdrE, sej, eta, and ica) remained associated with disease once hospital-acquired strains were removed. Although hlg was numerically more common in the community-acquired disease group than in the hospital-acquired group (98% versus 95%), comparison with the carriage isolates (88%) was no longer statistically significant.
Multivariate analysis. (i) Linkage disequilibrium between virulence-associated genes.
We examined the possibility that identification of one or more of these seven virulence-associated genes was actually the result of linkage disequilibrium within this group. Logistic regression modeling of the relationship of the seven factors to both disease and each other suggested that four of the factors (sej, fnbA, cna, and sdrE) were both independent of each other and independently associated with disease. Two of the remaining three factors, ica and hlg, were in linkage disequilibrium with fnbA, though each contributed significantly to virulence after taking this association into account. eta was in linkage disequilibrium with sej but also contributed independently to virulence. We thus concluded that all seven factors were associated with virulence.
(ii) Role of individual genes in disease.
Although upon univariate analysis the odds ratio for disease for fnbA was twice as high as that for any of the other virulence-associated genes, in the full multivariate model, the odds ratios for disease for each gene fell between 2.2 (eta) and 3.3 (fnbA). This suggests that each of the seven genes contributed approximately equally to virulence and that no single bacterial factor predominated as the major predictor of virulence.
(iii) Cumulative effect of genes.
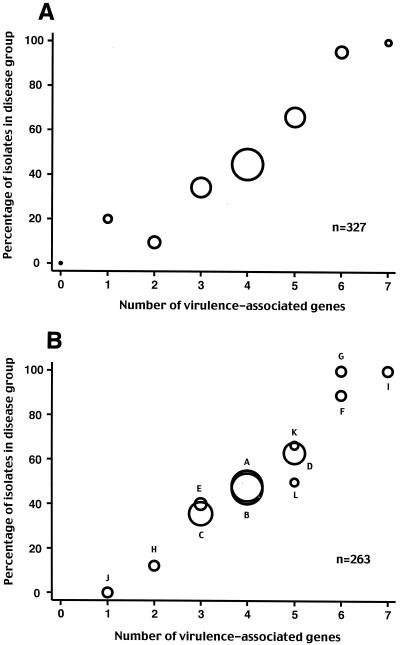
There was a linear trend between the number of these seven virulence determinants carried by particular isolates and the proportion of isolates causing disease (P < 0.0001) (Fig. 2A). This suggested that the effects of the factors on virulence were cumulative. The association between the number of determinants and the odds of disease was log linear, consistent with the logistic regression model in which there is a cumulative effect of each subsequent factor on virulence. None of the remaining 26 putative virulence determinants were associated with disease on univariate analysis, either separately or combined, as a count of the number of determinants in each organism.
FIG. 2.
(A) Association between the number of virulence-associated determinants and the proportion of isolates from cases of disease (rather than carriage). For example, 20% of isolates with one virulence-associated gene were from cases of disease and 80% were carriage strains. The area of each circle is proportional to the number of isolates with that number of determinants (ranging from zero genes to seven genes, with n = 2, 15, 21, 64, 140, 56, 22, and 7, respectively). (B) Association between the number of virulence-associated determinants in a gene combination and the proportion of isolates with that combination from cases of disease (rather than carriage). Each combination is identified by a letter; for the actual gene complement, refer to Table 4. The 12 most common combinations representing 80% of isolates are shown. The area of each circle is proportional to the number of isolates in that gene combination (n for each gene combination is given in Table 4).
Gene combinations and virulence.
Given that the effects of the seven genes on the odds of being a disease-causing strain are additive, we examined whether particular gene combinations were either (i) more commonly found together in an organism than by chance or (ii) more virulent.
In the study population, there were 46 (of a possible 128) different combinations of the seven genes, with more than 80% of the isolates accounted for by 12 of these gene combinations (Table 4). We compared the actual frequencies with the predicted frequencies of each gene combination, assuming independent segregation of the genes. One common combination (combination B [Table 4]) was overrepresented in both the carriage and disease groups (19.4% observed versus 11.7% predicted in carriage isolates and 18.6% observed versus 10% predicted in disease isolates), two virulent combinations (combinations F and G, each with six virulence-associated genes) were overrepresented in the disease group (5.2% observed versus 1.3% predicted and 5.2% observed versus 1.1% predicted, respectively), and one less virulent combination (combination J, with one virulence-associated gene) was overrepresented in the carriage group (4.1% observed versus 0.9% predicted). Comparison of observed and predicted frequencies demonstrated that no gene combinations were significantly underrepresented. Are these discrepancies due to the effect of clonality, or do particular combinations of genes have either less or greater virulence than expected? None of the 12 most common combinations were either more or less likely to cause disease than others with the same number of virulence-associated genes, suggesting the effect was due to the clonal population structure (Fig. 2B).
TABLE 4.
The 12 most common gene combinations
| Combi- nation | Complement
|
na | No. of isolates | Overall % of total | Cumu- lative % of total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| fnbA | cna | sdrE | sej | eta | hlg | ica | |||||
| A | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 4 | 64 | 19.6 | 19.6 |
| B | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 4 | 64 | 19.6 | 39.1 |
| C | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 3 | 39 | 11.9 | 51.1 |
| D | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 5 | 35 | 10.7 | 61.8 |
| E | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 3 | 10 | 3.1 | 64.8 |
| F | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 6 | 9 | 2.8 | 67.6 |
| G | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 6 | 8 | 2.4 | 70.0 |
| H | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 8 | 2.4 | 72.5 |
| I | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | 7 | 2.1 | 74.6 |
| J | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 7 | 2.1 | 76.8 |
| K | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 5 | 6 | 1.8 | 78.6 |
| L | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 5 | 6 | 1.8 | 80.4 |
Number of virulence-associated determinants within the gene combination (out of seven).
Effect of clonality on genes and gene combinations. (i) Distribution of individual genes between lineages.
Using contingency table analysis, we examined whether individual genes were apparently randomly distributed between the major MLST-derived clonal complexes or whether their distribution was significantly influenced by the underlying clonality of the population. clfB, sdrC, and see were excluded from this analysis, as they were present or absent in all isolates. The genes fell into two distinct groups. The distribution of the enterotoxins, tst, the exfoliatins (eta and etb), beta- and delta-toxins, the variable sdr genes (sdrD, sdrE, and bbp), cna, ebpS, and efb within the population were all highly significantly related to clonal complex (P < 0.001 in all cases). In contrast, the distributions of fnbA, clfA, coagulase, spa, map, pvl, hlg, alpha-toxin, ica, and V8 appeared to be unrelated to the underlying clonal structure (P > 0.1 in all cases). Most in this second group are genes which are either very common or very rare, and it is possible that the absence of a significant association with clonal complex is due to lack of power, leading to a type II error. However, the difference in the P value range between the two groups is striking, and even after removing from the second group those genes which are either present or absent in more than 95% of isolates, fnbA, spa, ica, map, and hlg remain apparently unrelated to underlying clonality (and three of these are virulence-associated genes).
Although there is strong statistical evidence for an association with clonality for the first group of genes listed above, there is also evidence to suggest that horizontal transfer of genes plays an important role in determining the distribution of these genes. All the genes with a prevalence of more than 30% were found in isolates from all 11 major lineages. The exceptions to this diversity are agr subgroups I to IV, which were very tightly (but not completely) linked to the underlying clonal structure (Table 5).
TABLE 5.
Background clonality and agr subgroups
| Clonal complex | No. of genes in subgroupa
|
Total | |||
|---|---|---|---|---|---|
| I | II | III | IV | ||
| CC30 | 2 | 2 | 90 (96) | 0 | 94 |
| CC39 | 0 | 0 | 24 (100) | 0 | 24 |
| CC15 | 0 | 31 (100) | 0 | 0 | 31 |
| CC45 | 28 (93) | 0 | 1 | 1 | 30 |
| CC22 | 25 (100) | 0 | 0 | 0 | 25 |
| CC25 | 23 (100) | 0 | 0 | 0 | 23 |
| CC5 | 3 | 18 (86) | 0 | 0 | 21 |
| CC1 | 3 | 0 | 13 (81) | 0 | 16 |
| CC8 | 16 (100) | 0 | 0 | 0 | 16 |
| CC12 | 0 | 11 (100) | 0 | 0 | 11 |
| CC51 | 1 | 3 | 1 | 5 (50) | 10 |
Numbers in parentheses are percentages of organisms within that clonal complex. The population genetic structure of S. aureus can be found on the MLST website (http://www.mlst.net).
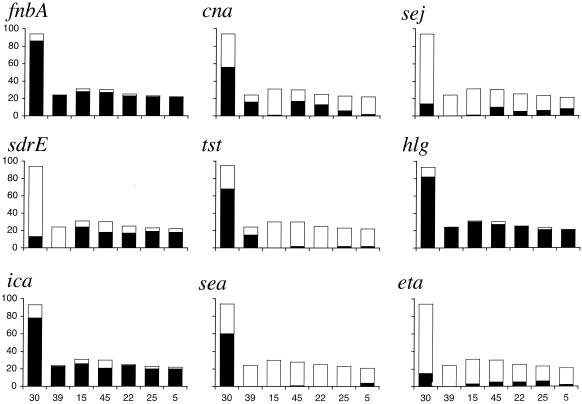
The most clonal of all the genes characterized as either present or absent was tst, with 92% of positive isolates occurring in clonal complexes CC30 or CC39. These two clonal complexes are closely related (the only two which are) and are probably derived from the same progenitor strain. One interpretation is that CC30 and CC39 represent a relatively recent but highly successful clonal expansion (perhaps fueled by tst) which has had less time to diversify through horizontal transfer than other lineages. That other genes (sea and bbp) were also heavily polarized with respect to this complex lends support to this view. The distributions of the seven virulence-related genes plus sea and tst are shown in Fig. 3; this selection includes genes which are chromosomally encoded (ica, fnbA, and sdrE) and associated with plasmids (sej), bacteriophage (sea), and the SaPI 1 pathogenicity island (tst).
FIG. 3.
Distribution of genes within the seven largest MLST-defined clonal complexes (denoted by numbers along the x axes in the bottom row). The overall height of each bar denotes the total number of isolates in the complex. The height of the shaded area represents the number of isolates positive for the determinant. The population genetic structure of S. aureus can be found on the MLST website (http://www.mlst.net).
(ii) Distribution of gene combinations.
Underlying clonality had an important effect on the distribution of certain combinations of genes (linkage disequilibrium). The overrepresentation in both disease and carriage populations of the virulence-associated gene combination B (containing ica, fnbA, hlg, and cna but lacking sdrE, sej, and eta) (Table 4) is entirely because it was present in 51% of the isolates from clonal complexes CC30 and CC39; its frequency in other clonal complexes was only 6%, not significantly different from its predicted frequency. However, this clonality was not the explanation for the association of these genes with virulence; combination B was not itself associated with disease, and three of the four genes (ica, fnbA, and hlg) were significantly associated with disease within CC30/C39, even after excluding those isolates with combination B. Despite the underlying clonality, there was still considerable variation within clonal complexes and even within MLST sequence types. The 22 isolates of nosocomial disease-causing EMRSA-16 (ST36, a single MLST) were represented by 10 different gene combinations, despite all being isolated from within the same city over a 2-year period (data not shown). ST36 was not unique in showing this diversity; the five most common combinations were all present in over half of the MLST-defined clonal complexes (data not shown), suggesting that horizontal transfer of genes between lineages is a relatively frequent occurrence.
DISCUSSION
These data represent a comprehensive analysis of virulence determinants in a large, natural population of S. aureus isolates of clearly defined clinical provenance. Our finding that several bacterial factors were associated with invasive disease may directly reflect their involvement in the pathogenesis of severe disease. It is unlikely that one or more of the determinants have become overrepresented due to the hitchhiker effect, as the positive associations remained despite stratification of the analysis by genetic lineage. This was demonstrated against a background in which many determinants were highly clonal, as shown here for several determinants.
The study was assumption free with respect to the presumed function and biological effect during infection of the bacterial factors examined. However, having identified seven virulence-associated genes, we now consider the biological plausibility of our findings. Fibronectin, the host ligand for the adhesin encoded by fnbA, is a ubiquitous host protein present in soluble form in the blood and in fibrillar form in cellular matrices, bacterial adhesion to which may be important during several steps of the disease process (31). Likewise, toxins that modulate the immune response through superantigen activity probably make a significant contribution to disease manifestations. However, it is also important to be somewhat circumspect when ascribing a given function to the pathogenic process for the following reasons. First, many bacterial determinants are multifunctional (for example, fibronectin binding protein has recently been found to be an adhesin for fibrinogen) (39) and the critical interaction of a given factor with the host may not yet have been defined. Second, the functions ascribed to a determinant by in vitro assays may not accurately reflect their behavior in vivo.
Our study design enriched the chances of finding virulence-associated determinants by examining those previously thought to be involved in disease and excluding those with purely housekeeping functions. However, it is unlikely that we have identified all the genes that are responsible for disease manifestations, an enterprise well suited to microarray analysis. In addition, some of the non-virulence-associated determinants as defined here may indeed play a role in pathogenesis but, because of their essential nature, were not flagged as virulence associated by our study design. For example, genes encoding the fibrinogen binding proteins ClfA and ClfB were ubiquitous, regardless of the origin of the strain.
Studies of staphylococcal pathogenesis often focus on the presence or absence of a given determinant. Our study results were analyzed on the same basis. However, the nature of the methodology used here means that we cannot differentiate between the presence or absence of the entire gene and the presence or absence of an allelic variant at a polymorphic locus. Southern hybridization may have been more sensitive than PCR in determining the presence or absence of a given gene. However, we believe that allelic variation may be important in defining virulence and that as such, PCR provides interesting clues. For example, bbp and sdrE appear to be allelic variants of each other yet only sdrE was associated with virulence. This observation is currently undergoing further study.
The variation in virulence determinants in a given clone (as demonstrated for EMRSA-16), together with the widespread nature of combinations of virulence determinants, suggests that horizontal transfer of genes is a common event. The preservation of the associations between certain genes and virulence after stratification for bacterial lineage is in itself strong evidence for either loss or gain through horizontal transfer of genetic elements. The mechanism of transfer can be readily explained for mobile determinants such as sej, which is plasmid mediated (40). However, the majority of the virulence-associated genes are chromosomal and it is not clear how such genes are being transmitted. In addition, not all mobile elements appeared to undergo frequent horizontal transmission. An example of this is tst, which resides on a pathogenicity island termed SaPI 1 (35). This gene was very common (>60%) in two closely related clonal complexes in our study (CC30 and CC39) but appeared either very infrequently or not at all in the remainder.
S. aureus usually behaves as a harmless human commensal, as reflected by the large proportion of the healthy population who sometimes or usually carry this organism, the transition to disease representing the exception rather than the rule. So what is the relationship between virulence-associated genes and bacterial fitness? The answer to this question is unknown but is likely to be complex and could vary depending on the factor in question. For example, a determinant that confers the ability to invade the host may be associated with a fitness disadvantage since invasion could lead to bacterial death following antibiotic treatment or death of the host. Conversely, the presence of genes that enhance the ability to cause conditions such as superficial skin infection could lead to enhanced host-to-host transmission. A further consideration is that accessory genes that are important to disease pathogenesis but which are not critical to survival may impose a cost to fitness through additional gene replication and protein secretion if expressed during periods of carriage. We postulate, therefore, that the proportion of strains circulating in the community which carry virulence determinants and the number of virulence-associated genes carried per strain are a product of the interplay between rates of gene acquisition, the cost to biological fitness, and the rate of decay of strains causing human disease.
Host factors for S. aureus disease are likely to include a genetic predisposition via one or more susceptibility genes and acquired factors such as the presence of intravenous devices, surgical wounds, and other events that perturb normal host defenses. It seems plausible to speculate that invasive disease occurs in two overlapping host populations. Individuals affected in the community (where the rate of disease is low) may represent those most genetically predisposed to S. aureus infection, while individuals infected in the hospital (where the burden of disease is higher) may represent a much larger at-risk group who may or may not carry susceptibility genes. Given this scenario, it is possible that the pattern or number of bacterial determinants associated with disease in the hospitalized host would differ from that seen in the community. However, we found little difference between strains associated with disease in the two settings, suggesting that bacterial factors play a role in causing disease, even in the compromised host. Whether this is the case for truly immunocompromised individuals such as neutropenics and those with AIDS requires further study. A larger study would also be required to address whether a given determinant(s) is associated with a particular clinical syndrome(s).
In conclusion, this study has demonstrated the variable presence of virulence genes in natural populations of S. aureus, providing evidence that bacterial factors play a role in determining invasive disease in both community and hospital settings. We have demonstrated that the effect of these genes was cumulative, each independently multiplying the odds of disease. There was also evidence of considerable horizontal transfer of genes on a background of clonality. This study also indicates that it may be an oversimplification to consider virulence in relation to the presence or absence of a given bacterial factor. Our findings suggest that allelic variants of a polymorphic locus can make different contributions to the disease process, further study of which is likely to expand our understanding of staphylococcal disease pathogenesis.
Acknowledgments
This project was supported by Royal Society grant 20955 and Wellcome Trust grant 059064 to S. Peacock and Wellcome Trust Career Development award 049310 to N. Day.
We are grateful to B. Cameron for technical assistance during pilot studies.
Editor: E. I. Tuomanen
REFERENCES
- 1.Arbuthnott, J. P. 1982. Bacterial cytolysins (membrane-damaging toxins), p. 107-129. In P. Cohen and S. van Heyningen (ed.), Molecular actions of toxins and viruses. Elsevier Biomedical Press, Amsterdam, The Netherlands.
- 2.Bhakdi, S., and J. Tranum-Jensen. 1991. Alpha-toxin of Staphylococcus aureus. Microbiol. Rev. 55:733-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boden, M. K., and J. I. Flock. 1994. Cloning and characterization of a gene for a 19 kDa fibrinogen-binding protein from Staphylococcus aureus. Mol. Microbiol. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 4.Boden, M. K., and J. I. Flock. 1992. Evidence for three different fibrinogen-binding proteins with unique properties from Staphylococcus aureus strain Newman. Microb. Pathog. 12:289-298. [DOI] [PubMed] [Google Scholar]
- 5.Boden Wastfelt, M. K., and J. I. Flock. 1995. Incidence of the highly conserved fib gene and expression of the fibrinogen-binding (Fib) protein among clinical isolates of Staphylococcus aureus. J. Clin. Microbiol. 33:2347-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohach, G. A., M. M. Dinges, D. T. Mitchell, D. H. Ohlendorf, and P. M. Schlievert. 1997. Exotoxins, p. 83-111. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 7.Bohach, G. A., D. J. Fast, R. D. Nelson, and P. M. Schlievert. 1990. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit. Rev. Microbiol. 17:251-272. [DOI] [PubMed] [Google Scholar]
- 8.Collee, J. G., A. G. Fraser, B. P. Marmion, and A. Simmons (ed.). 1996. Mackie & McCartney practical medical microbiology, 14th ed. Churchill Livingstone, New York, N.Y.
- 9.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Downer, R., F. Roche, P. W. Park, R. P. Mecham, and T. J. Foster. 2002. The elastin-binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. J. Biol. Chem. 277:243-250. [DOI] [PubMed] [Google Scholar]
- 11.Drapeau, G. R., Y. Boily, and J. Houmard. 1972. Purification and properties of an extracellular protease of Staphylococcus aureus. J. Biol. Chem. 247:6720-6726. [PubMed] [Google Scholar]
- 12.Emori, T. G., and R. P. Gaynes. 1993. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsgren, A., V. Ghetie, R. Lindmark, and J. Sjoquist. 1983. Protein A and its exploitation, p. 429-480. In C. S. F. Easmon and C. Adlams (ed.), Staphylococci and staphylococcal infections. Academic Press, London, England.
- 14.Greene, C., D. McDevitt, P. Francois, P. E. Vaudaux, D. P. Lew, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143-1152. [DOI] [PubMed] [Google Scholar]
- 15.Hartleib, J., N. Kohler, R. B. Dickinson, G. S. Chhatwal, J. J. Sixma, O. M. Hartford, T. J. Foster, G. Peters, B. E. Kehrel, and M. Herrmann. 2000. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood 96:2149-2156. [PubMed] [Google Scholar]
- 16.Hemker, H. C., B. M. Bas, and A. D. Muller. 1975. Activation of a pro-enzyme by a stoichiometric reaction with another protein. The reaction between prothrombin and staphylocoagulase. Biochim. Biophys. Acta 379:180-188. [DOI] [PubMed] [Google Scholar]
- 17.Hienz, S. A., T. Schennings, A. Heimdahl, and J. I. Flock. 1996. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J. Infect. Dis. 174:83-88. [DOI] [PubMed] [Google Scholar]
- 18.Jarraud, S., G. J. Lyon, A. M. S. Figueiredo, L. Gérard, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 20.Jonsson, K., D. McDevitt, M. H. McGavin, J. M. Patti, and M. Hook. 1995. Staphylococcus aureus expresses a major histocompatibility complex class II analog. J. Biol. Chem. 270:21457-21460. [DOI] [PubMed] [Google Scholar]
- 21.Josefsson, E., K. W. McCrea, D. Ni Eidhin, D. O'Connell, J. Cox, M. Hook, and T. J. Foster. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144:3387-3395. [DOI] [PubMed] [Google Scholar]
- 22.Kawabata, S., T. Morita, S. Iwanaga, and H. Igarashi. 1985. Enzymatic properties of staphylothrombin, an active molecular complex formed between staphylocoagulase and human prothrombin. J. Biochem. (Tokyo) 98:1603-1614. [DOI] [PubMed] [Google Scholar]
- 23.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 24.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 25.McGavin, M. H., D. Krajewska-Pietrasik, C. Ryden, and M. Hook. 1993. Identification of a Staphylococcus aureus extracellular matrix-binding protein with broad specificity. Infect. Immun. 61:2479-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLauchlin, J., G. L. Narayanan, V. Mithani, and G. O'Neill. 2000. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. 63:479-488. [DOI] [PubMed] [Google Scholar]
- 27.Moreillon, P., J. M. Entenza, P. Francioli, D. McDevitt, T. J. Foster, P. Francois, and P. Vaudaux. 1995. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect. Immun. 63:4738-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 29.Palma, M., A. Haggar, and J. I. Flock. 1999. Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J. Bacteriol. 181:2840-2845.10217776 [Google Scholar]
- 30.Park, P. W., J. Rosenbloom, W. R. Abrams, and R. P. Mecham. 1996. Molecular cloning and expression of the gene for elastin-binding protein (ebpS) in Staphylococcus aureus. J. Biol. Chem. 271:15803-15809. [DOI] [PubMed] [Google Scholar]
- 31.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 32.Patti, J. M., H. Jonsson, B. Guss, L. M. Switalski, K. Wiberg, M. Lindberg, and M. Hook. 1992. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J. Biol. Chem. 267:4766-4772. [PubMed] [Google Scholar]
- 33.Prevost, G., B. Cribier, P. Couppie, P. Petiau, G. Supersac, V. Finck-Barbancon, H. Monteil, and Y. Piemont. 1995. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect. Immun. 63:4121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenesis, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 35.Ruzin, A., J. Lindsay, and R. P. Novick. 2001. Molecular genetics of SaPI1—a mobile pathogenicity island in Staphylococcus aureus. Mol. Microbiol. 41:365-377. [DOI] [PubMed] [Google Scholar]
- 36.Steinberg, J. P., C. C. Clark, and B. O. Hackman. 1996. Nosocomial and community-acquired Staphylococcus aureus bacteremias from 1980 to 1993: impact of intravascular devices and methicillin resistance. Clin. Infect. Dis. 23:255-259. [DOI] [PubMed] [Google Scholar]
- 37.Supersac, G., G. Prevost, and Y. Piemont. 1993. Sequencing of leucocidin R from Staphylococcus aureus P83 suggests that staphylococcal leucocidins and gamma-hemolysin are members of a single, two-component family of toxins. Infect. Immun. 61:580-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tung, H., B. Guss, U. Hellman, L. Persson, K. Rubin, and C. Ryden. 2000. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem. J. 345:611-619. [PMC free article] [PubMed] [Google Scholar]
- 39.Wann, E. R., S. Gurusiddappa, and M. Hook. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275:13863-13871. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, S., J. J. Iandolo, and G. C. Stewart. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168:227-233. [DOI] [PubMed] [Google Scholar]