Abstract
Screening of 7,680 Salmonella enterica serovar Enteritidis mutants for attenuation in a chicken macrophage infection model yielded a series of mutants including several with defects in previously unrecognized Salmonella virulence genes. One of the newly identified genes was the pbpA2 gene, belonging to the penicillin binding protein gene family.
Salmonella strains pose a major problem in public health, causing diseases ranging from gastroenteritis to typhoid fever. In recent years, Salmonella enterica serovar Enteritidis has replaced serovar Typhimurium as the primary etiologic agent of Salmonella infections in many countries (15). A likely source of serovar Enteritidis is the consumption of infected poultry (6, 15). The molecular mechanisms that enable this serovar to persist in poultry are poorly defined. To address this topic, we generated a mini-Tn10 mutant library of serovar Enteritidis strain CVI-1 and searched for cellular-infection-impaired mutants in a chicken macrophage infection model.
The mutant library was constructed by transforming strain CVI-1 with plasmid pKO3 carrying the mini-Tn10 delivery system from plasmid pLOF/KM (7). Induction of the transposition event eventually yielded 7,680 kanamycin-resistant colonies that were individually collected and tested for their ability to invade or survive in the chicken macrophage HD-11 cell line (5). After several rounds of selection which involved plate counting of the number of intracellular bacteria recovered after 30 min of infection and 2 h of treatment with colistin (100 μg/ml) to kill the extracellular microorganisms, 37 clones (designated SEM1 to SEM37) were identified that consistently yielded reduced (5 to 50% of wild-type) levels of cellular infection.
Genetic characterization of the selected mutants with inverse PCR using outward-oriented transposon primers followed by DNA sequencing of the products revealed the transposon insertion sites for 36 of the 37 mutants (Table 1). Sequence analysis indicated that in 14 strains (38%, group I in Table 1) genes had been disrupted that shared homology with genes involved in the assembly of flagella or bacterial motility in serovar Typhimurium. Analysis of the 37 selected mutants for their behavior in U-shaped tubes containing 0.4% Luria-Bertani soft agar demonstrated that all mutants in group I indeed were nonmotile. Bacterial motility has previously been demonstrated to expedite Salmonella entry into cultured epithelial cells, probably by enabling more rapid contact with the monolayer (13, 21). Fluorescence-activated cell sorting analysis with the serovar Enteritidis strain CVI-1 and the mutant strains made fluorescent by introduction of the plasmid pEGFP, encoding the green fluorescent protein, yielded similar results for chicken macrophages (data not shown), explaining the organisms' poor recovery from the cells.
TABLE 1.
Identification and characterization of serovar Enteritidis mutants impaired in their ability to invade and/or survive in chicken macrophages
| Groupa | Serovar Enteritidis mutant(s) | BLAST identityb (%) | Insertion locus | Putative function |
|---|---|---|---|---|
| I | 7, 14, 15, 16 | 86 X | fliD | Flagellar hook-associated protein 2 |
| 5, 21, 28 | 98 | flhD | Transcriptional activator for motility genes | |
| 33 | 98 | flhC | Transcriptional activator for motility genes | |
| 36 | 97 | fliA | Flagellum-specific promoter | |
| 25 | 98 | fliL | Flagellar structure protein | |
| 41 | 97 | fliN | Flagellar switch protein | |
| 38 | 98 | fliP | Unknown | |
| 43 | 94 | motA | Flagellar motor protein | |
| 37 | 94 | flgI | Flagellar Pring | |
| II | 6 | 94 | invG | SPI-1, type III secretion system |
| 13 | 98 | invI | SPI-1, type III secretion system | |
| 27 | 90 | spaS | SPI-1, type III secretion system | |
| 8 | 95 | rfbI | LPS side chain synthesis | |
| 42 | 91 | rfaI | LPS outer core synthesis | |
| 2 | 97 | rfaJ | LPS outer core synthesis | |
| 1 | 98 | pmrB | Regulator of pmr gene expression | |
| 9, 30 | 92 | pmrF | Lipid A modification | |
| 19, 24 | 99 | phoP | Regulator of virulence genes | |
| 44 | 84 | oxyR | Sensor and regulator of oxidative stress | |
| 12 | 100 | barA | Sensor and regulator | |
| 35 | 97 | pbpA2 | Peptidoglycan synthesis | |
| 10, 11, 17, 23 | 97 | ugd | UDP-glucose dehydrogenase | |
| 20 | 92 | tdh | Threonine dehydrogenase | |
| 31 | 36 | cat2 | 4-Hydroxybutyrate coenzyme A transferase | |
| 3 | 90 | icd | Isocitrate dehydrogenase | |
| 26 | 81 | yegQ | Putative protease precursor | |
| 29c |
Group I are nonmotile mutants; group II are motile mutants.
Percentages of sequence identity were obtained by BLASTN analysis of obtained sequence flanking the mini-Tn10. X indicates that the BLASTX algorithm instead of BLASTN was used.
For mutant 29, only plasmid sequence was obtained, suggesting that in this mutant the vector carrying mini-Tn10 may have been integrated into the genome.
From the group of motile mutants (group II in Table 1), three carried the transposon in homologues of invG, invI, and spaS located in Salmonella pathogenicity island 1 (SPI-1). Six mutants had defects in genes involved in lipopolysaccharide (LPS) biosynthesis (rfaI, rfaJ, and rfbI) or lipid A modification (pmrB and pmrF), four had defects in homologues of global regulatory genes (phoP, oxyR, and barA), one had a defect in a peptidoglycan synthesis gene (pbpA), seven had defects in putative metabolic genes (ugd, tdh, icd, and cat2), and one had a defect in a homologue of the Escherichia coli yegQ gene. For serovar Typhimurium, SPI-1, which encodes a type III secretion system, and LPS have been demonstrated previously to facilitate bacterial invasion of mammalian cells (10) and intracellular survival (8, 11, 12), respectively. Similarly, the regulator proteins PhoP (8), OxyR (18), and BarA (2) contribute to serovar Typhimurium virulence. These data confirm the existence of considerable functional conservation of these pathogenic mechanisms among the serovars Enteritidis and Typhimurium and indicate that these mechanisms are also important in infection of chicken macrophages.
Several of the metabolic genes defective in our mutants have not been previously identified as Salmonella virulence determinants. Apart from the PhoP/PhoQ-regulated ugd (pagA) gene, which in serovar Typhimurium has been demonstrated to be transcriptionally active inside macrophages and to be necessary for growth in a low-magnesium environment (1, 19, 20), two genes were identified with homology to the E. coli tdh gene and the cat2 gene in Clostridium aminobutyricum. These genes encode a threonine dehydrogenase (3) and a 4-hydroxybutyrate coenzyme A transferase (16), respectively. In E. coli, the tdh homologue is regulated by the lrp gene (9), which is one of the regulators of virulence genes in serovar Typhimurium (14). The function of the tdh gene in Salmonella has never been established. An additional previously unrecognized gene important for cellular infection of chicken macrophages was the serovar Enteritidis icd gene, which may encode the enzyme isocitrate dehydrogenase. This gene is considered to have a housekeeping function and appears to be conserved among most Salmonella serovars (22). Together, these results strongly suggest that metabolic adaptation is an important feature in serovar Enteritidis infection of chicken macrophages. Whether the newly identified genes also contribute to virulence of other Salmonella serovars and/or in other hosts awaits further study.
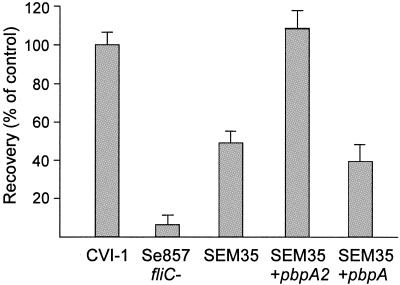
Another gene not previously implicated in Salmonella virulence was the pbpA2 gene defective in mutant SEM35. This gene exhibited approximately 60% homology with E. coli pbpA encoding penicillin binding protein 2 (4). Analysis of the serovar Typhimurium genome sequence indicated that this serovar carries two related pbpA sequences, one (designated pbpA) positioned adjacent to the putative rodA locus, and the other (designated pbpA2) flanking the argS locus. The latter gene was >99% identical in sequence to the gene disrupted in SEM35. To ascertain that the mutant phenotype of SEM35 was caused by disruption of pbpA2, intact copies of both the serovar Enteritidis pbpA and pbpA2 genes were PCR amplified, cloned onto plasmid pWSK29, and introduced into SEM35. Infection assays with HD-11 cells demonstrated that the intact pbpA2 gene completely restored the wild-type phenotype, while the pbpA gene was unable to complement the defect in SEM35 (Fig. 1). In serovar Typhimurium, the promoter of the pbpA2 gene is acid inducible (20), which could point to a role in intracellular survival. Furthermore, the homology of PbpA2 with the members of the penicillin binding protein family may indicate a role in cell wall synthesis and in the resistance against antimicrobial peptides (17). Thus, PbpA2 may contribute to intracellular survival of Salmonella by providing resistance to antimicrobial peptides in a low-pH environment. The finding that the pbpA gene was unable to complement pbpA2 function indicates that these two related genes have different functional properties.
FIG. 1.
Recovery of strains CVI-1 (positive control), Se857fliC mutant (nonflagellated, negative control) (21), SEM35 (pbpA2 mutant), and SEM35 carrying pWSK29::pbpA2 or SEM35 carrying pWSK29::pbpA, from the intracellular compartment of HD-11 macrophages after 30 min of infection and 2 h of colistin treatment. Data are the means ± standard deviations of three experiments.
Thus far, the molecular mechanisms that enable serovar Enteritidis to colonize and infect chicken macrophages are largely unknown and assumed to resemble those of the related serovar Typhimurium. Our experimental work largely confirms this notion, although a number of novel genes involved in cellular infection of chicken macrophages were identified. It can be expected that further detailed analysis of the isolated mutants will shed more light on the possible host, cell type, and/or pathogen specificity of these bacterial traits and their potential as targets for future infection intervention.
Acknowledgments
K. N. Timmis is gratefully acknowledged for providing plasmid mini-Tn10-pLOF/KM.
This work was supported in part by grants from the CVVM and TNO, The Netherlands.
Editor: V. J. DiRita
REFERENCES
- 1.Alpuche-Arande, C. M., J. A. Swanson, W. P. Loomis, and S. I. Miller. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. USA 89:10079-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:635-646. [DOI] [PubMed] [Google Scholar]
- 3.Aronson, B. D., R. L. Somerville, B. R. Epperly, and E. E. Dekker. 1989. The primary structure of Escherichia coli l-threonine dehydrogenase. J. Biol. Chem. 264:5226-5232. [PubMed] [Google Scholar]
- 4.Asoh, S., H. Matsuzawa, F. Ishino, J. L. Strominger, M. Matsuhashi, and T. Ohta. 1986. Nucleotide sequence of the pbpA gene and characteristics of the deduced amino acid sequence of penicillin-binding protein 2 of Escherichia coli K12. Eur. J. Biochem. 160:231-238. [DOI] [PubMed] [Google Scholar]
- 5.Beug, H., A. von Krichbach, G. Doderlein, J. F. Conscience, and T. Graf. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375-390. [DOI] [PubMed] [Google Scholar]
- 6.Cox, J. M. 1995. Salmonella enteritidis: the egg and I. Aust. Vet. J. 72:108-115. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 8.Ernst, R. K., T. Guina, and S. I. Miller. 1999. How intracellular bacteria survive: surface modifications that promote resistance to host innate immune responses. J. Infect. Dis. 179(Suppl. 2):S326-S330. [DOI] [PubMed] [Google Scholar]
- 9.Ernsting, B. R., M. R. Atkinson, A. J. Ninfa, and R. G. Matthews. 1992. Characterization of the regulon controlled by the leucine-responsive regulatory protein in Escherichia coli. J. Bacteriol. 174:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galan, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 13.Khoramian-Falsafi, T., S. Harayama, K. Kutsukake, and J. C. Pechere. 1990. Effect of motility and chemotaxis on the invasion of Salmonella typhimurium into HeLa cells. Microb. Pathog. 9:47-53. [DOI] [PubMed] [Google Scholar]
- 14.Marshall, D. G., B. J. Sheehan, and C. J. Dorman. 1999. A role for the leucine-responsive regulatory protein and integration host factor in the regulation of the Salmonella plasmid virulence (spv) locus in Salmonella typhimurium. Mol. Microbiol. 34:134-145. [DOI] [PubMed] [Google Scholar]
- 15.Rabsch, W., H. Tschäpe, and A. J. Bäumler. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3:237-247. [DOI] [PubMed] [Google Scholar]
- 16.Scherf, U., and W. Buckel. 1991. Purification and properties of 4-hydroxybutyrate coenzyme A transferase from Clostridium aminobutyricum. Appl. Environ. Microbiol. 57:2699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Signoretto, C., F. Di Stefano, and P. Canepari. 1996. Modified peptidoglycan chemical composition in shape-altered Escherichia coli. Microbiology 142:1919-1926. [DOI] [PubMed] [Google Scholar]
- 18.Storz, G., and S. Altuvia. 1994. OxyR regulon. Methods Enzymol. 234:217-223. [DOI] [PubMed] [Google Scholar]
- 19.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 20.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 21.van Asten, F. J., H. G. Hendriks, J. F. Koninkx, B. A. M. Van der Zeijst, and W. Gaastra. 2000. Inactivation of the flagellin gene of Salmonella enterica serotype Enteritidis strongly reduces invasion into differentiated Caco-2 cells. FEMS Microbiol. Lett. 185:175-179. [DOI] [PubMed] [Google Scholar]
- 22.Wang, F. S., T. S. Whittam, and R. K. Selander. 1997. Evolutionary genetics of the isocitrate dehydrogenase gene (icd) in Escherichia coli and Salmonella enterica. J. Bacteriol. 179:6551-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]