Abstract
The human fungal pathogen Candida albicans contains a close homologue of yeast siderophore transporters, designated Sit1p/Arn1p. We have characterized the function of SIT1 in C. albicans by constructing sit1 deletion strains and testing their virulence and ability to utilize a range of siderophores and other iron complexes. sit1 mutant strains are defective in the uptake of ferrichrome-type siderophores including ferricrocin, ferrichrysin, ferrirubin, coprogen, and triacetylfusarinine C. A mutation of FTR1 did not impair the use of these siderophores but did affect the uptake of ferrioxamines E and B, as well as of ferric citrate, indicating that their utilization was independent of Sit1p. Hemin was a source of iron for both sit1 and ftr1 mutants, suggesting a pathway of hemin uptake distinct from that of siderophores and iron salts. Heterologous expression of SIT1 in the yeast Saccharomyces cerevisiae confirmed the function of Sit1p as a transporter for ferrichrome-type siderophores. The sit1 mutant was defective in infection of a reconstituted human epithelium as a model for human oral mucosa, while the SIT1 strain was invasive. In contrast, both sit1 and SIT1 strains were equally virulent in the mouse model of systemic infection. These results suggest that siderophore uptake by Sit1p/Arn1p is required in a specific process of C. albicans infection, namely epithelial invasion and penetration, while in the blood or within organs other sources of iron, including heme, may be used.
The supply of iron limits the growth of pathogens within the human host. Body proteins including transferrin and lactoferrin sequester iron, and pathogens grow successfully only if they are able to acquire such bound iron. A frequent strategy used by microorganisms for iron acquisition is the secretion of siderophore iron chelators, which are able to withdraw iron from human protein-iron complexes because of their high affinities (18, 34). Siderophore-iron complexes are taken up by specific transport systems; however, microorganisms have also developed transport systems for heterologous siderophores produced in other species. For example, Escherichia coli possesses transporters not only for the homologous enterobactin (EB) and aerobactin siderophores but also for hydroxamate siderophores including ferrichrome (FCH), which are produced by fungi (34). Clearly, cross-species utilization of siderophores may be of ecological and medical importance, for example, in case of mixed infections in humans. The human host counteracts such microbial strategies, e.g., by lowering the iron content in circulating iron proteins such as transferrin (44).
Siderophore production and uptake has been demonstrated with many fungal species (18, 25, 45). However, the yeast Saccharomyces cerevisiae does not secrete siderophores, but it is able to take up ferric ions by a reduction system, which includes the Fre1p and Fre2p proteins (3, 6, 11); reduction at the cell surface is followed by the uptake of iron via the high- and low-affinity iron uptake systems. The multicopper oxidase Fet3p accepts ferrous ions generated by the Fre proteins, oxidizes them, and transfers the ferric ions to the membrane transporter encoded by FTR1 (2, 42). The low-affinity uptake system consists of the FET4 gene product (8). Alternatively, S. cerevisiae is able to take up siderophores produced by other microorganisms (27). Cell wall proteins Fit1p to Fit3p are required for the uptake of the siderophores FCH and ferrioxamine B (FoxB) but not triacetylfusarinine C (TAFC) and EB (32). Four transporters in the cytoplasmic membrane belonging to the major facilitator superfamily are relatively specific in their abilities to transport siderophores, e.g., Enb1p transports EB and Arn2p/Taf1p transports triacetylfusarinine and fusigen (15, 17, 48); other transporters are less specific, e.g., Sit1p transports ferrioxamine and FCH (26, 47) and Arn1p transports a wide range of hydroxamate-type siderophores (16, 48). The expression of genes encoding siderophore transporters is repressed by iron and by the high activity of the Tpk2p protein kinase A isoform (27, 35, 47, 48); the general transcriptional repressors Tup1p and Ssn6p contribute to the silencing of these loci (27). In contrast, in the absence of iron, the transcription factors Aft1p and Aft2p (4, 46) activate genes encoding siderophore transporters (47, 48).
Elements of a reductive iron uptake system similar to that of S. cerevisiae have been identified in the human fungal pathogen Candida albicans. The product of the CaCFL95 gene, which is homologous to S. cerevisiae Fre proteins, was able to restore the growth of fre mutants, while an FET3 homologue, CaFET99, was not functional in the heterologous yeast (23). Two C. albicans homologues of FTR1 were identified, of which FTR1 was shown to be necessary for growth under iron-limiting conditions and for virulence (33). An FET3 homologue was also identified, but it was not relevant for virulence (9). The standard strain used for gene disruptions in C. albicans, CAI4, has been reported to have a deletion of a gene next to the URA3 locus, which is able to complement an S. cerevisiae aft1 mutation but whose function in iron metabolism in C. albicans is unclear (13). Some reports have described the production of siderophores, especially of the hydroxamate type, by C. albicans; however, the nature of such siderophores has not been elucidated (20, 43). The uptake of siderophores, including FCH, has also been documented (1, 28, 30). Recently, a homologue of siderophore transporters in C. albicans, named CaArn1p/CaSit1p, has been characterized (1, 28). Its expression in S. cerevisiae was shown by Ardon et al. (1) to mediate uptake of FCH, but Lesuisse et al. (28) did not find transport activity and even demonstrated inhibition of FCH uptake in S. cerevisiae by CaArn1/CaSit1p. As in S. cerevisiae, the expression of genes encoding components of the reductive iron system and of SIT1/ARN1 is induced in low-iron conditions and is repressed by Tup1p in C. albicans (28, 31).
The previously reported data on the Sit1p/Arn1p transporter left several questions unresolved, including (i) whether Sit1p/Arn1p was the only siderophore transporter in C. albicans and to what degree it contributed to the overall pattern of siderophore utilization and (ii) what the phenotype of a SIT1/ARN1 deletion strain was with respect to C. albicans virulence. Both of these questions are addressed in the present study. We show that a group of siderophores of the FCH-type require Sit1p/Arn1p for uptake, while others are independent of this transporter. The iron chelate hemin (HEM), which is found in the host in large amounts, is used as an iron source by C. albicans independent of Sit1p/Arn1p. Importantly, siderophore uptake mediated by Sitp/Arn1p is required in a specific process during the infection of humans, namely the infection of epithelial layers, while infections of the bloodstream cause disease independent of Sit1p/Arn1p.
MATERIALS AND METHODS
Strains and growth conditions.
Yeast strains (Table 1) were grown in yeast extract-peptone-dextrose (YPD) medium or on synthetic dextrose (SD) or synthetic galactose (SGal) minimal medium at 30°C (41). SD medium contained 2% glucose and SGal medium contained 2% galactose and 0.1% glucose as carbon sources. The solid and liquid media used to test hyphae formation of strains were as described previously (5). Transformation of C. albicans was carried out by the spheroplast method and that of S. cerevisiae was carried out by the salt method (41).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| C. albicans strains | ||
| CAF2-1 | URA3/ura3Δ::imm434 | 10 |
| CAI4 | ura3Δ::imm434/ura3Δ::imm434 | 10 |
| C4-SB1, C4-SB4 | as CAI4, but SIT1/sit1Δ::hisG-URA3-hisG | This study |
| C4-SH1.1, C4-SH4.1 | as CAI4, but SIT1/sit1Δ::hisG | This study |
| C4-SHB1.1 | as CAI4, but sit1Δ::hisG-URA3-hisG/sit1Δ::hisG | This study |
| C4-SHH1.1 | as CAI4, but sit1Δ::hisG/sit1Δ::hisG | This study |
| C4-SHB4.21 | as CAI4, but SIT1/sit1Δ::hisG-URA3-hisG/sit1Δ::hisG | This study |
| C4-SHH4/21-VII | as CAI4, but SIT1/sit1Δ::hisG/sit1Δ::hisG | This study |
| C4-SHHB4/21-VII | as CAI4, but sit1Δ::hisG-URA3-hisG/sit1Δ::hisG/sit1Δ::hisG | This study |
| C4-SHHH4/21-VII | as CAI4, but sit1Δ::hisG/sit1Δ::hisG/sit1Δ::hisG | This study |
| SM1813C | integration of p1367-CaSITpORF/ClaI in C4-SHH1.1 | This study |
| SM1814C | integration of p1367-CaSITp/ClaI in C4-SHH1.1 | This study |
| ΔCaftr1 (ura+) | as CAI4, but ftr1Δ::hisG ftr1Δ::hisG ftr1Δ::hisG-URA3-hisG | 33 |
| Fet- | as CAI4, but fet3Δ::hisG fet3Δ::hisG-URA3-hisG | 9 |
| S. cerevisiae strains | ||
| DEY1394 | MATα ade6 can1 his3 leu2 trp1 ura3 fet3-2::HIS3 | 2 |
| PHY14 | as DEY1394, but arn1Δ::loxP arn2Δ::loxP sit1Δ::loxP enb1Δ::loxP-kanMX-loxP | This study |
| C. albicans plasmids | ||
| p1367/1 | URA3-marked integrative vector | 29 |
| p1367-CaSITpORF | SIT1 promoter and ORF in p1367 | This study |
| p1367-CaSITp | SIT1 promoter in p1367 | This study |
| S. cerevisiae plasmids | ||
| YCp22Gal | GAL1/10 promoter in YCplac22 | This study |
| YCp22Gal/SIT1 | SIT1 transcribed by GAL1 promoter, in YCp22 | This study |
To construct an S. cerevisiae strain lacking all known siderophore transporters, successive deletions in the fet3 mutant DEY1394 (2) were made with the loxP-kanMX-loxP module by using PCR fragments as described previously (14). ARN1 (16, 48), ARN2/TAF1 (15, 48), SIT1 (26, 47), and ENB1 (17) were deleted, with the kanMX-loxP sequence removed in the first three cases by the Cre recombinase, to allow subsequent disruptions. The correct insertion of the disruption fragments was verified by diagnostic PCRs on genomic DNA, with primers external and internal to the disruption fragments (data not shown).
Siderophores and siderophore biotests.
Siderophores were isolated from microbial cultures as described previously, and the purity of the siderophores was checked by high-pressure liquid chromatography (15, 16, 17). In addition, FCH was purchased from Sigma.
The solid media used in the C. albicans siderophore growth tests were Lee's medium containing 1% agar (24) and 550 μM bathophenanthroline disulfonic acid (BPDA) as an iron chelator. Twenty to forty microliters of an overnight culture, in Lee's medium, of the yeast strain to be tested was added to 10 ml of molten medium, mixed, and allowed to solidify in petri dishes. Ten microliters of siderophore solution (100 μM) or of other iron chelates (1 mM ferric citrate [CIT], 300 μM HEM) was dropped on sterile filter disks, which were placed on the agar surface. Plates were incubated for 2 to 4 days at 37°C. Siderophore growth tests for S. cerevisiae were performed similarly, except that SGal medium was used for pregrowth of cells and SGal agar containing 200 μM BPDA was used and siderophores were at a concentration of 10 μM, with the exception of EB and coprogen (COP) (100 μM).
Disruption of SIT1.
SIT1 alleles were deleted by the URA blaster protocol (10). To generate a disruption fragment for SIT1, the 5′ and 3′ untranslated regions of SIT1 were first produced by genomic PCR using DNA of strain CAI4. The 5′ region (1.533 kb) was amplified by using primers SITdis1/for (5′-ATTAGATCTTTGATACGCAGGAG-3′) and SITdis1/rev (5′-ATTGGATCCTAAGATGTCATGCTAGC-3′), and the 3′ region (595 bp) was amplified by using primers SITdis2/for (5′-ATTAGATCTCAAACTTGGTTGTAAC-3′) and SITdis2/rev (5′-ATTGGATCCTGTAAGTAGTGATTACTG-3′) (newly introduced BglII and BamHI sites are underlined; regions of homology are in italic type). Both fragments were introduced into the BglII or BamHI site flanking the URA blaster module (hisG-URA3-hisG) of plasmid p5921 (10) to generate plasmid pSITdisBlaster. From this plasmid, a 6.14-kb BglII-BamHI disruption fragment was excised, which was used for transformations of ura3 C. albicans strains. A 1.161-kb KpnI fragment of a pUC18 subclone of the 5′ untranslated fragment (plasmid pSIT-disI) was used as a probe in Southern blottings.
Reconstitution of SIT1 in the sit1 mutant.
Reconstituted strains were constructed in which the sit1 mutation in one chromosomal allele of strain C4-SHH1.1 was reconstituted (SM1813C) or not reconstituted (SM1814C) and which contained the URA3 allele in the same position, within the SIT1 locus. An SpeI fragment containing the SIT1 promoter and open reading frame (ORF) regions was generated first by genomic PCR using primers contig prom+SpeI (5′-TTAACTAGTCATCTACGAGACTG-3′) and contig stop+SpeI (5′-TTAACTAGTTCTAAACAGCTACTC-3′) (introduced SpeI sites are underlined; regions of homology are in italic type); the SpeI site at the 5′ end of the promoter segment is at position −1057 relative to the ATG start sequence of the ORF. This fragment was introduced into pUC18 to generate pPromSit1 (1). The 2.873-kb SpeI fragment of this plasmid was inserted into the single XbaI site of plasmid p1367/1 (29) to generate the URA3-containing integration vector p1367-CaSITpORF. pPromSit1 (1) was also doubly digested with SpeI/NheI, and the 1.051-kb SpeI/NheI ARN1 promoter fragment was isolated (the NheI site is 6 bp upstream of the ATG translation initiation sequence of SIT1) and inserted into the XbaI site of p1367/1 to generate the control plasmid p1367-CaSITp. Both p1367/1 derivatives were cut within the SIT1 promoter at the single ClaI site (situated at position −851 relative to the ATG) and transformed into the sit1 mutant C4-SHH1.1. The correct integration of plasmid p1367-CaSITpORF at the SIT1 locus was verified by Southern blottings by using genomic DNA digested with EheI, which cuts genomic DNA at position −1766 relative to the ATG, within pUC18 sequences of the p1367/1 derivatives and within the hisG sequence. As a probe, the 640-bp HindIII-BglII fragment of plasmid pSIT-disI was used. In cases of correct integration of p1367-CaSITpORF in one arn1 allele (about 60% of transformants), the reconstituted SIT1 gene was apparent as two fragments of 5.123 and 4.419 kb (the latter fragment appeared weak on blots because of low homology to the probe), while the hisG-disrupted allele yielded a 2.63-kb fragment. The correct integration of the control plasmid p1367-CaSITp (lacking the SIT1 ORF) was verified by Southern blottings by using genomic DNA digested with SspI and by using a 515-bp SspI/HindIII probe derived from plasmid pSIT-disI. The starting mutant p1367-CaSITp generated a single fragment of 3.5 kb, while in the case of an integrated plasmid, two fragments of 1.814 and 5.2 kb were observed because of an SspI site located within pUC18 sequences; in CAI4, the expected genomic fragment of 3.9 kb was verified.
Construction of an S. cerevisiae expression vector for CaSIT1.
The CaSIT1 ORF was amplified by PCR on genomic DNA of strain CAI4 by using primers Contig4-3057/for (5′-TTAGGATCCTAGCATGACATCTTACCAGTC-3′) and Contig4-3057/rev (5′-TTAGGATCCTAGTTCTAAACAGCTACTC-3′) (newly introduced BamHI sites are in boldface type, start and stop sequences are underlined, and regions of homology are in italic type). The 1.842-kb BamHI fragment was inserted into the transformation vector YCplac22Gal (BamHI cut), placing CaSIT1 in the resulting plasmid YCp22Gal/SIT1 under the control of the GAL1 promoter. YCplac22Gal was constructed by inserting the BamHI-EcoRI fragment carrying GAL1 (22) into YCplac22 (12).
In vitro model of oral candidiasis.
The human epithelium for the in vitro model of oral candidiasis was supplied by Skinethic Laboratory (Nice, France). Human keratinocytes derived from a squamous cell carcinoma of the buccal mucosa (cell line TR146) were cultured on an inert supporting membrane (37). A defined medium based on the MCDB-153 medium (Clonetics, San Diego, Calif.) supplemented with 5 μg of insulin ml−1 was deposited below a 0.5-cm2 microporous polycarbonate filter (pore size, 0.5 μm) and incubated for 7 days at 37°C with 100% humidity and 5% CO2 to generate a human reconstituted epithelium (RHE). No antibiotics and antimycotics were used.
C. albicans strains were cultured for 24 h at 37°C on Sabouraud dextrose agar (Difco). A part of the culture was suspended in a 0.9% sodium chloride solution and centrifuged. After the suspension was washed three times, 2 × 105 cells were suspended in 10 ml of YPD medium. The suspension was cultured for 16 h at 25°C, 4 × 106 cells were suspended in fresh medium, and the suspension was incubated again for 24 h at 37°C with shaking. After the suspension was washed three times with phosphate-buffered saline (PBS) solution, the desired inoculum size was adjusted with PBS.
RHE (0.5 cm2) was infected with 2 × 106 cells of each C. albicans strain in 50 μl of PBS; as a control, 50 μl of PBS alone was used. RHE was incubated at 37°C with 5% CO2 at 100% humidity for 6, 12, and 22 h. Infected RHE samples were examined by light microscopy as described previously (39).
Mouse infections.
Six-week-old BALB/c male mice were housed 10 per cage and given food and water ad libitum. The C. albicans strains to be tested were subcultured on SD agar medium for 18 h at 30°C before inoculation; strain identities were encoded to secure unbiased experimentations. The yeast colonies were scraped off the plates and washed three times in sterile saline. The inoculum was adjusted to a concentration of 107 cells/ml by microscopy countings, and 100 μl of the suspension was injected into the lateral tail vein of each mouse. The viability of the yeast cells was assessed by plating appropriate dilutions of the inoculum onto Sabouraud dextrose agar. Survival was monitored once daily up to day 68 after inoculation. Relevant results were confirmed thrice.
RESULTS
Disruption of SIT1/ARN1.
The sequence of the C. albicans genome (obtained from the Stanford Genome Technology Center website [http://sequence-www.stanford.edu/group/candida]) revealed an ORF (Contig6-2443 complementary strand, coordinates 14150 to 15964) encoding a theoretical protein of 604 amino acids with high homology to S. cerevisiae siderophore transporters. Computer predictions suggested a membrane protein of nine transmembrane regions which is 51% identical to Arn1p, 46% identical to Sit1p, 44% identical to Arn2p/Taf1p, and 32% identical to Enb1p of S. cerevisiae. While this present study was in progress, Ardon et al. (1) and Lesuisse et al. (28) reported that this gene, designated CaARN1 or CaSIT1, respectively, is regulated by iron in C. albicans and that its expression permits or inhibits FCH uptake in S. cerevisiae. Here, this gene is referred to as SIT1 (alias ARN1), because (i) the designation SIT (siderophore iron transport) is descriptive of its function, (ii) a close homologue of AFT1 is lacking in C. albicans genomic sequences and thus makes the designation ARN (Atf1 regulated) meaningless, and (iii) it is the only close homologue in C. albicans of the four known siderophore transporters of S. cerevisiae.
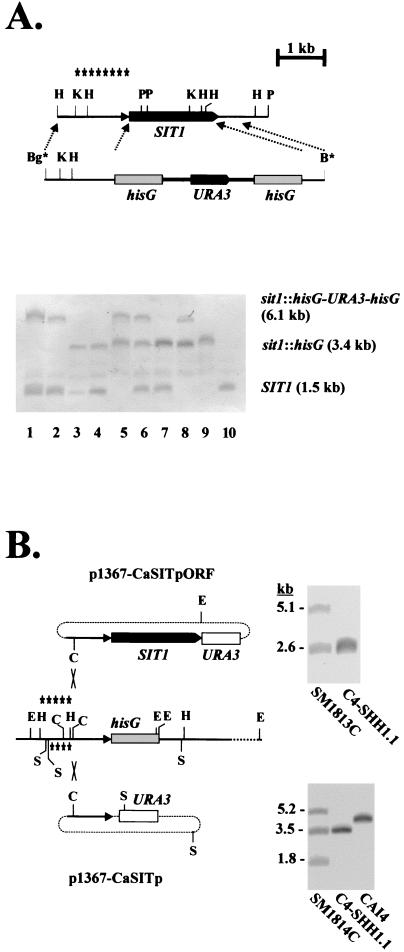
We successively disrupted SIT1 alleles of C. albicans by the URA blaster method (10). The 5′ and 3′ untranslated regions of ARN1 were produced by genomic PCRs and inserted next to the hisG-URA3-hisG blaster module to construct a disruption fragment, which was able to integrate into the SIT1 locus by homologous recombination (Fig. 1A, top). ura3 derivatives of disruptants were isolated and used for subsequent rounds of gene disruptions with the disruption fragment, as described previously (10). Correct insertion and disruption of SIT1 was verified by Southern blottings able to distinguish the wild-type allele (SIT1), the URA blaster-disrupted allele (sit1Δ::hisG-URA3-hisG), and the URA3-free allele (sit1Δ::hisG) (Fig. 1A, bottom) (see Materials and Methods). This procedure led to the disrupted strain C4-SHB1.1, which was disrupted in two SIT1 alleles (Table 1) (Fig. 1A, bottom, lane 5). Surprisingly, in another lineage, the presence of a third SIT1 allele was detected, becoming apparent after disruption of the presumed second SIT1 allele in strain C4-SHB4.21; in this case, all three possible alleles, SIT1/sit1Δ::hisG-URA3-hisG/sit1Δ::hisG, occurred (Fig. 1A, bottom, lane 6). Because a regular diploid disruptant was obtained, we consider the occurrence of three SIT1 alleles in strain C4-SHB4.21 as an artifact generated during the disruption processes.
FIG. 1.
Disruption of SIT1. (A) The wild-type genomic structure of SIT1 and the disruption fragment containing the URA blaster module (hisG-URA3-hisG) are shown (top). The relevant diagnostic restriction sites at the examination of the genomic configuration of SIT1 are HindIII (H), KpnI (K), and PvuII (P); the BglII (Bg*) and BamHI (B*) sites at the ends of the disruption fragment were generated by PCR. Asterisks indicate the region used as a probe in the Southern blot (bottom). Genomic DNAs of the following strains were digested by KpnI and PvuII and analyzed by Southern blotting: C4-SB1 (lane 1)and C4-SB4 (lane 2) (SIT1/sit1Δ::hisG-URA3-hisG); C4-SH1.1 (lane 3) and C4-SH4.1 (lane 4) (SIT1/sit1Δ::hisG); C4-SHB1.1 (lane 5) (sit1Δ::hisG-URA3-hisG/sit1Δ::hisG); C4-SHB4.21 (lane 6) (SIT1/sit1Δ::hisG-URA3-hisG/sit1Δ::hisG); C4-SHH4/21-VII (lane 7) (SIT1/sit1Δ::hisG/sit1Δ::hisG); C4-SHHB4/21-VII (lane 8) (sit1Δ::hisG-URA3-hisG/sit1Δ::hisG/sit1Δ::hisG); C4-SHHH4/21-VII (lane 9) (sit1Δ::hisG/sit1Δ::hisG/sit1Δ::hisG); and CAI4 (lane 10) (SIT1/SIT1). (B) Strategy to reconstitute the SIT1 gene in the sit1 mutant C4-SHH1.1. Plasmid p1367-CaSITpORF was cut with ClaI (C) and integrated into the SIT1 promoter region by homologous recombination. As a control, plasmid p1367-CaSITp was integrated similarly. The diagnostic restriction sites used in Southern blots were EheI (E) and SspI (S); the respective probes for the plasmids are indicated above and below the sit1 region. Examples of Southern blots verifying correct integrations are shown next to the respective plasmids (see Materials and Methods).
To ensure that any potential phenotype observed in the sit1 mutant was due to the lack of SIT1 and not to other details of genetic configurations (e.g., any locus-dependent differences in URA3 expression between the mutant and wild-type strains, which can occur [L. L. Sharkley, W.-L. Liao, and W. A. Fonzi, Abstr. EuroConference on Fungal Virulence Factors and Disease, abstr. p. 21, 2001]), we constructed two closely related strains either containing or lacking SIT1 (strains SM1813C and SM1814C, respectively). To accomplish this, plasmid p1367-CaSITpORF (containing the SIT1 promoter and coding region) or plasmid p1367-CaSITp (containing only the SIT1 promoter) was cut at the single ClaI site within the promoter to stimulate integration into the genomic SIT1 promoter in strain C4-SHH1.1 (sit1Δ::hisG/sit1Δ::hisG) (Fig. 1B). The correct integration of both plasmids in single copy at the SIT1 locus was verified by Southern blottings (Fig. 1B). Thus, the resulting strains SM1813C and SM1814C both contain URA3 at the SIT1 locus and differ only in the presence of the SIT1 coding region.
No growth defect was observed in the homozygous sit1 mutant C4-SHB1.1 compared with the wild-type or reconstituted SIT1 strain. Similarly, the formation of hyphae on solid media (Lee's medium, Spider medium, and serum-containing medium) or in liquid inducing media (in the presence of GlcNAc or serum) was not affected. The only exception was the triple allelic sit1 mutant C4-SHHB4/21-VII, which in YPD and SD media had a growth rate reduced by about 20% compared to those of SIT1 strains and mutant C4-SHB1.1 and which failed to form hyphae. Thus, these results suggest that an increased gene dosage or other genetic rearrangements interfere with growth and hyphal morphogenes in the triple allelic mutant but that these phenotypes are not related to the lack of SIT1.
Siderophore uptake by C. albicans sit1 mutants.
To establish the specificity of the theoretical Sit1p transporter protein, we compared the ability of SIT1+ and sit1 mutant strains to take up various siderophores or other iron-containing compounds. Strains were embedded in Lee's medium containing 550 μM BPDA to make remaining traces of iron unavailable to C. albicans cells; growth on these plates was stimulated by the presence of siderophores or other iron compounds within the filter disks (Fig. 2).
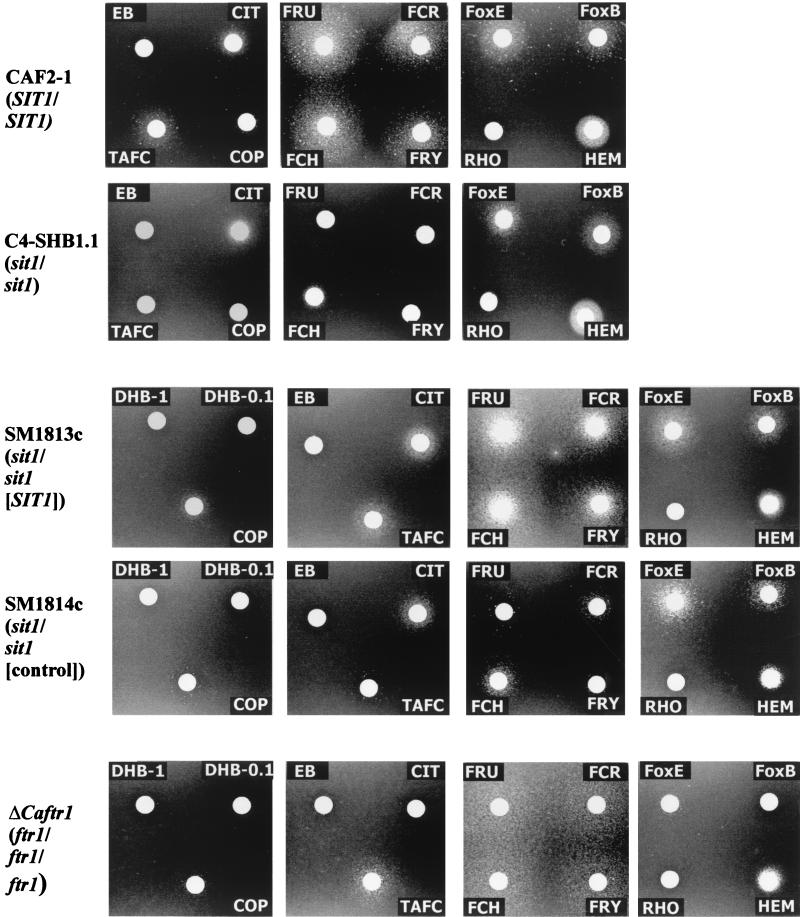
FIG. 2.
Utilization of siderophores by C. albicans strains. The indicated strains were embedded in solid Lee's medium containing 550 μM BPDA, and sterile filter paper disks containing siderophores (10 μl) were placed on the agar surface. Plates were incubated at 37°C for 2 to 3 days and photographed. The concentrations of siderophores and other compounds are listed in Table 2.
In the first set of experiments, we compared the wild-type strain CAF2-1 with the sit1 mutant C4-SHB1.1. The hydroxamate siderophores FCH, ferricrocin (FCR), ferrichrysin (FRY), and ferrirubin (FRU) were excellent iron sources for the SIT1+ strain, but these siderophores were hardly used by the sit1 mutant. Compared to these siderophores, the hydroxamates TAFC and COP were less efficiently used by the wild-type strain, but in the presence of an sit1 mutant background, siderophore utilization declined further. In contrast, ferrioxamine E (FoxE) and to a lesser extent FoxB were used as iron sources, but the uptake of these hydroxamates was not affected by the sit1 mutation. Similarly, the uptakes of CIT and of HEM were not affected by the SIT1 status of cells. Other (iron-free) siderophores used in these assays, including EB, dihydroxybenzoic acid (DHB), or rhodotorulic acid (RHO), did not stimulate growth. An identical iron chelate utilization pattern was obtained for strain C4-SHHB4/21-VII, which contains three sit1 alleles (data not shown). Thus, these results suggested that the Sit1p transporter is essential for the uptake of a group of hydroxamate siderophores, especially of the FCH type, while the uptake of ferrioxamines, CIT, and HEM occurs irrespective of the presence of Sit1p.
This conclusion was confirmed by analysis of a carefully matched pair of strains, SM1813c and SM1814c, which differ only in the presence of the SIT1 coding region, while all other genetic markers including the URA3 gene are in identical locations. The above-mentioned patterns of siderophore utilization were again obtained with respect to (i) FCH, FCR, FRY, and FRU; (ii) TAFC and COP; (iii) FoxE and FoxB, CIT and HEM; and (iv) EB, DHB, and RHO (Fig. 2).
C. albicans ftr1 and fet3 mutants defective in the reductive iron uptake system have been described previously (9, 33) but have not yet been tested for uptake of the range of siderophores as described here. Siderophore and iron chelate uptake in the fet3 mutant was indistinguishable from that in any SIT1 or sit1 mutant strain (data not shown). In contrast, in the ftr1 mutant, the uptake of the apparent Sit1p substrates FCH, FCR, FRY, FRU, TAFC, and COP still occurred as in the wild type, but the uptake of FoxE, FoxB, and CIT was prevented. Thus, the results summarized in Table 2 indicate that the uptake of the latter substrates depends on Ftr1p and is independent of the Sit1 protein, which we can consider as the main hydroxamate siderophore transporter of C. albicans.
TABLE 2.
Siderophore uptake by C. albicans strains
| Siderophore or other compounda | Growth of C. albicans strainsb
|
|||||
|---|---|---|---|---|---|---|
| CAF2-1 (SIT1/SIT1) | C4-SHB1.1 (sit1/sit1) | SM1813c (sit1/sit1 [SIT1]) | SM1814c (sit1/sit1 [control]) | ΔCaftr1 (ftr1/ftr1/ftr1) | Fet- (fet3/fet3) | |
| COP | + | −/+ | + | −/+ | −/+ | + |
| EB | − | − | − | − | − | − |
| FCH | +++ | −/+ | +++ | −/+ | +++ | +++ |
| FCR | +++ | −/+ | +++ | −/+ | +++ | +++ |
| FRY | +++ | −/+ | +++ | −/+ | +++ | +++ |
| FRU | +++ | −/+ | +++ | −/+ | +++ | +++ |
| FoxB | + | + | + | + | − | + |
| FoxE | ++ | + | ++ | ++ | −/+ | ++ |
| RHO | − | − | − | − | − | − |
| TAFC | + | −/+ | + | −/+ | + | + |
| CIT | + | + | + | −/+ | − | + |
| DHB | ND | ND | − | − | − | − |
| HEM | + | + | + | + | + | + |
The concentrations were 100 μM for the siderophores, 1 mM for DHB, 1 mM for CIT, and 300 μM for HEM. Siderophore solutions (10 μl) were added to sterile filter disks and placed on solid Lee's medium containing 550 μM BPDA.
Growth zone diameters in agar diffusion tests: +++, >30 mm; ++, 20 to 30 mm; +, 10 to 20 mm; −/+, up to 10 mm. ND, not determined.
Heterologous expression of CaSIT1 in S. cerevisiae.
While this present study was in progress, two reports on the expression of CaARN1/SIT1 in S. cerevisiae appeared, one of which described the functional expression of ARN1/SIT1 (1), which is at variance with the results of the second report (28). In our study, we prevented the significant activity of resident iron transporters by constructing an S. cerevisiae host strain (PHY14) defective in elements of reductive iron uptake (fet3) and lacking all four known transporters of siderophores, Arn1p, Arn2p/Taf1p, Sit1p, and Enb1p. This strain was unable to grow on low-iron media, even in the presence of most siderophores, while the parental strain DEY1394 (2) appeared to utilize siderophores as an iron source. PHY14 was transformed with an expression vector for CaSIT1 (pYCp22Gal/SIT1) in which the coding region of this gene was transcribed by the GAL1 promoter. Transformants were tested for their utilization of siderophores in the plate assay by using galactose as the carbon source to induce expression of the GAL1-SIT1 fusion. The results are shown in part in Fig. 3 and summarized in Table 3.
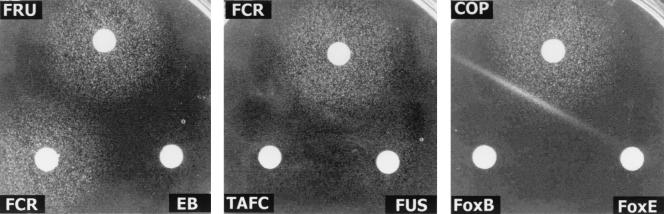
FIG. 3.
Utilization of siderophores by CaSIT1 transformant of S. cerevisiae. Strain PH14(YCp22Gal/SIT1) was embedded in solid SGal medium containing 200 μM BPDA, and sterile filter paper disks containing siderophores (10 μl) were placed on the agar surface. Plates were incubated at 30°C for 2 to 3 days and photographed. See Table 2 for concentrations. FUS, fusigen.
TABLE 3.
Siderophore uptake by S. cerevisiae strains
| Siderophorea | Growth of S. cerevisiae strainsb
|
||
|---|---|---|---|
| DEY1394 | PHY14 | PHY14 (YCp22Gal/SIT1) | |
| FoxB | ++ | − | − |
| FoxE | ++ | − | − |
| EB | ++ | − | − |
| TAFC | ++ | − | +/− |
| FRY | ++ | − | ++ |
| FCR | ++ | − | ++ |
| FCH | ++ | − | ++ |
| FRU | ++ | − | ++ |
| Ferrirhodin | ++ | − | ++ |
| COP | + | − | ++ |
Siderophore concentrations were 10 μM, except for EB and COP (100 μM).
Growth zone diameters in agar diffusion tests: ++, 30 to 35 mm; +, 25 to 30 mm; +/−, up to 10 mm.
Hydroxamate siderophores of the FCH type, including FCH, FCR, FRY, and FRU, as well as COP and TAFC, allowed the growth of S. cerevisiae PHY14(pYCp22Gal/SIT1) on galactose-containing media but not on glucose media, which repressed the GAL1-SIT1 fusion gene. In addition, the growth of ferrioxamine-type siderophores such as FoxE or FoxB was not improved by expression of CaSIT1. These results reflected the above-mentioned siderophore specificities obtained for CaSit1p in C. albicans and strongly suggested that these specificities were due to CaSit1p itself and not to another component, such as a resident transporter stimulated by CaSit1p.
In vitro model of oral candidiasis.
The keratinocyte cell line TR146 was allowed to differentiate on an inert supporting membrane, and as a result, a multilayered keratinocyte tissue was generated, which is strikingly similar to human oral mucosa (Fig. 4). This RHE was infected with 2 × 106 cells of C. albicans strains (yeast form) to explore any function of Sit1p in the infection process.
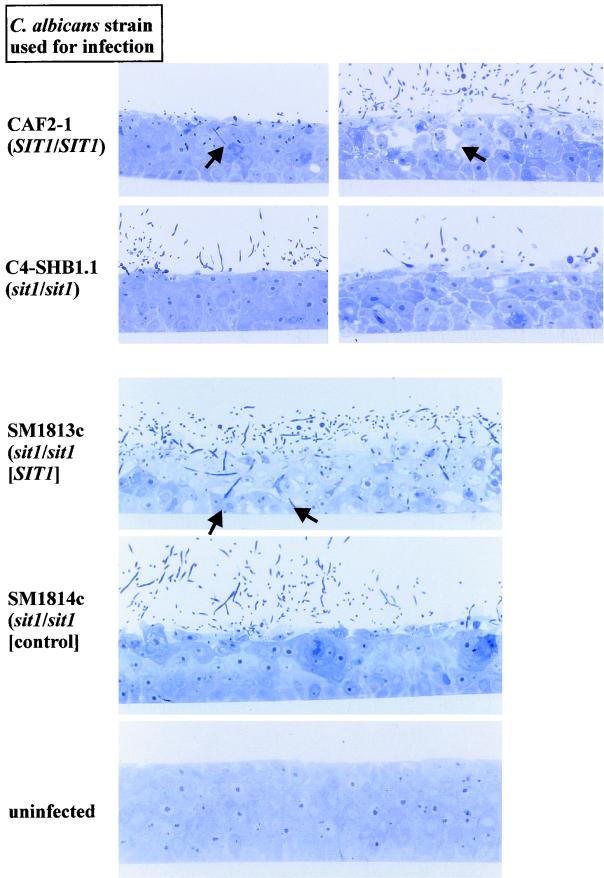
FIG. 4.
Infection of an RHE model by 2 × 106 C. albicans cells. RHE infected by strains CAF2-1 and C4-SHB1.1 are shown at 6 and 22 h of incubation; RHE infected by strains SM1813c and SM1814c are shown at 12 h of incubation. Note the deep penetration and formation of vacuoles by SIT1+ strains (arrows).
Inspection of the RHE by light microscopy revealed that infection by the sit1 mutant C4-SHB1.1 at an early time point (6 h) did not result in any noticeable damage to the RHE, and even at later time points, few C. albicans cells were observed in the upper layer of keratinocytes. In contrast, the SIT1+ strain CAF2-1 had entered the upper cellular layers at 6 h and both hyphae and yeast form cells were visible. At 22 h after infection, the SIT1+ strain had caused extensive damage to the RHE, which was apparent as large cavities in the keratinocyte tissue. These results were confirmed by use of the matched pair of transformants, strains SM1813c (SIT1) and SM1814c (sit1). While SM1814c had hardly damaged the RHE at 12 h after infection, hyphal forms of SM1813c had penetrated to the deepest keratinocyte layers at this time point (Fig. 4).
Thus, these results suggest that in a specific phase of infection of the human host by C. albicans, i.e., the invasion and penetration of the mucosal epithelium, siderophore uptake by Sit1p is required.
Virulence tests with mice.
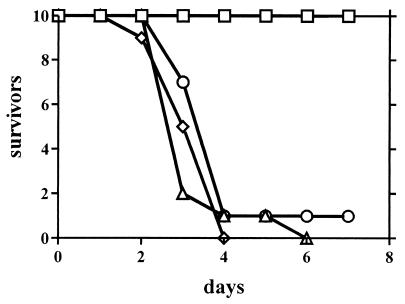
The virulence of sit1 mutants was compared with that of SIT1+ strains in the standard mouse model of systemic infection. BALB/c mice were injected in their tail veins with 106 yeast form cells, and the number of surviving mice was recorded. In a representative experiment, the sit1 mutant C4-SHB1.1 was able to kill mice as fast as the wild-type strain CAF2-1 or the SIT1/sit1 half-disruptant strain C4-SB1 (Fig. 5). In contrast, the abnormal strain C4-SHH4/21-VII, containing three disrupted SIT1 alleles, was completely avirulent, and mice survived for 68 days following their inoculation. However, because the sit1 mutant is virulent, it appears that the Sit1 siderophore transporter is not essential for survival and virulence if C. albicans cells are injected into the bloodstream. HEM and other iron-containing compounds such as transferrin may be used as sources of iron in the blood and organs (Fig. 2). The avirulence of the triple-allele mutant must be explained by genetic alterations unrelated to the absence of SIT1, such as the effects on virulence caused by increased gene dosages.
FIG. 5.
Virulence of C. albicans in mice. C. albicans cells (106; yeast form) were injected into the tail veins of BALB/c mice (n = 10), and the survival of mice was monitored daily. The strains used were CAF2-1 (SIT1/SIT1) (Δ), C4-SHB1.1 (sit1/sit1) (⋄), C4-SH1.1 (SIT1/sit1) (○), and C4-SHHB4/21-VII (sit1/sit1/sit1) (□).
DISCUSSION
The construction of C. albicans sit1 mutants and their comparison to SIT1+ strains allowed us to verify the role of the encoded protein Sit1p in siderophore utilization and virulence. With respect to siderophore utilization, we found by using a range of iron chelates including siderophores that Sit1p is needed for the uptake of hydroxamate-type siderophores, not only for that of FCH as previously suggested (1) but also for that of related siderophores including FCR, FRY, FRU, COP, and TAFC. The latter two siderophores were taken up less efficiently than the former siderophores, but their utilization was clearly dependent on Sit1p. Unexpectedly, sit1 mutants were still able to the use FoxE and FoxB, suggesting the presence of a siderophore transporter other than Sit1p in C. albicans. Analysis of the recently reported genomic sequences of C. albicans (contig 6) revealed two distantly related homologues of Sit1p encoding putative facilitators; the theoretical transporter proteins encoded by ORF6.8902 and ORF6.1908 have 25 and 22% identity, respectively, to Sit1p. Interestingly, the Sit1p-independent uptake of ferrioxamine siderophores was dependent on the function of the reductive iron transport system, since an ftr1 mutant (33) was unable to utilize ferrioxamines as sources of iron. In S. cerevisiae, reductive iron uptake requires both Ftr1 and Fet3 proteins (2, 42). Since a C. albicans fet3 mutant lacking the second possible element of reductive iron import (9) was not blocked in ferrioxamine import, it appears that homologues of Fet3p, which are indeed predicted from the C. albicans genomic sequences, have functions that are redundant to those of Fet3p. The ftr1 mutant was also defective in the uptake of CIT, while HEM utilization was not blocked in either the sit1 or ftr1 mutant.
The specificity of Sit1p for FCH-type siderophores was confirmed by the expression of Sit1p in the heterologous yeast S. cerevisiae. In agreement with the results of a recent study (1), we found that an Sit1p-producing S. cerevisiae transformant lacking known components of reductive and siderophore-mediated iron uptake was able to utilize FCH and related siderophores, but not ferrioxamines, as sources of iron. Possibly, the lack of transport function of Sit1p, which was reported in another recent study (28), was due to accidental mutations in the expression plasmid or in the host. Thus, it appears that C. albicans has at least the following four uptake systems for iron chelates: (i) Sit1p, as an uptake system for FCH-type siderophores; (ii) a ferrioxamine uptake system including elements of the reductive iron import such as Ftr1p; (iii) conceivably, an uptake system for CIT, which also requires Ftr1p; and (iv) an uptake system for HEM, which does not depend on either Sit1p or Ftr1p.
The effect of the sit1 mutation on virulence was tested (i) in vitro by using RHE, which is a model of human oral mucosa (39), and (ii) in the standard intravenous model of systemic infection in mice. In the mucosal model, sit1 mutants were clearly compromised in invading keratinocyte layers and caused less damage than SIT1 cells, which penetrated deeply and caused the formation of cavities within the epithelium. The mutant phenotype was absent in the sit1/SIT1 heterozygous strain and was restored by reintegrating a single copy of SIT1 at its genomic locus. On the other hand, the virulence of the sit1 mutant in the systemic model of infection was comparable to that of the wild-type and reconstituted SIT1 strains, indicating that siderophore transport is not of importance once the fungal pathogen is in the bloodstream. This assumption is supported by the pattern of iron chelate uptake in C. albicans discussed above, since HEM is a source of iron even in sit1 and ftr1 mutants. Presumably, the secretion of proteinases by C. albicans, which are able to degrade heme-containing proteins including hemoglobin (36), helps to liberate the heme moiety and to provide a source of iron; in addition, transferrin is likely to be another source of iron, permitting growth in blood and possibly within cells. Other examples of the differential effects of C. albicans mutations on infectivity in the mucosal and systemic models are known and indicate the importance of virulence traits that depend on specific host niches. For example, phr2 mutants were virulent in the systemic mouse model of infection but were avirulent in a vaginal infection model; in the case of the phr1 mutant, the phenotype was the inverse (7). Phospholipase D1 and Sap4p to Sap6p were required for virulence in the systemic model but were not required in the mucosal model (19, 38, 40). Interestingly, during the course of SIT1 disruption, we isolated a mutant containing three alleles of sit1 (strain C4-SHHB4/21-VII) which with respect to siderophore utilization had the same phenotype as an sit1/sit1 mutant. However, the three-allele mutant strain was completely avirulent in the systemic model of infection and showed reduced growth and defective hyphal morphogenesis. Thus, its virulence phenotype is not caused by a lack of Sit1p but is related to other unknown genetic alterations occurring in this strain. For example, it is known that the increased gene dosage of a repressor protein may prevent utilization of certain carbon sources in C. albicans (21). The three sit1 alleles we have found are reminiscent of the ftr1 mutant, which has also been reported as avirulent (33).
Acknowledgments
We are grateful to R. Eck and Y. Wang for distributing the strains, and we acknowledge the excellent technical assistance of J. Laude (Munich) and L. Improvisi (Paris).
Sequencing of C. albicans was accomplished with the support of the NIDCR and the Burroughs Wellcome Fund.
Editor: T. R. Kozel
REFERENCES
- 1.Ardon, O., H. Bussey, C. Philpott, D. McVey Ward, S. Davis-Kaplan, S. Verroneau, B. Jiang, and J. Kaplan. 2002. Identification of a Candida albicans ferrichrome transporter and its characterization by expression in Saccharomyces cerevisiae. J. Biol. Chem. 276:43049-43055. [DOI] [PubMed] [Google Scholar]
- 2.Askwith, C., D. Eide, A. van Ho, P. S. Bernhard, L. Li, S. Davis-Kaplan, D. M. Sipe, and J. Kaplan. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Askwith, C. C., D. de Silva, and J. Kaplan. 1996. Molecular biology of iron acquisition in Saccharomyces cerevisiae. Mol. Microbiol. 20:27-34. [DOI] [PubMed] [Google Scholar]
- 4.Blaiseau, P. L., E. Lesuisse, and J. M. Camadro. 2001. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 276:34221-34226. [DOI] [PubMed] [Google Scholar]
- 5.Bockmühl, D. P., S. Krishnamurthy, M. Gerads, A. Sonneborn, and J. F. Ernst. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243-1257. [DOI] [PubMed] [Google Scholar]
- 6.Dancis, A., D. G. Roman, G. J. Anderson, A. G. Hinnebusch, and R. D. Klausner. 1992. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. USA 89:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Bernardis, F., F. A. Mühlschlegel, A. Cassone, and W. A. Fonzi. 1998. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect. Immun. 66:3317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dix, D. R., J. T. Bridgham, M. A. Broderius, C. A. Byersdorfer, and D. J. Eide. 1994. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 269:26092-26099. [PubMed] [Google Scholar]
- 9.Eck, R., S. Hundt, A. Härtl, E. Roemer, and W. Kuenkel. 1999. A multicopper oxidase gene from Candida albicans: cloning, characterization and disruption. Microbiology 145:2415-2422. [DOI] [PubMed] [Google Scholar]
- 10.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgatsou, E., and D. Alexandraki. 1994. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 13.Gómez García, M., J.-E. O'Connor, L. Latorre García, S. I. Martínez, E. Herrero, and L. del Castillo Agudo. 2001. Isolation of a Candida albicans gene, tightly linked to URA3, coding for a putative transcription factor that suppresses a Saccharomyces cerevisiae aft1 mutation. Yeast 18:301-311. [DOI] [PubMed] [Google Scholar]
- 14.Güldener, U., S. Heck, T. Fiedler, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed]
- 15.Heymann, P., J. F. Ernst, and G. Winkelmann. 1999. Identification of a fungal triacetylfusarinine C siderophore transport gene (TAF1) in Saccharomyces cerevisiae as a member of the major facilitator superfamily. Biometals 12:301-306. [DOI] [PubMed] [Google Scholar]
- 16.Heymann, P., J. F. Ernst, and G. Winkelmann. 2000. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 186:221-227. [DOI] [PubMed] [Google Scholar]
- 17.Heymann, P., J. F. Ernst, and G. Winkelmann. 2000. A gene of the major facilitator superfamily encodes a transporter for enterobactin (Enb1) in Saccharomyces cerevisiae. Biometals 13:65-72. [DOI] [PubMed] [Google Scholar]
- 18.Howard, D. H. 1999. Acquisition, transport, and storage of iron by pathogenic fungi. Clin. Microbiol. Rev. 12:394-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hube, B., D. Hess, C. A. Baker, M. Schaller, W. Schäfer, and J. W. Dolan. 2001. The role and relevance of phospholipase D1 during growth and dimorphism of Candida albicans. Microbiology 147:879-889. [DOI] [PubMed] [Google Scholar]
- 20.Ismail, A., G. W. Bedell, and D. M. Lupan. 1985. Siderophore production by the pathogenic yeast, Candida albicans. Biochem. Biophys. Res. Commun. 130:885-891. [DOI] [PubMed] [Google Scholar]
- 21.Janbon, G., F. Sherman, and E. Rustchenko. 1998. Monosomy of a specific chromosome determines l-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 95:5150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, M., and R. W. Davis. 1984. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1440-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight, S. A. B., E. Lesuisse, R. Stearman, R. D. Klausner, and A. Dancis. 2002. Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology 148:29-40. [DOI] [PubMed] [Google Scholar]
- 24.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 25.Leong, S., and G. Winkelmann. 1998. Molecular biology of iron transport in fungi, p. 147-186. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems, vol. 35. Marcel Dekker Inc., New York, N.Y. [PubMed]
- 26.Lesuisse, E., M. Simon-Casteras, and P. Labbe. 1998. Siderophore-mediated iron uptake in Saccharomyces cerevisiae: the SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144:3455-3462. [DOI] [PubMed] [Google Scholar]
- 27.Lesuisse, E., P.-L. Blaiseau, A. Dancis, and J.-M. Camadro. 2001. Siderophore uptake and use by the yeast Saccharomyces cerevisiae. Microbiology 147:289-298. [DOI] [PubMed] [Google Scholar]
- 28.Lesuisse, E., S. A. B. Knight, J.-M. Camadro, and A. Dancis. 2002. Siderophore uptake by Candida albicans: effect of serum treatment and comparison with Saccharomyces cerevisiae. Yeast 19:329-340. [DOI] [PubMed] [Google Scholar]
- 29.Losberger, C., and J. F. Ernst. 1989. Sequence and transcript analysis of the C. albicans gene (URA3) encoding orotidine-5′-phosphate decarboxylase. Curr. Genet. 16:153-157. [DOI] [PubMed] [Google Scholar]
- 30.Minnick, A. A., L. E. Eizember, J. A. McKee, E. K. Dolence, and M. J. Miller. 1991. Bioassay for siderophore utilization by Candida albicans. Anal. Biochem. 194:223-229. [DOI] [PubMed] [Google Scholar]
- 31.Morrissey, J. A., P. H. Williams, and A. M. Cashmore. 1996. Candida albicans has a cell-associated ferric-reductase activity which is regulated in response to levels of iron and copper. Microbiology 142:485-492. [DOI] [PubMed] [Google Scholar]
- 32.Protchenko, O., T. Ferea, J. Rashford, J. Tiedeman, P. O. Brown, D. Botstein, and C. C. Philpott. 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:49244-49250. [DOI] [PubMed] [Google Scholar]
- 33.Ramanan, N., and Y. Wang. 2000. A high-affinity iron permease essential for Candida albicans. Science 288:1062-1064. [DOI] [PubMed] [Google Scholar]
- 34.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 35.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory functions. Proc. Natl. Acad. Sci. USA 97:5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rüchel, R., R. Tegeler, and M. A. Trost. 1982. Comparison of secretory proteinases from different strains of Candida albicans. Sabouraudia 20:233-244. [DOI] [PubMed] [Google Scholar]
- 37.Rupniak, H. T., C. Rowlatt, E. B. Lane, J. G. Steele, L. K. Trejdosiewicz, B. Laskiewicz, S. Povey, and B. T. Hill. 1985. Characteristics of four new human cell lines derived from squamous cell carcinomas of the head and neck. JNCI 75:621-635. [PubMed] [Google Scholar]
- 38.Sanglard, D., B. Hube, M. Monod, F. C. Odds, and N. A. Gow. 1997. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect. Immun. 65:3539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaller, M., W. Schäfer, H. C. Korting, and B. Hube. 1998. Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol. Microbiol. 29:605-615. [DOI] [PubMed] [Google Scholar]
- 40.Schaller, M., H. C. Korting, W. Schäfer, J. Bastert, W. C. Chen, and B. Hube. 1999. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol. Microbiol. 34:169-180. [DOI] [PubMed] [Google Scholar]
- 41.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 43.Sweet, S. P., and L. J. Douglas. 1991. Effect of iron concentration on siderophore synthesis and pigment production by Candida albicans. FEMS Microbiol. Lett. 80:87-92. [DOI] [PubMed] [Google Scholar]
- 44.Weinberg, E. D. 1999. The role of iron in protozoan and fungal infectious diseases. J. Eukaryot. Microbiol. 46:231-238. [DOI] [PubMed] [Google Scholar]
- 45.Winkelmann, G., and H. Drechsel. 1997. Microbial siderophores, p. 200-246. In H.-J. Rehm and P. Stadlers (ed.), Bio/Technology, 2nd ed. VCH, Weinheim, Germany.
- 46.Yamaguchi-Iwai, Y., R. Stearman, A. Dancis, and R. D. Klausner. 1996. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15:3377-3384. [PMC free article] [PubMed] [Google Scholar]
- 47.Yun, C.-W., T. Ferea, J. Rashford, O. Ardon, P. Brown, D. Botstein, J. Kaplan, and C. C. Philpott. 2000. Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake. J. Biol. Chem. 275:10709-10715. [DOI] [PubMed] [Google Scholar]
- 48.Yun, C.-W., J. S. Tiedeman, R. E. Moore, and C. C. Philpott. 2000. Siderophore-iron uptake in Saccharomyces cerevisiae. J. Biol. Chem. 275:16354-16359. [DOI] [PubMed] [Google Scholar]