Abstract
The outer membrane (OM) of the mammalian pathogen Leptospira kirschneri was isolated in the form of membrane vesicles by alkaline plasmolysis and separated from the protoplasmic cylinder by sucrose density gradient ultracentrifugation. All four components of the alkaline plasmolysis buffer, including 1.0 M NaCl, 27% sucrose (wt/vol), 2 mM EDTA, and 10 mM Tris (pH 9), were required for efficient OM release, as judged by recovery of leptospiral lipopolysaccharide. Two populations of OM vesicles (OMVs) were recovered, with peak concentrations found in the sucrose gradient at densities of 1.16 and 1.18 g/ml. Transmission electron microscopy revealed that the more buoyant OMV population was smaller (<0.1 μm in diameter) than the denser OMV population (0.2 to 0.3 μm in diameter). The densities of both populations of OMVs were distinct from that of the protoplasmic-cylinder material, which was found in the sucrose gradient at a density of 1.20 g/ml. The OMV fractions were free of protoplasmic-cylinder material, as judged by immunoblotting with antibodies to the endoflagellar sheath protein, heat shock protein GroEL, and two novel cytoplasmic membrane proteins, lipoprotein LipL31 and transmembrane protein ImpL63. The protein components of the OMVs were characterized by one- and two-dimensional immunoblotting and found to include previously described OM proteins (OMPs), including the porin OmpL1; the lipoproteins LipL32, LipL36, and LipL41; and the peripheral membrane protein P31LipL45. A number of less well-characterized OMPs were also identified, including those with molecular masses of 16, 21, 21.5, 22, 31, 36, 44, 48, 90, and 116 kDa. The 48-kDa OMP was identified as a novel OM lipoprotein designated LipL48. The use of membrane-specific markers in OM isolation techniques facilitates an accurate description of the leptospiral OM and its components.
Identification and characterization of outer membrane (OM) components are essential in the development of a molecular understanding of bacterial structure and function. Development of techniques for isolation of the OM from Leptospira species and other spirochetes has been difficult because of their unique architecture and a lack of sensitive markers for the cytoplasmic membrane. Like enteric gram-negative bacteria, spirochetes have both an OM and a cytoplasmic membrane, separated by a periplasmic space. However, spirochetal architecture differs significantly from that of gram-negative bacteria in that the peptidoglycan layer of spirochetes is associated with the cytoplasmic membrane rather than the OM (22, 24). For this reason, techniques developed for isolation of the gram-negative OM from components of the underlying cell wall and cytoplasmic membrane are unlikely to be useful in spirochetes.
Several approaches have been used in the isolation of the leptospiral OM, including sodium dodecyl sulfate (SDS) treatment of salt-altered cells (2, 7, 25, 28), Sarkosyl (27), and Triton X-114 extraction and phase partitioning (20, 45). On the basis of these approaches, three classes of leptospiral OM proteins (OMPs) have been described: transmembrane, lipoprotein, and peripheral membrane proteins. OmpL1 was the first leptospiral OMP to be described (16). The structure of OmpL1 is predicted to contain at least 10 β-sheet transmembrane segments, which probably accounts for its heat-modifiable electrophoretic mobility. Its transmembrane structure is also supported by evidence that purified OmpL1 creates porin channels in the planar lipid bilayer assay (36). A second class of leptospiral OMPs are the lipoproteins that are anchored to the OM by fatty acids attached to an amino-terminal cysteine (15). Analysis of the detergent phase of Triton X-114 extracts of Leptospira kirschneri intrinsically labeled with tritiated palmitate indicates that the leptospiral OM contains at least five lipoproteins (37). The genes encoding three leptospiral OM lipoproteins, LipL32, LipL36, and LipL41, have been described (17, 18, 37). LipL32 is known to be the most prominent protein in the leptospiral protein profile and is an immunodominant antigen during human leptospirosis (10, 14, 17). LipL41 is a surface-exposed lipoprotein that provides synergistic immunoprotection with OmpL1 (19, 37). Recently, the peripheral membrane protein P31LipL45 was described, which represents a third type of leptospiral OMP (J. Matsunaga, T. A. Young, J. Croda, J. Lima, A. I. Ko, and D. A. Haake, 101st Gen. Meet. Am. Soc. Microbiol. 2001, abstr. D-229, p. 324, 2001). P31LipL45 is exported as a 45-kDa lipoprotein and processed to a 31-kDa C-terminal form that is associated with the OM. Evidence that P31LipL45 is a peripheral OMP, rather than an integral OMP, comes from studies showing that it is released from membranes by urea and that it partitions into the Triton X-114 detergent and aqueous phases.
In the absence of a detergent-independent technique for isolation of the leptospiral OM, it has not been possible to confirm that proteins, such as lipoproteins, are actually OM components or rather cytoplasmic membrane proteins selectively extracted by detergents. An example of the aberrant behavior of spirochetal cytoplasmic membrane lipoproteins in detergent extraction methods is TpN47 of Treponema pallidum, which is readily extracted by Triton X-114 (32) yet has been shown to be virtually absent from T. pallidum OM vesicles (OMVs) isolated and purified by isopycnic sucrose gradient ultracentrifugation (4, 34). Another caveat about detergent-based methods of isolating spirochetal OMs was brought to light by the finding that LipL32 is selectively degraded during Triton X-114 phase partitioning (17, 45).
Notably lacking from previous studies of the leptospiral OM are controls to assess contamination of the OM fraction with cytoplasmic membrane components. We have developed antisera to two leptospiral cytoplasmic membrane proteins that serve as sensitive markers of the cytoplasmic membrane. Leptospiral lipopolysaccharide (LPS) serves as a specific marker for the OM. By using these cytoplasmic membrane and OM detection tools, we have developed a method for isolation of the leptospiral OM in the form of membrane vesicles. Our method is a modification of techniques that use buffers that destabilize the spirochetal OM and facilitate its release in the form of OMVs (4, 5, 33, 34, 38). These OM isolation techniques take advantage of the structural lability of the spirochetal OM to produce OMVs that can be separated from protoplasmic-cylinder (PC)components by isopycnic sucrose gradient density ultracentrifugation. We believe that the method presented here provides the most accurate assessment of a protein's representation in the leptospiral OM.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
L. kirschneri strain RM52 (39) and other Leptospira species were received from C. A. Bolin (National Animal Disease Center, Agricultural Research Service, U.S. Department of Agriculture, Ames, Iowa) and passaged in bovine serum albumin-Tween 80 medium (bovuminar PLM-5 microbiological media; Intergen) (23). Escherichia coli DH5α [supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used as the host strain for transformations of recombinant DNA. E. coli PLK-F′ {recA lac mcrA mcrB hsdR gal supE [F′ proAB lacIqZ ΔM15 Tn10(Tetr)]} was used as the host strain for infection with the Lambda Zap II vector (Stratagene). E. coli strain PLK-F′ and the ExAssist helper phage were used for in vivo excision of the pBluescript phagemid (Stratagene). E. coli SOLR [e14− mcrA Δ(mcrCB-hsdSMR-mrr)171 sbcC recB recJ umuC::Tn5(Kanr) uvrC lac gyrA96 relA1 thi-1 endA1 λr (F′ proAB lacIqZ ΔM15) Su− (nonsuppressing)] was used as the host strain for replication of the excised pBluescript phagemid from the Lambda Zap II vector (Stratagene). E. coli BLR(DE3)pLysS [F− ompT hsdSB (rB−mB−) gal dcm Δ(srl-recA)306::Tn10(Tcr) (DE3) pLysS(Cmr)] (Novagen) was used as the host strain for the pRSET expression vector (Invitrogen). E. coli strains were grown in LB (Luria broth) supplemented with 100 μg of ampicillin per ml, 100 μg of carbenicillin per ml, or 25 μg of chloramphenicol per ml when appropriate. Antibiotics were purchased from Sigma.
L. kirschneri OM isolation in sucrose-NaCl.
L. kirschneri bacteria (1.6 × 1011) were washed once in phosphate-buffered saline (PBS) plus 5 mM MgCl2 and once in PBS. The pellet was resuspended in 4 ml of TNSEDR buffer consisting of 20 mM Tris (pH 9), 1 M NaCl, 27% (wt/vol) sucrose, and 2 mM EDTA. The leptospiral suspension was placed in a 2059 tube (Falcon) with a magnetic microstir bar (12 by 2 mm). After 90 min of mixing at room temperature, 50 μg of DNase per ml and 0.25 μg of RNase per ml were added and the combination was mixed for 30 min. The suspension was centrifuged for 10 min at 10,000 × g to remove aggregated material. The supernatant was loaded onto a continuous 27 to 55% (wt/vol) sucrose gradient (in 10 mM Tris [pH 9], 1 M NaCl, and 2 mM EDTA) in Beckman Ultraclear tubes and centrifuged overnight at 28,000 rpm in an SW28 rotor. The entire gradient was separated into 0.75-ml fractions by pipetting from the top of the gradient. The refractive index of each fraction was measured with a refractometer. The protein concentration was determined on the fractions with the bicinchoninic acid protein assay system of Pierce Chemical Co. (Rockford, Ill.). Material for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and/or immunoblot analysis was obtained by trichloroacetic acid precipitation performed by incubation with 4 volumes of 5% trichloroacetic acid for 16 h at 4°C. Precipitated material was collected by centrifugation for 15 min at top speed in a Microfuge, washed with acetone, centrifuged for 5 min at top speed in a Microfuge, and then resuspended in sample buffer.
Electron microscopy.
Whole-mount transmission electron microscopy was performed by placing 10-μl samples of alkaline plasmolysis-treated L. kirschneri and sucrose gradient-purified OMVs on Parlodion (Mallinckrodt, Inc., St. Louis, Mo.)-coated 300-mesh copper grids (Ted Pella Inc., Tustin, Calif.) for 1 min, followed by staining with 1% uranyl acetate. Grids were examined in an electron microscope (JEOL 100 CX) at an accelerating voltage of 80 kV.
Gel electrophoresis and immunoblotting.
For one-dimensional (1D) SDS-PAGE, samples were solubilized in a final sample buffer composed of 62.5 mM Tris hydrochloride (pH 6.8), 10% glycerol, 5% 2-mercaptoethanol, and 2% SDS and separated on a discontinuous buffer system (26). Two-dimensional (2D) gel electrophoresis was performed by the method of O'Farrell (29), as modified by Görg et al. (12, 13). Samples for 2D gel electrophoresis were solubilized in a rehydration solution composed of 8 M urea, 2% Triton X-100, 20 mM dithiothreitol, and 2% carrier ampholyte mixture (IPG buffer; Pharmacia). Immobiline DryStrips (Pharmacia) were rehydrated overnight in rehydration solution containing leptospiral material. Isoelectric focusing was performed with a Pharmacia Multiphor II System. After isoelectric focusing, SDS-PAGE was performed as described above. Gels were stained with Coomassie brilliant blue or silver (SilverXpress; Invitrogen) or transferred to 0.45-μm-pore-size Immobilon-P membranes (Millipore) for immunoblotting. For antigenic detection on immunoblots, the nitrocellulose was blocked with 5% nonfat dry milk in PBS-0.1% Tween 20, incubated for 1 h with antiserum diluted 1:5,000 (unless otherwise noted) in PBS-0.1% Tween 20, and probed with donkey anti-rabbit antiserum conjugated to horseradish peroxidase (Amersham). Antigen-antibody binding was detected with the enhanced-chemiluminescence system (Amersham). Blots were incubated in enhanced-chemiluminescence system reagents for 1 min and then exposed to Hyperfilm (Amersham).
Cloning and sequencing of the lipL48 gene.
Work by Alves et al. found the N-terminal amino acid sequence of a 24-kDa clostripain proteolytic fragment of LipL48 to be AAAQNTEGGTGLQYNSGAND (1). A 23-bp oligonucleotide probe, LipL48a, with 1,024-fold degeneracy, GCWGCNGCNCARAAYACNGARGG, was designed on the basis of the AAAQNTEG portion of the sequence. On the basis of the low GC content of Leptospira spp., codon bias was used for the first alanine residue.
L. kirschneri genomic DNA was prepared by the method of Yelton and Charon (44). Leptospiral DNA was digested with EcoRI and electrophoresed in a 1.0% agarose gel. Following denaturation and neutralization, the DNA was transferred to a nylon filter (Zeta-Probe; Bio-Rad) by the method of Southern (35). Filters were prehybridized for 3 h at 37°C in buffer containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1x Denhardt's solution, 0.05% sodium PPi, 0.5% SDS, and 100 μg of denatured salmon sperm DNA per ml. Oligonucleotide probe LipL48a was end labeled with [32P]dATP by using T4 polynucleotide kinase (Promega). Unincorporated label was removed with a NucTrap Probe Purification Column (Stratagene). The filters were then hybridized overnight at 37°C with radiolabeled oligonucleotides at a concentration of 106 cpm/ml. Filters were washed at 45°C in 3.0 M tetramethylammonium chloride (Aldrich)-50 mM Tris (pH 8.0)-2.0 mM EDTA-1.0% SDS as previously described (16). Washed filters were exposed to XAR-5 film (Kodak) at −80°C.
EcoRI fragments of L. kirschneri genomic DNA were ligated into the Lambda Zap II vector (Stratagene). The ligated DNA was packaged with Gigapack II Gold packaging extract (Stratagene) and stored in 0.3% chloroform at 4°C. The plaque titer was determined by infecting E. coli PLK F′ (Stratagene). Plaques were plated, transferred to filters in duplicate, and processed as previously described (16). Filters were probed with the end-labeled LipL48a probe with the same hybridization and washing conditions as in Southern hybridization. Recombinant pBluescript SK(−) clones were recovered from phage producing positive plaques by in vivo excision as described by the manufacturer. After restriction mapping, appropriate DNA fragments were subcloned into pBluescript KS and sequenced at the University of California at Los Angeles Core DNA Sequencing Facility by the dideoxy-chain termination method with fluorescein-labeled dideoxy nucleotides (Applied Biosystems).
Cloning and sequencing of the impL63 gene.
The impL63 gene was isolated from an L. kirschneri library constructed with the pMG plasmid. pMG is an expression plasmid that includes the tac promoter, a polylinker with multiple cloning sites, and an alkaline phosphatase-encoding gene lacking the region encoding a signal peptide (3). L. kirschneri genomic DNA was partially digested with Sau3A, agarose gel purified, and ligated into pMG, which had been digested with BamHI and dephosphorylated with calf intestinal alkaline phosphatase (Stratagene). E. coli KS330r− was transformed with the ligation mixture and spread onto LB plates containing 50 μg of ampicillin per ml, 100 μM isopropyl β-D-thiogalactopyranoside (IPTG), and 40 μg of 5-bromo-4-chloro-3-indoyl-phosphate (XP) per ml. Recombinant pMG clones were recovered from alkaline phosphatase-expressing recombinants and tested for the ability to form colonies with blue halos as previously described (11). One of these pMG clones, encoding the 5′ end of the impL63 gene, was selected for further study. The insert DNA was used as a probe to isolate the remainder of the impL63 gene from a Lambda Zap II library of L. kirschneri DNA fragments as described above.
Cloning and sequencing of the lipL31 gene.
Isolation of the lipL31 gene from a λTriplEx expression library has been described previously (Matsunaga et al., 101st Gen. Meet. Am. Soc. Microbiol.). In brief, L. kirschneri DNA was partially digested with AluI, modified with EcoRI methylase, ligated to an EcoRI linker (pCGGATTCCG), ligated to λTriplEx arms (Clontech), and packaged into λ heads with the Gigapack III Gold Packaging Extract (Stratagene). The library was plated with E. coli XL1-Blue, transferred to nitrocellulose membranes soaked with IPTG, processed, and probed with rabbit antiserum to virulent L. kirschneri as previously described (Matsunaga et al., 101st Gen. Meet. Am. Soc. Microbiol.). Reactive λ clones were purified, and the recombinant plasmid was excised by infecting E. coli BM25.8.
Antisera.
Antisera to OmpL1, LipL36, LipL41, and LipL32 was prepared as previously described (16-18, 37). Antiflagellar monoclonal antibody 1H8 was a generous gift of Richard Zuerner, National Animal Disease Center, Animal Research Service, U.S. Department of Agriculture, Ames, Iowa. Rabbit antiserum to leptospiral GroEL was a generous gift of Ben Adler, Department of Microbiology, Monash University, Clayton, Victoria, Australia. Murine monoclonal antibody F71C2 against serovar grippotyphosa (21) was a generous gift of Rudy Hartskeerl (Royal Tropical Institute, Amsterdam, The Netherlands).
Antiserum to LipL48 was prepared as follows. A PCR was used to amplify the portion of the lipL48 gene encoding the mature protein beginning with the first residue after the amino-terminal cysteine. The 5′ oligonucleotide contained the nucleotide sequence coding for the six amino acids following the amino-terminal cysteine of mature LipL48, including an XhoI restriction endonuclease site (underlined): 5′-TTT TAT CTC GAG TAA AGA AGA CAA AGA CGA-3′. The 3′ oligonucleotide contained the nucleotide sequence coding for the five carboxy-terminal amino acids and the lipL48 stop codon, including an SmaI restriction endonuclease site (underlined): 5′-TAA TGT CCC GGG TTA TCT TGC TCT ATA AAC-3′. L. kirschneri genomic DNA was used as the template. The 1,321-bp amplified lipL48 gene was digested with XhoI and SmaI and ligated into pRSETb (Invitrogen) digested with XhoI and PvuII. The resulting construct, pRSETb-LipL48, was transformed into E. coli BLR(DE3)pLysS (Novagen). Expression of the His6-LipL48 fusion protein was achieved by IPTG (Sigma) induction. Bacteria were lysed by sonication, and the His6-LipL48 fusion protein was purified by affinity chromatography with Ni2+-nitrilotriacetic acid-agarose in accordance with the manufacturer's (Qiagen) instructions. The purified His6-LipL48 fusion protein was loaded onto an SDS-PAGE gel. After electrophoresis, the His6-LipL48 band containing approximately 150 μg of protein was cut out of the acrylamide gel, desiccated, ground to powder, mixed with Freund's complete adjuvant, and inoculated subcutaneously and intramuscularly into a male New Zealand White rabbit. Additional immunizations with approximately 150 μg of His6-LipL48 fusion protein in Freund's incomplete adjuvant were given at 4 and 8 weeks after the primary immunization. The rabbit was bled 10 weeks after the primary immunization.
Antiserum to LipL31 was prepared as follows. A PCR was used to amplify the portion of the lipL31 gene encoding the mature protein beginning with the first residue after the amino-terminal cysteine. The 5′ oligonucleotide contained the nucleotide sequence coding for the seven amino acids following the amino-terminal cysteine of mature LipL31, including an XhoI restriction endonuclease site (underlined): 5′-CG ACT CGA GGA GAT AAT TCC GAA GTG ATT GA-3′. The 3′ oligonucleotide contained the nucleotide sequence coding for the five carboxy-terminal amino acids and the lipL48 stop codon, including a HindIII restriction endonuclease site (underlined): 5′-T CAA AGC TTA TTA CTG CCC GGT AGT TTT C-3′. The cloned lipL31 gene was used as the template. The 0.65-kb amplified lipL31 gene was digested with XhoI and HindIII and ligated into pRSETc (Invitrogen) digested with XhoI and HindIII. The resulting construct, pRSETb-LipL31, was transformed into E. coli BLR(DE3)pLysS (Novagen). His6-LipL31 expression and purification and immunization of rabbits were carried out as described above for antiserum to LipL48.
Antiserum to ImpL63 was prepared as follows. The pBluescript SK(−) plasmid 86-4A was recovered from a Lambda Zap II clone encoding the 3′ 1,558 bp of the impL63 gene. Plasmid 86-4A was digested at the BamHI site of the pBluescript polylinker and then partially digested with ClaI to obtain a 1,700-bp BamHI-ClaI DNA fragment encoding all except the first 21 amino acids of the ImpL63 protein. The 1,700-bp BamHI-ClaI DNA fragment was ligated to the pRSETc (Invitrogen) plasmid, which had been digested with BamHI and Csp45I. The resulting plasmid, pRSETc-ImpL63, was used to produce a His6-ImpL63 fusion protein and generate ImpL63 antiserum as described above for LipL48, except that special measures were needed to recover the full-length His6-ImpL63 protein, presumably because of proteolytic breakdown in E. coli. After transformation of the pRSETc-ImpL63 plasmid into E. coli BLR(DE3)pLysS (Novagen), cells were grown at 30°C. When the optical density at 600 nm reached 0.7, 0.1 mM IPTG was added and the cells were incubated at 30°C for an additional 2 h. EDTA (1 mM) was added, and the culture was rapidly cooled to 4°C in an ice bath. All subsequent steps were carried out at 4°C.
Nucleotide sequence accession numbers.
The nucleotide sequences of the impL63, lipL48, and lipL31 genes from L. kirschneri serovar grippotyphosa strain RM52 have been deposited in the GenBank database and assigned accession numbers AF394741, AF394742, and AF394743, respectively.
RESULTS
Cloning and sequence analysis of the lipL48 gene.
Southern hybridization studies revealed binding of a degenerate oligonucleotide, LipL48a, whose design was based on the partial amino acid sequence of LipL48 (1), to a single 2.7-kb band in digests of total genomic L. kirschneri serovar grippotyphosa strain RM52 DNA with the restriction enzyme EcoRI. The 2.7-kb DNA fragment was isolated from a Lambda Zap II library of L. kirschneri EcoRI DNA fragments. DNA sequencing revealed that the 2.7-kb DNA fragment encodes two intact open reading frames (ORFs), ORF1 and the lipL48 gene, and the 5′ end of a third ORF, ORF3. The TAA stop codon of ORF1 is 70 bp upstream of the lipL48 gene. There are no transcription terminators or promoter elements between ORF1 and the lipL48 gene. The absence of these elements suggests that these genes are cotranscribed. Analysis of the lipL48 sequence indicates that it encodes a membrane lipoprotein with a 25-amino-acid signal peptide, a lipoprotein signal peptidase cleavage site, and an amino-terminal cysteine. The mature protein would be 436 amino acids long, with a predicted molecular mass of 44.1 kDa, similar to the observed molecular mass of 48 kDa. Clostripain is known to cleave peptides following basic amino acids. The predicted amino acid sequence following arginine residue 159 is identical to that obtained by N-terminal amino acid sequencing of proteolytic fragments of the native protein. Immediately following the lipL48 gene, there is an inverted repeat that may function as a rho-independent transcription terminator.
Cloning and sequence analysis of the impL63 gene.
L. kirschneri DNA Sau3A fragments were screened in KS330r− for sequences permitting secretion of alkaline phosphatase. Clone L2.086 was identified as a blue-halo colony. The DNA insert of clone L2.086 was sequenced and found to have a 24-amino-acid transmembrane domain that allows the export of alkaline phosphatase. The remainder of the gene belonging to the L2.086 insert was cloned on a 3.0-kb EcoRI fragment. Restriction endonuclease mapping was performed, and appropriate DNA fragments were subcloned into pBluescript KS for sequencing. An ORF of 1,740 bp was found, which would encode a 540-amino-acid polypeptide with a predicted molecular mass of 63 kDa. PSORT analysis predicted two α-helical transmembrane domains (amino acids 8 to 24 and 463 to 479), indicating that ImpL63 is a transmembrane cytoplasmic membrane protein. Clone L2.086 contains only the first of these two putative transmembrane domains. The blue-halo phenotype of clone L2.086 implies that the first transmembrane domain is part of a cleaved export signal. A BLAST search of the GenBank database revealed 48% amino acid sequence identity with a partial ORF in L. meyeri upstream of the metYX operon (GenBank accession number Y10744).
Cloning and sequence analysis of the lip31 gene.
Fourteen of 75 clones isolated from a lambda expression library of L. kirschneri genomic DNA fragments contained the lipL31 gene. One of these 14 clones, clone 8, was selected for further analysis. The sequence of clone 8 revealed two intact ORFs, lipL31 and 8orf2. The deduced amino acid sequence of LipL31 would encode a 239-amino-acid polypeptide with a 17-amino-acid signal peptide, followed by a lipoprotein signal peptidase cleavage site (Table 1). The processed protein has a calculated molecular weight of 25,492. Eighteen percent of the amino acids of LipL31 are either glutamates or aspartates. The high percentage of acidic amino acids may reduce SDS binding, which could contribute to the discrepancy between the calculated molecular weight of LipL31 and its observed mobility on SDS-PAGE. 8orf2 begins 7 bp after the lipL31 stop codon, suggesting that the two genes are cotranscribed. 8orf2 would begin with a UUG start codon. A BLASTP search of the GenBank database revealed 34% amino acid sequence identity with the undecaprenol kinase (BacA) of Pseudomonas aeruginosa.
TABLE 1.
Relationship between the charge of N-terminal amino acids and membrane destinations of lipoproteinsa
| Protein | Amino acid at position:
|
Presence in:
|
|||
|---|---|---|---|---|---|
| +1 | +2 | +3 | CM | OM | |
| LipL48 | Cys | Lys | Glu | − | + |
| LipL41 | Cys | Ala | Ala | + | + |
| LipL36 | Cys | Lys | Ser | − | + |
| LipL32 | Cys | Gly | Ala | + | + |
| LipL31 | Cys | Gly | Asp | + | − |
| E. coli murein lipoprotein | Cys | Ser | Ser | − | + |
| Inouye's mutant | Cys | Asp | Ser | + | − |
LipL31 is the only leptospiral lipoprotein with a negatively charged amino acid in the +2 or +3 position after the N-terminal cysteine without a counterbalancing positively charged residue. LipL31 is the only leptospiral lipoprotein that is found exclusively in the cytoplasmic membrane (CM). Site-specific mutagenesis of the E. coli murein lipoprotein suggests that charged amino acids in this location function as a lipoprotein-sorting signal (42).
Isolation of the leptospiral OM.
When the hypotonic citrate and hypertonic sucrose techniques, developed for OM isolation from Treponema species and Borrelia burgdorferi, were applied without modification, there was poor recovery of the leptospiral OM, as judged by recovery of LPS. Initially we found that addition of 1 M NaCl greatly improved the release of leptospiral LPS. Eventually, four components of the alkaline plasmolysis buffer were tested in isolation and found to be essential for efficient release of the leptospiral OM: hypertonic sucrose, 1 M NaCl, 2 mM EDTA, and an alkaline pH. For brevity, only the data on the effect of pH on leptospiral OM release are presented here. Figure 1 shows that leptospiral LPS release increased as the pH of the plasmolysis buffer was increased from 3 to 9. At pH 9, most of the LPS had been removed from the PC material. It is useful to note (see Fig. 4) that the PC recovered with this technique is relatively free of OM material in order to judge the relative distribution of proteins in the two leptospiral membranes. While it is possible that a more rapid OM release could be achieved at a more alkaline pH than 9, there is also the potential for hydrolysis of the linkage between fatty acids and the N-terminal cysteine of lipoproteins. Hydrolysis was not observed at pH 9, as judged by the recovery of all five known leptospiral lipoproteins in either the OM or the PC fraction.
FIG. 1.
Effect of pH on release of OMVs. L. kirschneri bacteria were treated with hypertonic sucrose buffers ranging from pH 3 to pH 9, followed by sucrose density gradient ultracentrifugation. Peak OMV fractions were probed with monoclonal antibody F71C2, which is specific for serovar grippotyphosa LPS. Maximal OM release occurred at pH 9. Locations of molecular size standards (M) are shown (in kilodaltons) on the left.
FIG. 4.
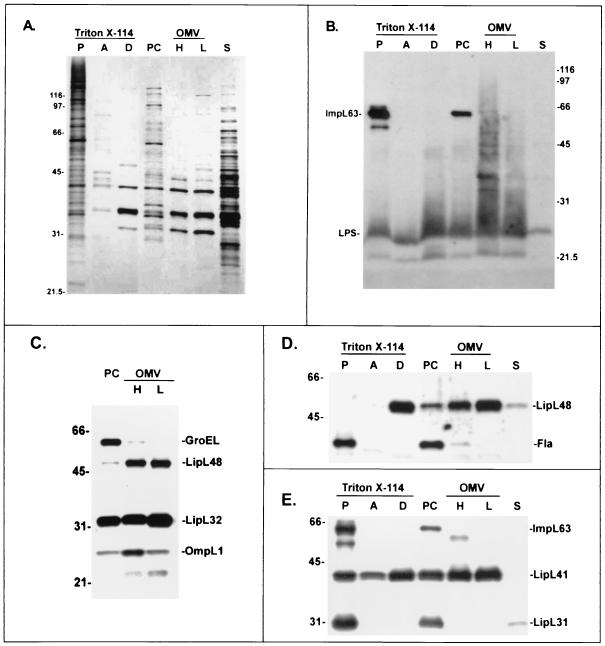
Comparison of leptospiral fractionation with Triton X-114 and alkaline plasmolysis. Fractions were separated by SDS-PAGE and silver stained (A) or probed with LPS monoclonal antibody F71C2 and antiserum to ImpL63 (B); antisera to GroEL, LipL48, LipL32, and OmpL1 (C); flagellar monoclonal antibody 1H8 and antiserum to LipL48 (D); or antisera to LipL31, LipL41, and ImpL63 (E). The Triton X-114 fractions analyzed were the insoluble pellet (lane P), the aqueous phase (lane A), and the detergent phase (lane D). The alkaline plasmolysis fractions analyzed were the PC, high (lane H)- and low (lane L)-density OMV, and soluble supernatant (lane S) fractions. There was no evidence of contamination of the OM fractions by any of the markers of the PC, including GroEL, flagella, LipL31, or ImpL63. The values on the left of panels A, C, D, and E and on the right of panel B are molecular sizes in kilodaltons.
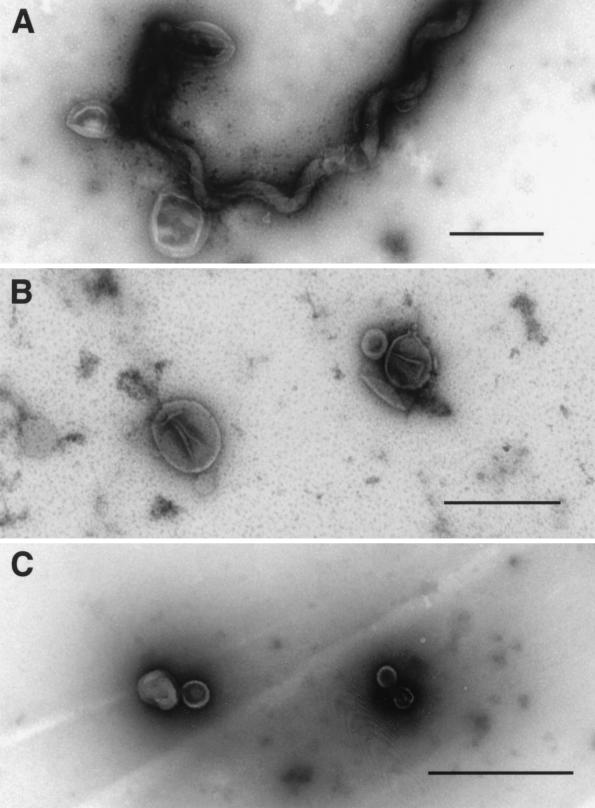
Electron microscopy revealed that treatment of L. kirschneri with the alkaline plasmolysis buffer resulted in disruption of the OM and formation of OMVs that adhered to the PC (Fig. 3A). Mechanical mixing was found to be important for release of OMVs from the PC. Simple rocking of the bacterial suspension in the alkaline plasmolysis buffer was insufficient to release the leptospiral OMVs. Stirring with a magnetic microstir bar (2-mm diameter) provided the proper amount of mechanical agitation for OM release. The size of the microstir bar and the rate of mixing were important. It was found that contamination of the OM fraction with PC components could occur if the bacterial suspension was mixed too rapidly with a larger-diameter (5/16-in.) microstir bar. Released nucleic acids were found to cause aggregation of membrane vesicles. When added prior to loading onto the sucrose gradient, DNase and RNase also improved separation of the OM and PC components.
FIG. 3.
Whole-mount electron microscopy of OMVs isolated from L. kirschneri. (A) Organisms shortly after exposure to alkaline plasmolysis buffer showing release of OMVs. (B) Purified OMVs obtained from the high-density OMV fraction of the sucrose gradient. (C) Purified OMVs obtained from the low-density OMV fraction of the sucrose gradient. Bars, 0.5 μm.
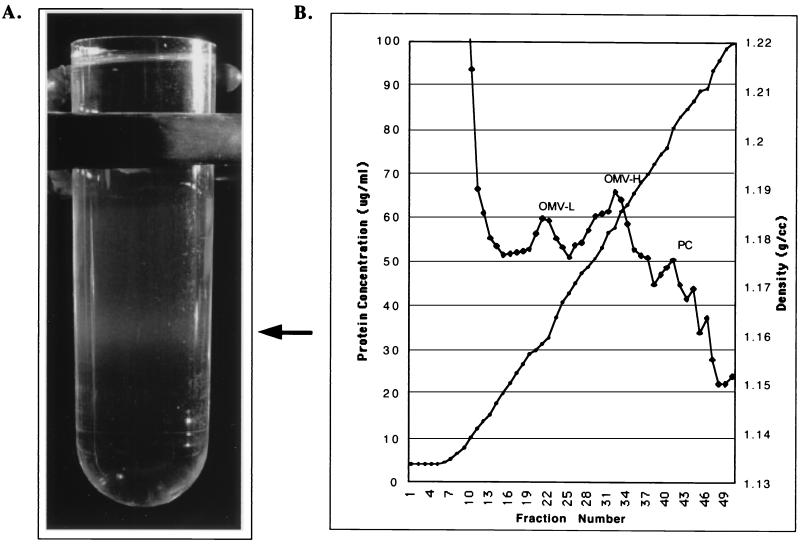
The protein concentration of the sucrose gradient fractions peaked in four places (Fig. 2B). The first protein peak occurred in the most buoyant portion of the gradient containing leptospiral proteins that were unable to enter the gradient. The second and third protein peaks contained high concentrations of leptospiral LPS, as determined by immunoblotting (Fig. 4B), and were designated the light (OML) and heavy (OMH) OM fractions. Examination of the OML and OMH fractions by electron microscopy revealed membrane vesicles free of PCs. The membrane vesicles in the OMH fraction were generally larger (0.2 to 0.3 μm in diameter; Fig. 3B) than the membrane vesicles in the OML fraction (<0.1 μm in diameter; Fig. 3C). The density of OMVs in the OMH fraction was sufficient to form a discrete band visible in the sucrose density gradient shown in Fig. 2A. A fourth peak contained the cytoplasmic membrane proteins LipL31 (Fig. 4E) and ImpL63 (Fig. 4B and E) and PC components GroEL (Fig. 4C) and the endoflagellar sheath protein (Fig. 4D).
FIG. 2.
Isolation of leptospiral OMVs. (A) Tube showing banding of OMVs in the sucrose density gradient after ultracentrifugation. (B) Fractions were collected from the top of the gradient and tested for protein concentration (♦) and density (•). The locations of the protein concentration peaks containing low-density OMVs (OMV-L), high-density OMVs (OMV-H), and PC fractions are shown.
The results of refractive-index analysis were as follows. The protein profile of the four sucrose gradient peaks was compared with those of the pellet, aqueous, and detergent phase fractions obtained by Triton X-114 extraction and phase partitioning (Fig. 4A). There is overall similarity between the protein profile of the Triton X-114 pellet fraction and that of the sucrose gradient PC fraction. Likewise, there is similarity between the protein profile of the Triton X-114 detergent fraction and those of the OMH and OML fractions. The four bands common to the Triton X-114 detergent fraction and the OMH and OML fractions represent, in order of increasing molecular mass, the OM lipoproteins LipL32, LipL36, LipL41, and LipL48. This result confirms that, as in other spirochetes, the most abundant proteins in the leptospiral OM are lipoproteins. The transmembrane porin OmpL1 is known to be a relatively minor component of the leptospiral OM (16, 36) and was not visualized in the protein silver stain (Fig. 4A), but its presence in the OM was demonstrated by immunoblotting (Fig. 4C). OMPs with molecular masses of 44, 90, and 116 kDa are better represented in either the OMH or the OML fraction than in the Triton X-114 detergent fraction (Fig. 4A). The 44-kDa band may correspond to an OMP previously identified on the leptospiral surface (20). Several differences between the OMH and OML fractions were noted. The OMH fraction has a higher concentration of LPS, OmpL1, and the 44-kDa OMP. In contrast, the OML fraction has a higher concentration of LipL32, LipL36, and the 90- and 116-kDa proteins. The significance of the two OM fractions is uncertain. The greater density of the OMH fraction is not due to contamination with heavier PC material, as indicated by the absence of cytoplasmic membrane or PC markers (please refer to the previous paragraph). We intentionally avoided the use of a French press or other techniques that employ high shear forces, which are known to generate OM-cytoplasmic membrane hybrids (30).
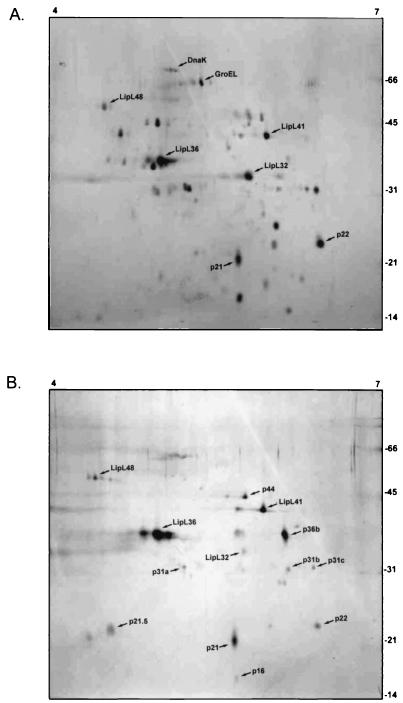
Proteins isolated in the OMH fraction were analyzed by 2D gel electrophoresis and compared with whole-cell proteins (Fig. 5). Isoelectric focusing was initially performed with 3-to-10 and 4-to-7 Immobiline Drystrips. Essentially all leptospiral proteins were found to have isoelectric points between 4 and 7. At least 30 separate protein spots were visualized on silver-stained 2D gels of whole L. kirschneri cells. In contrast, at least 12 protein spots were visualized on silver-stained 2D gels of the OMH fraction at concentrations equal to or greater than those seen in whole cells. Five of these 13 protein spots correspond to the known proteins P31LipL45, LipL32, LipL36, LipL41, and LipL48 (Table 2). A recently reported study of L. interrogans serovar Lai OMPs included tandem electrospray mass spectrometry peptide sequencing of several of the unidentified spots (9).
FIG. 5.
Comparison of silver-stained 2D gels of whole cells (A) and high-density OMV fractions (B) of L. kirschneri obtained by alkaline plasmolysis and sucrose density gradient ultracentrifugation. The locations of molecular mass markers (in kilodaltons) and reference isoelectric points are indicated on the right and top, respectively.
TABLE 2.
Proteins observed in leptospiral OM fractions by 1D and/or 2D gel electrophoresis at concentrations equal to or greater than the concentrations found in whole cellsa
| Protein | Identity | Antigenicity |
|---|---|---|
| p116 | − | |
| p90 | − | |
| p48 | LipL48 | − |
| p44b | + | |
| p41 | LipL41 | + |
| p36a | LipL36 | − |
| p36b | − | |
| p32 | LipL32 | + |
| p31a | P31LipL45 | ± |
| p31b | − | |
| p31c | − | |
| p22 | + | |
| p21.5b | − | |
| p21b | LipL21c | − |
The identities of known OMPs are shown in the middle column. Comparisons with 1D and 2D immunoblots using clinical leptospirosis sera allowed the identification of some of these proteins as antigens recognized by the humoral immune response during human leptospirosis (14), as indicated in the right-hand column.
Protein for which peptide sequencing has been performed in a recently reported study of L. interrogans serovar Lai OMPs (9).
The pL21 protein has been identified as an OM lipoprotein designated LipL21 (P. A. Cullen, B. Adler, D. M. Bulach, D. A. Haake, and R. L. Zuerner, unpublished data).
DISCUSSION
A major focus of current leptospiral research is the identification and characterization of leptospiral OMPs. OMPs are of great interest in understanding how bacteria adapt to and interact with their environment. Assignment of proteins to the leptospiral OM previously relied on one of three detergent-based approaches. The first of these leptospiral OM isolation methods involves treatment of Leptospira spp. with hypertonic saline, resulting in salt-altered cells in which the OM is dissociated from the underlying PC, followed by release of the OM with the ionic detergent SDS (2, 7, 25, 28). Another study applied a technique developed for isolation of the Haemophilus influenzae OM involving sonication and sodium-N-lauroylsarcosinate (27). Both of these approaches result in recovery of membrane vesicles containing leptospiral LPS. However, in neither case was an attempt made to evaluate the selectivity of these detergent-based approaches by assessing contamination of the OM fraction with PC components. Because of the unique architecture of spirochetes and the fragility of the OM, it cannot be assumed that there is differential solubility of leptospiral membranes to SDS or sodium-N-lauroylsarcosinate.
Previously, we examined the use of Triton X-114 for solubilization of the leptospiral OM (20, 45). This approach appears to be somewhat selective, based upon a lack contamination of the Triton X-114 detergent phase with flagella and penicillin-binding proteins (20). However, some spirochetal cytoplasmic membrane proteins may be more susceptible to solubilization by Triton X-114 than others. For example, the nonionic detergent Triton X-114 solubilizes the TpN47 lipoprotein of T. pallidum, which has been shown to be a cell wall-associated penicillin-binding protein (42). Another flaw in the Triton X-114 approach is that generation of the Triton X-114 detergent phase may result in loss of certain leptospiral OMPs. For example, degradation of the leptospiral lipoprotein LipL32 occurs during Triton X-114 phase partitioning (17, 45). The purpose of the present study was to develop a detergent-independent technique for isolation of the leptospiral OM to confirm that proteins thought to be located in the leptospiral OM are actually OM components rather than cytoplasmic membrane proteins selectively extracted by detergents.
Spirochetal OM isolation has been advanced by developments in two areas. Firstly, a number of antibody reagents have been developed that recognize components of specific leptospiral cellular compartments. In this study, we utilized antisera to the inner membrane proteins LipL31 and ImpL63. LipL31 is presumed to be a lipoprotein, while ImpL63 is a transmembrane cytoplasmic membrane protein. Since these proteins interact with the cytoplasmic membrane in different ways, utilizing antisera to both proteins to detect cytoplasmic membrane contamination of OM fractions is complementary and increases confidence in the specificity of the OM isolation technique. Other PC components that could potentially contaminate the leptospiral OM fractions are the endoflagella, and we have included immunoblots probed with antibody specific for endoflagellar sheath protein (40). Probing of immunoblots with antisera to these proteins is a more rapid and sensitive approach to detection of the PC than the use of radiolabeled or digoxigenin-labeled penicillin binding proteins (6, 20).
Secondly, new techniques have been developed for isolation of the spirochetal OM that do not involve the use of detergents or mechanical disruption. It was discovered that exposure of T. pallidum and T. vincentii to a hypotonic sodium citrate buffer, pH 3.2, resulted in the release of the OM as unilamellar vesicles (4). Once released, the OMVs could then be isolated by sucrose gradient centrifugation. The hypotonic citrate technique has also been applied to the isolation of the B. burgdorferi OM (38). The second new technique involves the use of hypertonic sucrose to release the T. pallidum and B. burgdorferi OMs (33, 34). When these techniques were applied without modification, OM release was inefficient and only small amounts of OM material were obtained (data not shown). However, by taking into account the unique features of leptospiral architecture, we were successful in modifying the hypertonic sucrose techniques for use in the isolation of sufficient amounts of leptospiral OM material for analysis of its components.
The findings presented here shed light on the architecture of the leptospiral OM. In addition to hypertonic sucrose, three other components of the alkaline plasmolysis buffer were found to be essential for efficient release of leptospiral OM material: 1 M NaCl, 2 mM EDTA, and a pH of 9. The roles of salt and an alkaline pH in releasing the leptospiral OM indicate that salt bridging or hydrogen bonding between charged amino acids plays an important role in stabilizing the leptospiral OM. The finding that EDTA facilitates LPS release suggests that, as in E. coli, divalent cations are essential for interactions between leptospiral LPS molecules (8). In addition to its buffering function, Tris may also act to weaken interactions between LPS molecules (41).
An advantage of the alkaline plasmolysis technique is that the distribution of proteins in the OM and PC fractions may more accurately reflect the actual distribution in leptospiral membranes than Triton X-114 fractionation. LPS, OmpL1, LipL36, and LipL48 are largely, or exclusively, found in the OMH and OML fractions, while LipL32 and LipL41 are significantly represented in both membrane fractions. This resolves a previously poorly understood finding that OmpL1 is incompletely extracted by Triton detergents. Porins, such as OmpL1, are unlikely to be found in the cytoplasmic membrane of bacteria because of the adverse effect this would have on the electrochemical gradient across the cytoplasmic membrane. Incomplete extraction is most likely due to the relative insolubility of OmpL1 in nonionic Triton detergents.
Two novel leptospiral lipoproteins are described in this report, LipL31 and LipL48. Comparison of their sequences with those of the other three known leptospiral lipoproteins, LipL32, LipL36, and LipL41, makes it possible to form a hypothesis regarding membrane targeting of leptospiral lipoproteins. As shown in Table 1, there is a close correlation between the charge of the amino acids in the +2 and +3 positions and the membrane destination of the protein. LipL31 is the only one of the five leptospiral lipoproteins that is restricted to the inner membrane, and the first charged residue of the mature protein is the Glu at +3. In contrast, LipL36 and LipL48 are found exclusively in the OM, and in both cases, the first charged amino acid following the Cys at +1 is a positively charged residue in the +2 position. LipL32 and LipL41 have neutral amino acids in both the +2 and +3 positions, consistent with their presence in both the cytoplasmic membrane and the OM as determined by the alkaline plasmolysis membrane fractionation technique. These data indicate strongly that sorting of leptospiral lipoproteins is governed by mechanisms that are similar to those in E. coli. Experiments show that mutagenesis of the Ser at +2 to Asp results in sorting of E. coli murein lipoprotein to the inner membrane instead of the OM (43). Information relevant to prediction of the membrane destination of lipoproteins will be useful in the interpretation of leptospiral genomic sequence data. Validation of this hypothesis requires gene inactivation and complementation studies, an approach that has already been used with L. biflexa (31).
In summary we have presented an alkaline plasmolysis method for isolation of the leptospiral OM. The cytoplasmic membrane markers ImpL63 and LipL31 and the PC markers GroEL and the endoflagellar sheath protein were used to demonstrate complete separation of the OM from PC contamination. A number of novel OMPs were identified by 1D and 2D gel electrophoresis in amounts sufficient for identification by mass spectroscopy analysis once the leptospiral genomic sequence is eventually released. The significance of finding two populations of OMVs, OMH and OML, is uncertain. However, it is interesting that all three of the antigens found in greater abundance in the OMH fraction are known or suspected to be surface exposed, while at least one of the proteins (LipL36) found in greater abundance in the OML fraction has been shown to be restricted to the inner leaflet of the OM. On the basis of this evidence, we hypothesize that the OMH fraction is enriched for components of the outer leaflet of the OM while the OML fraction is enriched for components of the inner leaflet of the OM. We believe that this work will lead to improved understanding of the leptospiral OM.
Acknowledgments
This work was supported by VA Medical Research Funds (to J.M.) and Public Health Service grant AI-34431 (to D.A.H.).
We thank Paul Cullen for critical review of the manuscript. We thank Garlo Chao for expert technical assistance in isolation of the gene encoding LipL48. We thank R. Hartskeerl for monoclonal antibody F71C2.
Editor: B. B. Finlay
REFERENCES
- 1.Alves, S. F., R. B. Lefebvre, and W. Probert. 2000. Amino acid sequences of proteins from Leptospira serovar pomona. Mem. Inst. Oswaldo Cruz 95:503-504. [DOI] [PubMed] [Google Scholar]
- 2.Auran, N. E., R. C. Johnson, and D. M. Ritzi. 1972. Isolation of the outer sheath of Leptospira and its immunogenic properties in hamsters. Infect. Immun. 5:968-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco, D. R., M. Giladi, C. I. Champion, D. A. Haake, G. K. Chikami, J. N. Miller, and M. A. Lovett. 1991. Identification of Treponema pallidum subspecies pallidum genes encoding signal peptides and membrane-spanning sequences using a novel alkaline phosphatase expression vector. Mol. Microbiol. 5:2405-2415. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, D. R., K. Reimann, J. Skare, C. I. Champion, D. Foley, M. M. Exner, R. E. Hancock, J. N. Miller, and M. A. Lovett. 1994. Isolation of the outer membranes from Treponema pallidum and Treponema vincentii. J. Bacteriol. 176:6088-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bledsoe, H. A., J. A. Carroll, T. R. Whelchel, M. A. Farmer, D. W. Dorward, and F. C. Gherardini. 1994. Isolation and partial characterization of Borrelia burgdorferi inner and outer membranes by using isopycnic centrifugation. J. Bacteriol. 176:7447-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenot, A., D. Trott, I. Saint Girons, and R. Zuerner. 2001. Penicillin-binding proteins in Leptospira interrogans. Antimicrob. Agents Chemother. 45:870-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, J. A., R. B. LeFebvre, and M. J. Pan. 1991. Protein and antigen profiles of prevalent serovars of Leptospira interrogans. Infect. Immun. 59:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coughlin, R. T., S. Tonsager, and E. J. McGroarty. 1983. Quantitation of metal cations bound to membranes and extracted lipopolysaccharide of Escherichia coli. Biochemistry 22:2002-2007. [DOI] [PubMed] [Google Scholar]
- 9.Cullen, P. A., S. J. Cordwell, D. M. Bulach, D. A. Haake, and B. Adler. 2002. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 70:2311-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flannery, B., D. Costa, F. P. Carvalho, H. Guerreiro, J. Matsunaga, E. D. Da Silva, A. G. Ferreira, L. W. Riley, M. G. Reis, D. A. Haake, and A. I. Ko. 2001. Evaluation of recombinant Leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J. Clin. Microbiol. 39:3303-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giladi, M., C. I. Champion, D. A. Haake, D. R. Blanco, J. F. Miller, J. N. Miller, and M. A. Lovett. 1993. Use of the “blue halo” assay in the identification of genes encoding exported proteins with cleavable signal peptides: cloning of a Borrelia burgdorferi plasmid gene with a signal peptide. J. Bacteriol. 175:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Görg, A., W. Postel, and S. Gunther. 1988. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 9:531-546. [DOI] [PubMed] [Google Scholar]
- 13.Görg, A., W. Postel, S. Gunther, and J. Weser. 1985. Improved horizontal two-dimensional electrophoresis with hybrid isoelectric focusing in immobilized pH gradients in the first dimension and laying-on transfer to the second dimension. Electrophoresis 6:599-604. [Google Scholar]
- 14.Guerreiro, H., J. Croda, B. Flannery, M. Mazel, J. Matsunaga, M. Galvao Reis, P. N. Levett, A. I. Ko, and D. A. Haake. 2001. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 69:4958-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haake, D. A., C. I. Champion, C. Martinich, E. S. Shang, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1993. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J. Bacteriol. 175:4225-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haake, D. A., G. Chao, R. L. Zuerner, J. K. Barnett, D. Barnett, M. Mazel, J. Matsunaga, P. N. Levett, and C. A. Bolin. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 68:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haake, D. A., C. Martinich, T. A. Summers, E. S. Shang, J. D. Pruetz, A. M. McCoy, M. K. Mazel, and C. A. Bolin. 1998. Characterization of leptospiral outer membrane lipoprotein LipL36: downregulation associated with late-log-phase growth and mammalian infection. Infect. Immun. 66:1579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haake, D. A., M. K. Mazel, A. M. McCoy, F. Milward, G. Chao, J. Matsunaga, and E. A. Wagar. 1999. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect. Immun. 67:6572-6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haake, D. A., E. M. Walker, D. R. Blanco, C. A. Bolin, M. N. Miller, and M. A. Lovett. 1991. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect. Immun. 59:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrmann, J. L., P. Bakoss, H. Korver, A. A. Bulu, E. Bellenger, W. J. Terpstra, I. Saint Girons, and G. Baranton. 1994. A new serovar in the Grippotyphosa serogroup comprising leptospiral isolates from different regions. Int. J. Syst. Bacteriol. 44:362-364. (Erratum, 44:597, 1994.) [DOI] [PubMed] [Google Scholar]
- 22.Holt, S. C. 1978. Anatomy and chemistry of spirochetes. Microbiol. Rev. 42:114-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, R. C., and V. G. Harris. 1967. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J. Bacteriol. 94:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joseph, R., S. C. Holt, and E. Canale-Parola. 1973. Peptidoglycan of free-living anaerobic spirochetes. J. Bacteriol. 115:426-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kida, H., H. Watanabe, S. Yamamoto, and R. Yanagawa. 1976. Immunological and morphological analysis of sodium dodecyl sulfate extract of Leptospira. Zentbl. Bakteriol. Orig. A 236:328-335. [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson, V. M., and J. F. Prescott. 1993. Outer membrane proteins of three pathogenic Leptospira species. Vet. Microbiol. 36:123-138. [DOI] [PubMed] [Google Scholar]
- 28.Nunes-Edwards, P. L., A. B. Thiermann, P. J. Bassford, Jr., and L. V. Stamm. 1985. Identification and characterization of the protein antigens of Leptospira interrogans serovar hardjo. Infect. Immun. 48:492-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 30.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 247:3962-3972. [PubMed] [Google Scholar]
- 31.Picardeau, M., A. Brenot, and I. Saint Girons. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40:189-199. [DOI] [PubMed] [Google Scholar]
- 32.Radolf, J. D., N. R. Chamberlain, A. Clausell, and M. V. Norgard. 1988. Identification and localization of integral membrane proteins of virulent Treponema pallidum subsp. pallidum by phase partitioning with the nonionic detergent triton X-114. Infect. Immun. 56:490-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radolf, J. D., M. S. Goldberg, K. Bourell, S. I. Baker, J. D. Jones, and M. V. Norgard. 1995. Characterization of outer membranes isolated from Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 63:2154-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radolf, J. D., E. J. Robinson, K. W. Bourell, D. R. Akins, S. F. Porcella, L. M. Weigel, J. D. Jones, and M. V. Norgard. 1995. Characterization of outer membranes isolated from Treponema pallidum, the syphilis spirochete. Infect. Immun. 63:4244-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Shang, E. S., M. M. Exner, T. A. Summers, C. Martinich, C. I. Champion, R. E. Hancock, and D. A. Haake. 1995. The rare outer membrane protein, OmpL1, of pathogenic Leptospira species is a heat-modifiable porin. Infect. Immun. 63:3174-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang, E. S., T. A. Summers, and D. A. Haake. 1996. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 64:2322-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skare, J. T., E. S. Shang, D. M. Foley, D. R. Blanco, C. I. Champion, T. Mirzabekov, Y. Sokolov, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1995. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J. Clin. Investig. 96:2380-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiermann, A. B., R. D. McClellan, and H. T. Hill. 1984. Improved techniques for the isolation of leptospires from swine abortion cases. Annu. Proc. Am. Assoc. Vet. Lab. Diagn. 27:233-244. [Google Scholar]
- 40.Trueba, G. A., C. A. Bolin, and R. L. Zuerner. 1992. Characterization of the periplasmic flagellum proteins of Leptospira interrogans. J. Bacteriol. 174:4761-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaara, M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigel, L. M., J. D. Radolf, and M. V. Norgard. 1994. The 47-kDa major lipoprotein immunogen of Treponema pallidum is a penicillin-binding protein with carboxypeptidase activity. Proc. Natl. Acad. Sci. USA 91:11611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi, K., F. Yu, and M. Inouye. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53:423-432. [DOI] [PubMed] [Google Scholar]
- 44.Yelton, D. B., and N. W. Charon. 1984. Cloning of a gene required for tryptophan biosynthesis from Leptospira biflexa serovar patoc into Escherichia coli. Gene 28:147-152. [DOI] [PubMed] [Google Scholar]
- 45.Zuerner, R. L., W. Knudtson, C. A. Bolin, and G. Trueba. 1991. Characterization of outer membrane and secreted proteins of Leptospira interrogans serovar pomona. Microb. Pathog. 10:311-322. [DOI] [PubMed] [Google Scholar]