Abstract
Streptococcus iniae causes meningoencephalitis and death in cultured fish species and soft-tissue infection in humans. We recently reported that S. iniae is responsible for local tissue necrosis and bacteremia in a murine subcutaneous infection model. The ability to cause bacteremia in this model is associated with a genetic profile unique to strains responsible for disease in fish and humans (J. D. Fuller, D. J. Bast, V. Nizet, D. E. Low, and J. C. S. de Azavedo, Infect. Immun. 69:1994-2000, 2001). S. iniae produces a cytolysin that confers a hemolytic phenotype on blood agar media. In this study, we characterized the genomic region responsible for S. iniae cytolysin production and assessed its contribution to virulence. Transposon (Tn917) mutant libraries of commensal and disease-associated S. iniae strains were generated and screened for loss of hemolytic activity. Analysis of two nonhemolytic mutants identified a chromosomal locus comprising 9 genes with 73% homology to the group A streptococcus (GAS) sag operon for streptolysin S (SLS) biosynthesis. Confirmation that the S. iniae cytolysin is a functional homologue of SLS was achieved by PCR ligation mutagenesis, complementation of an SLS-negative GAS mutant, and use of the SLS inhibitor trypan blue. SLS-negative sagB mutants were compared to their wild-type S. iniae parent strains in the murine model and in human whole-blood killing assays. These studies demonstrated that S. iniae SLS expression is required for local tissue necrosis but does not contribute to the establishment of bacteremia or to resistance to phagocytic clearance.
Streptococcus iniae is a hemolytic, gram-positive coccus that causes meningoencephalitis and significant mortality in a variety of commercial fish species, creating a substantial economic burden for the aquaculture industry (12, 13, 41, 42). S. iniae also causes fulminant soft-tissue infection in humans, with associated bacteremia, following percutaneous injury acquired while handling whole fish (48). Although our understanding of the pathogenicity of this organism is poor, pulsed-field gel electrophoresis (PFGE) has revealed that strains causing disease in fish and strains causing disease in humans are clonally related, whereas commensal isolates from nondiseased fish are genetically diverse (48). We recently reported that this unique genetic profile was associated with virulence in experimental model systems. Disease-associated strains caused significant weight loss and bacteremia in a murine model of subcutaneous infection, exhibited resistance to phagocytic clearance in human whole blood, and were cytotoxic to human endothelial cells in vitro (19). From these findings we hypothesized that this unique genetic profile indicated the presence of one or more virulence determinants important to disease.
One of the few distinguishing phenotypes of S. iniae is the zone of beta-hemolysis surrounding colonies cultured on blood agar (42). Hemolysins, or cytolysins, are well-recognized features of many bacterial species, including streptococci, and are generally associated with damage to the membranes of a variety of mammalian cell types. Examples of streptococcal cytolysins include streptolysin O (SLO) and streptolysin S (SLS) of group A streptococci (GAS), the beta-hemolysin/cytolysin of group B streptococci (GBS), and the pneumolysin of Streptococcus pneumoniae, each of which has been implicated as a virulence factor in animal models of infection (3, 4, 33, 49). In this investigation, we describe a molecular approach used to identify the genetic locus responsible for cytolysin production in S. iniae. The contribution of the S. iniae cytolysin to disease pathogenesis was examined by using a murine model of subcutaneous infection.
MATERIALS AND METHODS
Bacterial strains, media, and transformation.
Bacterial strains used in this study are listed in Table 1. S. iniae and other gram-positive bacterial strains were grown in Todd-Hewitt broth (THB) or on Columbia base agar (Difco, Detroit, Mich.) supplemented with 5% defibrinated sheep erythrocytes (SBA) (Quelab). Mid-log-phase cells were prepared from overnight cultures that were diluted 1/25 in THB and incubated at 37°C for 3 to 6 h. An optical density measurement of 0.35 to 0.40 at 620 nm, by use of a spectrophotometer (Beckman Instruments, Fullerton, Calif.), corresponded to mid-log-phase growth (108 CFU). For anaerobic conditions, cultures were incubated in anaerobic jars with an AnaeroGen pack (Oxoid). Escherichia coli was grown in Luria-Bertani (LB) broth or on LB agar (Difco). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; erythromycin (ERY), 5 (for GAS) or 300 (for E. coli MC1061) μg/ml; chloramphenicol (CHL), 2 to 4 (for GAS) or 2.5 (for S. iniae) μg/ml. Gram-positive strains were rendered competent for transformation by growth in THB plus 0.6% glycine as described for GBS (18). E. coli MC1061 was rendered competent by standard methods (11), and E. coli TOP10 competent cells were obtained from Invitrogen Life Technologies. Plasmids were introduced by electroporation (at 1,500 V; Eppendorf model 2510) and immediately transferred into 1 ml of recovery medium (for GAS, THB plus 0.25 M sucrose; for E. coli, SOC) for 1 to 3 h, at permissive temperatures, prior to antibiotic selection on agar media. Genomic and plasmid DNAs were extracted by previously described methods (5, 18, 46).
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Description | Reference or source |
|---|---|---|
| Bacterial strains | ||
| S. iniae | ||
| 9117 | Disease-associated blood culture isolate from patient with cellulitis | 19 |
| 9066 | Commensal isolate from surface of healthy fish | 19 |
| 9174 | Commensal isolate from surface of healthy fish | This study |
| SI sagB.KO.erm | Nonhemolytic insertion/deletion mutant of 9117 | This study |
| SI sagB.KO.Tn917 | Nonhemolytic Tn917 mutant of 9174 | This study |
| GAS | ||
| NZ131 | T14/M49 clinical isolate from patient with glomerulonephritis; SLS+ | 44 |
| NZ131 sagAΔcat | Nonhemolytic allelic replacement mutant of NZ131 | Datta et al., Abstr. 6th Ann. Streptococcal Genetics Conf. abstr. 132 |
| E. coli | ||
| MC1061 | F−araD139 Δ(araABC-leu)7696 ΔlacX74 galU galK hsdR2(rK− mK+) mcrB1 rpsL (Strr) | 50 |
| TOP10 | F−mcA Δ(mrr-hsd RMS-mcrBC) φ80 dlacZ ΔM15 Δ(lacZ)X74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr)end A1 nup G | 22 |
| Plasmids | ||
| pTV1OK | repA(Ts)-pWV01(Ts) aphA3 Tn917 (erm) | 23 |
| pUC19 | lacZα bla | Pharmacia |
| pVE6007Δ | Temperature-sensitive derivative of pWV01; Cmr | 35 |
| pCR2.1-TOPO | ColE ori Ampr KmrlacZα | Invitrogen |
| pSIsagB.KO.erm | pVE6007Δ + 2,782-bp ermB allelic replacement construct | This study |
| pDC123 | E. coli/streptococcal shuttle vector, JS-3 replicon; Cmr | 8 |
| pSIsagA | pDC123 + 196-bp PCR amplicon of S. iniae sagA | This study |
Primers and DNA sequencing.
Oligonucleotide primers are listed in Table 2. DNA sequences were obtained by using an automated sequencer (ABI Prism; Applied Biosystems, Oakville, Ontario, Canada) at the Center for Applied Genomics, DNA Sequencing Facility, Hospital for Sick Children (Toronto, Ontario, Canada). The S. iniae sequence was compared with those in the GenBank databases by using the BLAST local alignment program of the National Center for Biotechnology Information, National Institutes of Health (1), and was aligned and annotated by using Vector NTI (InforMax, Inc.) software.
TABLE 2.
Primer list
| Primer | Sequence | Description |
|---|---|---|
| ermB F | 5′ TGCGTCTGACATCTATCTGATTG 3′ | Tn917 left, ermB intragenic primer |
| ermB R | 5′ TTATCTGGAACATCTGTGGTATGG 3′ | Tn917 left, ermB intragenic primer |
| ltp-1 | 5′ AGAGAGATGTCACCGTCAAG 3′ | Tn917 left, outward-reading primer |
| ltp-2 | 5′ AATGTACAAAATAACAGCGAA 3′ | Tn917 left, outward-reading primer |
| rtp-1 | 5′ CTAAACACTTAAGAGAATTG 3′ | Tn917 right, outward-reading primer |
| rtp-2 | 5′ TAGGCCTTGAAACATTGGTT 3′ | Tn917 right, outward-reading primer |
| p1 | 5′ TGATTTCGTAGTCGCACCTCCG 3′ | psagB.KO.erm primer |
| p2-ascI | 5′ GGCGCGCCCCCGCTTCATCTTGGCTGATTGAC 3′a | psagB.KO.erm primer |
| Erm-PA | 5′ GGCGCGCCCCGGGCCCAAAATTTGTTTGAT 3′a | psagB.KO.erm primer |
| Erm-PB | 5′ GGCCGGCCAGTCGGCAGCGACTCATAGAAT 3′b | psagB.KO.erm primer |
| p3-ecoRI | 5′ GGCCGGCCCACATTAGTAGGAAACAAGGAGTCG 3′b | psagB.KO.erm primer |
| p4 | 5′ TAACTGGTTGCGTCGTTGG 3′ | psagB.KO.erm primer |
| SI sagA 1156 | 5′ ATTTGTATAAGGAGGTAAGC 3′ | S. iniae sagA primer |
| SI sagA 1352 | 5′ AGTGAATTACTTTGGAGCTG 3′ | S. iniae sagA primer |
| SK+ | 5′ GCTCTAGAACTAGTGG 3′ | pVE6007Δ primer |
| T7 | 5′ TAATACGACTCACTATAGGG 3′ | pVE6007Δ primer |
| sagA 1530 | 5′ ATGTTAAAATTTACTTC 3′ | GAS sagA primer |
| sagA 1688 | 5′ TTTACCTGGCGTATAACTTC 3′ | GAS sagA primer |
Underlining indicates AscI site.
Underlining indicates EcoRI site.
Tn917 mutagenesis and identification of transposon insertion sites.
Insertional mutagenesis of S. iniae strains 9117 and 9174 was performed as described for Streptococcus mutans by using the temperature-sensitive plasmid pTV1OK, which harbors the conjugative transposon Tn917 (23). Insertional libraries were screened on SBA for nonhemolytic (NH) transconjugants, and the genomic junction sites of Tn917 insertions were identified as follows. The junction site containing the transposon-associated ermB gene in SINH3, derived from the disease-associated strain 9117, was cloned as a HindIII fragment in pUC19, transformed into E. coli MC1061, selected on ERY, and sequenced by using the ermB R and M13 universal primers. DNA from the insertion sites of the mutants SINH1, derived from the disease-associated strain 9117, and SI sagB.KO.Tn917, derived from the commensal strain 9174, was isolated by using a novel PCR-based method described by Karlyshev and colleagues (26). Transposon-specific, outward-reading primers ltp-1 and rtp-1 were used for the single-primer PCR amplification of up- and downstream Tn917-junction sites, respectively, from the genomic DNA template. Reactions utilized Taq DNA polymerase (Invitrogen Life Technologies) under the following cycling conditions: 2 min at 94°C; 20 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 3 min; 30 cycles of 94°C for 30 s, 45°C for 30 s, and 72° for 2 min; 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 2 min; and 72°C for 7 min. Unique amplicons were gel purified with the QIAEX gel extraction kit (Qiagen) and sequenced by using the corresponding nested outward-reading primers, ltp-2 and rtp-2.
Southern localization of Tn917-junction fragments.
Genomic DNA (∼1 μg) was digested with HindIII, resolved by 0.6% Tris-borate-EDTA agarose gel electrophoresis, and transferred to a Nytran membrane. DNA probes, generated by PCR, were labeled by using the ECL direct nucleic acid labeling and detection system (Amersham Pharmacia Biotech, Quebec, Canada), and positive bands were visualized by chemiluminesence. SagA through SagHI probes were amplified from GAS NZ131 genomic DNA by using primers sagA 1530 and sagA 1688 (Table 2), as well as the sagB, -C, -D, -E, -F, -G, and -HI primer pairs described previously (38).
Targeted mutagenesis of S. iniae sagB.
Targeted mutagenesis of S. iniae sagB was based on a recently described PCR ligation mutagenesis technique, which promotes insertion of heterologous DNA and deletion of target DNA (30). The chromosomal regions upstream (P12) and downstream (P34) of S. iniae sagB were amplified and ligated to the ERY resistance gene ermB (9) via AscI and EcoRI restriction endonuclease sites, respectively, incorporated onto the 5′ ends of appropriate primers (Table 2). P12 (946 bp) was amplified by using a primer 630 bp upstream of S. iniae sagA (p1) and 65 bp downstream of S. iniae sagB (p2-ascI). Likewise, P34 (1,049 bp) was amplified by using a primer pair 23 bp upstream of the S. iniae sagB stop site (p3-ecoRI) and 807 bp downstream of the S. iniae sagC start codon (p4). The ermB product was amplified from the gene cassette (9) template and primers Erm-PA and Erm-PB (30), which have been described previously (Table 2). All three products were digested with AscI and/or EcoRI and ligated. The 2,782-bp construct (P12::erm::P34) was amplified from the ligation mixture by using primers p1 and p4 and was cloned into the TA vector pCR2.1-TOPO. The insert was directionally cloned (XhoI/HindIII) into the temperature-sensitive shuttle vector pVE6007Δ, and the resultant plasmid, pSIsagB.KO.erm, was transformed into S. iniae 9117. CHL-resistant transformants were identified at the permissive temperature for plasmid replication (30°C). Single-crossover Campbell-type genomic insertions were selected by incubation at a nonpermissive temperature (37°C) while maintaining CHL selection. Selection was relaxed by serial passage at 30°C without antibiotics, and double-crossover events were identified by negative CHL selection (CHL-susceptible) in broth and the NH phenotype on SBA. Genomic DNA was examined for ermB, pVE6007Δ, and sagB by PCR and Southern hybridization. During mutant selection, it was apparent that SI sagB.KO.erm was sensitive to ERY selection. Sequencing of ermB from plasmid pSIsagB.KO.erm revealed a nucleotide deletion, downstream of the start site, which had created a premature TAA stop codon.
Complementation of SLS-negative GAS.
By using primers SI sagA 1156 and SI sagA 1352, the entire S. iniae sagA locus, exclusive of the putative promoter, was amplified from S. iniae 9117 genomic DNA and TA-cloned into pCR2.1-TOPO. The insert was digested (EcoRV/BamHI) and directionally cloned into the pDC123 shuttle vector, in the same orientation as the upstream cat promoter. The resultant construct, pSIsagA, was transformed into competent NZ131 sagAΔcat. NZ131 sagAΔcat is an NH, CHL resistant mutant of GAS strain NZ131 and harbors a precise, in-frame allelic replacement of the GAS sagA gene with the CHL acetyltransferase (cat) gene. This mutant was constructed by the same methodology described for mutagenesis of the Streptococcus agalactiae beta-hemolysin (43) with one exception: the temperature-sensitive targeting vector contained a GAS sag locus sequence where sagA was replaced by cat through in vivo PCR recombination. The nonpolarity of this mutant was confirmed by single-gene complementation to full wild-type SLS activity when GAS sagA was returned on a plasmid vector (V. Datta, R. G. Kansal, and V. Nizet, Abstr. 6th Ann. Streptococcal Genetics Conf. 2002, abstr. 132, p. 109, 2002).
Transformation of GAS strain NZ131 (wild type) and NZ131 sagAΔcat with pDC123 alone provided appropriate experimental controls. Transformants were selected on SBA containing CHL (4 μg/ml), incubated both aerobically and anaerobically, and screened for a restored beta-hemolytic phenotype. Anaerobic incubation prevented alpha-hemolysis and potential misinterpretation of SLS-negative control strains. Plasmid DNA recovered from beta-hemolytic isolates was analyzed by PCR using primers SI sagA 1156 and SI sagA 1352.
Murine model of subcutaneous infection.
The virulence of S. iniae strains was assessed by using a murine model of subcutaneous inoculation as previously described for GAS (4, 19). Mid-log-phase cultures were washed twice in phosphate-buffered saline (PBS), and 106 CFU was mixed with an equal volume of sterilized Cytodex beads (Sigma Chemical Co., St. Louis, Mo.) that were suspended in PBS to a concentration of 20 μg/ml. This mixture (200 μl) was injected into the right flanks of five to seven hairless, outbred female SKH1 mice (Charles River Laboratories, Wilmington, Mass.) aged 4 to 5 weeks, weighing 15 to 20 g. Mice were weighed prior to injection and every 24 h for 3 days. Blood was collected from a single mouse at 24 h and from the remaining mice at the end of each trial (72 h) by cardiac puncture and was mixed with citrate-buffered saline. Representative tissue samples at the injection site were removed from mice and homogenized in PBS. Viable counts were determined from both sample types. The experimental procedures performed on the mice were conducted according to the principles of the Animal Care Committee of Mount Sinai Hospital, Toronto, Ontario, Canada.
Phagocytosis assay.
Resistance to phagocytosis in whole blood was examined by using a modified protocol of the Lancefield bactericidal assay for GAS (29). Mid-log-phase bacteria (108 CFU) were washed and serially diluted in PBS. Bacterial suspensions (100 μl) containing 102 to 104 CFU were added to 1 ml of fresh, heparinized human blood in sterile glass tubes and incubated on an orbital shaker for 1 h at 37°C. The survival index was calculated as the CFU recovered after 1 h of incubation divided by the initial inoculum added prior to incubation.
Hemolytic activity assay.
The nature of S. iniae cytolysin activity was examined using a hemolysis assay based on the method previously described for SLS activity in GAS (4). Whole-cell and supernatant samples were collected from 1 ml of mid-log-phase cultures by centrifugation. Whole-cell fractions were washed three times and resuspended in 1 ml of PBS. A 1% sheep erythrocyte solution was prepared in PBS by using erythrocytes washed three times in the same buffer. Cell and supernatant samples (200 μl) were serially diluted across a 96-well round-bottom microtiter plate in THB, and 100 μl of sheep erythrocytes was added to each well. Plates were incubated for 1 h and centrifuged to pellet both erythrocytes and bacteria, and 100-μl aliquots of supernatant were transferred to a replica plate. The A450 was measured to determine the release of hemoglobin. Erythrocytes suspended in PBS plus 0.1% sodium dodecyl sulfate (complete lysis) or PBS alone (no lysis) were used as controls. Trypan blue (Sigma) at a final concentration of 13 μg/ml was used as an inhibitor of SLS activity.
Nucleotide sequence accession number.
The nucleotide sequence data for the S. iniae sag gene cluster has been submitted to the DDBJ/EMBL/GenBank databases and was assigned accession number AF465842.
RESULTS
Cytolysin expression in S. iniae.
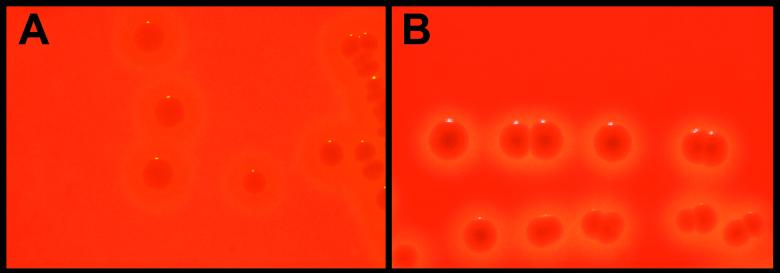
Commensal and disease-associated strains were cultured on SBA under aerobic and anaerobic conditions. Regardless of oxygen tension, all 32 commensal strains exhibited pronounced zones of beta-hemolysis surrounding individual colonies. However, comparable zones of beta-hemolysis were observed with disease-associated strains (21) only when they were cultured anaerobically. Under aerobic conditions, disease-associated strains were largely alpha-hemolytic, with a weak zone of beta-hemolysis surrounding the colonies. Incubation beyond 24 h yielded a thin halo of beta-hemolysis between the colony and the zone of alpha-hemolysis, similar to the pattern originally described by Pier and Madin (42) (Fig. 1). Cytolytic activity, examined by using a hemolysis assay, was found to be associated with whole-cell preparations and could be inhibited by addition of the dye trypan blue (13 μg/ml), indicating SLS activity. Hemolytic activity was not observed in supernatants (data not shown).
FIG. 1.
Photograph depicting the hemolytic phenotypes exhibited by the disease-associated strain 9117 (A) and the commensal strain 9066 (B) of S. iniae cultured on SBA under aerobic conditions. Panel B is representative of the hemolysis exhibited by both disease-associated and commensal strains cultured anaerobically.
Tn917 mutagenesis and genomic sequence recovery.
To identify the genomic region responsible for hemolysin expression in S. iniae, the single Tn917 insertion sites in two NH mutants, SINH1 and SINH3, were mapped by Southern blot analysis. In SINH3, a 405-bp cloned S. iniae DNA fragment representing the transposon insertion site shared significant nucleotide similarity with the junction of the sagG and sagH genes from the SLS biosynthetic operon in GAS (Fig. 2). For SINH1, single-primer PCR analysis of flanking DNA revealed sequences with homology to the complete GAS sagA gene and part of the sagB gene, localizing the Tn917 insertion to the region immediately downstream of the start codon of the homologue (Fig. 2).
FIG. 2.
Map of S. iniae SLS-associated gene cluster. Tn917 insertion points of characterized mutants (SINHI, SINH3, and SI sagB.KO.Tn917), and the putative promoter (P) and rho-independent terminator (T) regions, are indicated.
Characterization of the sag-like gene cluster in S. iniae.
Southern hybridization with GAS-derived sag probes against HindIII-digested S. iniae genomic DNA indicated the presence of sequences in S. iniae homologous to each of the nine genes in the GAS sag operon. Additionally, the S. iniae genes appeared to be placed in the same genetic order as those in GAS (data not shown). The genomic DNA between the S. iniae sagB and sagG open reading frames (ORFs) (∼4.5 kb) was amplified and sequenced through successive chromosomal walking steps. Sequence downstream of the S. iniae sagG ORF was obtained from a bacteriophage λ clone, generated in a concurrent study (C. Duncan, unpublished data) using the Lambda FIX II/XhoI Partial Fill-In Vector kit (Stratagene, La Jolla, Calif.), as identified by Southern hybridization with an S. iniae sagG probe. Oligonucleotide primers designed specifically for each of the nine sag ORFs of the disease-associated S. iniae strain 9117 yielded products of expected sizes from the representative commensal strain 9066. This finding indicated that conservation of the sag-like gene cluster in S. iniae was independent of the PFGE genetic profile associated with the capacity of the organism to produce invasive disease (48).
Sequence analysis of the S. iniae sag-like gene cluster.
Table 3 lists the ORF sizes and homology with each of the corresponding GAS sag genes. The S. iniae sagA ORF encoded a predicted 54-amino-acid (aa) peptide exhibiting 73% sequence identity with SagA of GAS, which contains several features reminiscent of a bacteriocin prepropeptide (Fig. 3) (38). Conserved features of the two predicted SagA proteins include a putative leader peptide with a glycine-glycine cleavage site and a number of target residues (cysteine, serine, and glycine) in the proposed propeptide, which may serve as targets for posttranslational modification (Fig. 3).
TABLE 3.
Predicted Sag proteins in S. iniae
| Gene | Protein length (aa) | % Identity (% similarity) to GAS Sag products |
|---|---|---|
| sagA | 54 | 73 (80) |
| sagB | 316 | 77 (85) |
| sagC | 354 | 78 (87) |
| sagD | 453 | 82 (87) |
| sagE | 220 | 60 (75) |
| sagF | 229 | 52 (70) |
| sagG | 307 | 75 (84) |
| sagH | 375 | 81 (89) |
| sagI | 372 | 74 (84) |
FIG. 3.
Amino acid sequence similarity between SagA proteins of S. iniae and GAS. Putative Gly-Gly cleavage sites are underlined, and residues thought to undergo posttranslational modification are shaded.
The S. iniae sagB ORF encoded a predicted 316-aa product with 77% identity to SagB of GAS, and both of these products shared weak homology with the cytoplasmic modifying enzyme McbC of E. coli (20). McbC is involved in the posttranslational modification of McbA, to produce the mature bacteriocin microcin B17, and uses flavin mononucleotide (FMN) as a cofactor (36). A conserved domain in the SagB ORF of S. iniae (positions 125 to 298) was identified by using the conserved domain database (RPS-BLAST) in GenBank. By use of default parameters, the CDD search tool (GenBank) aligned 97% of this region to a group of nitroreductases (flavoproteins) that bind FMN noncovalently (Fig. 4). The NADH oxidase (NO) of Thermus thermophilus and the nitroreductase (NR) of Enterobacter cloacae align most closely with S. iniae SagB (7, 40). Several residues, Ser46, Pro156, Leu158, and Gly159 of T. thermophilus, are directly associated with FMN binding (24) and, along with neighboring residues, are relatively conserved in this family of flavoproteins (47). These putative FMN-binding regions are also relatively conserved in the S. iniae and GAS SagB proteins and E. coli McbC, specifically Ser46 and Gly159 residues (Fig. 4).
FIG. 4.
Amino acid alignment of putative FMN-binding regions of bacterial flavoproteins and SagB. Sequences are derived from the NR of E. cloacae, NO of T. thermophilus, and bacteriocin-processing enzymes McbC and EpiD from E. coli and S. epidermidis, respectively. Asterisks indicate conserved residues. Residues directly bound to FMN in T. thermophilus NO are underlined. Boldfaced residues represent putative FMN-binding regions.
The sagC, sagD, sagE, and sagF ORFs of S. iniae encoded predicted products with significant amino acid identity to corresponding sag products of GAS yet shared no homology with other proteins in the GenBank database. As reported for GAS, the SagG, SagH, and SagI predicted proteins of S. iniae exhibited homology to the components of a family of ATP-binding cassette (ABC)-type transport systems. These products showed the greatest homology (47, 22, and 23% amino acid identity, respectively) to the ABC transport system expressed by Clostridium acetobutylicum (39). By use of the CDD search tool (RPS-BLAST) in GenBank, 58% (positions 30 to 210) of the predicted SagG-like protein structure was found to be completely conserved with the ATPase component of the ABC transporter protein family mentioned above. Inverted-repeat stem sequences consistent with rho-independent terminators were found between the sagA and sagB ORFs and immediately downstream of the sagI ORF of S. iniae (Fig. 2), similar to those reported for GAS (38).
Targeted mutagenesis of the S. iniae sagB locus.
Each of the nine sag operon genes appears to be required for SLS expression in GAS (38). We chose targeted mutagenesis of S. iniae sagB to confirm the role of the S. iniae sag locus in SLS expression and to accurately assess the role of SLS in S. iniae virulence. A plasmid was constructed for deletion of S. iniae sagB and insertion of ermB, by double-crossover recombination, using the temperature-sensitive vector pVE6007Δ. The plasmid, pSIsagB.KO.erm, was transformed into S. iniae 9117, and cultures were monitored for evidence of plasmid integration (i.e., isolates growing on CHL at the nonpermissive temperature for pVE6007Δ). Single-crossover events were identified by PCR using ermB and pVE6007Δ primers (SK+ and T7). Insertional mutants were continually subcultured at permissive temperatures to enable pVE6007Δ excision from the chromosome during homologous recombination. Following ∼6 subcultures, a CHL-sensitive, NH isolate was recovered. PCR indicated that the mutant, SI sagB.KO.erm, had retained the ermB gene but had lost the pVE6007Δ vector backbone. The ermB probe hybridized with the expected 6.0-kb region in both single- and double-crossover mutants (Fig. 5A). Conversely, pVE6007Δ hybridized with the expected 6.0-kb region in the single-crossover mutant only, indicating pVE6007Δ excision from SI sagB.KO.erm. Moreover, only the single-crossover mutant hybridized with a probe generated from S. iniae sagB (Fig. 5C). Collectively, these results indicated that the loss of the hemolytic phenotype in SI sagB.KO.erm could be attributed to the replacement of S. iniae sagB by ermB.
FIG. 5.
Southern blot analysis of pSIsagB.KO.erm recombination into the S. iniae 9117 genome. Each lane represents HindIII-digested genomic or plasmid DNA. Lanes: 1, wild type; 2, single-crossover mutant; M, marker; 3, double-crossover mutant; 4, pVE6007Δ (control); 5, pSIsagB.KO.erm (control). Probes were ermB (A), pVE6007Δ (B), and sagB from S. iniae (C). Molecular sizes (in kilobases) are indicated to the left of each image.
Restoration of hemolysis in SLS-negative GAS by use of the S. iniae sagA locus.
To determine whether the S. iniae sagA gene encodes a functional homologue of the putative GAS SLS precursor, the NH GAS allelic replacement mutant, NZ131 sagAΔcat, was transformed with S. iniae sagA (pSIsagA). Transformants exhibited a restored beta-hemolytic phenotype whether grown aerobically or anaerobically (Fig. 6), while the mutant strain transformed with the vector alone remained NH. The extent of beta-hemolysis exhibited by the complemented mutant NZ131 sagAΔcat(pSIsagA) was comparable to that observed for the wild-type GAS strain, NZ131, transformed with the vector alone. Thus, the sagA gene of S. iniae was sufficient to restore the SLS phenotype in a sagA mutant of GAS.
FIG. 6.
Photograph illustrating restoration of the SLS phenotype in the NH GAS mutant NZ131 sagAΔcat (left) following complementation with pSIsagA, harboring the sagA ORF from the disease-associated S. iniae strain 9117 (right). Cultures were incubated anaerobically at 37°C.
Virulence of SI sagB.KO.erm in a murine model.
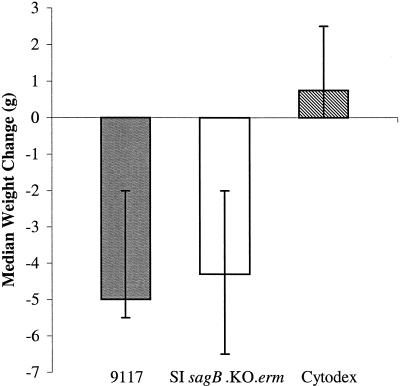
Mice infected with 106 CFU of SI sagB.KO.erm showed signs of lethargy and wasting similar to those of mice infected with the wild type at 24 h postinoculation. Similarly, a median weight loss of 4.3 g (range, 2.0 to 6.5 g), comparable to that of mice infected with the wild type (5.0 g; range, 2.0 to 5.5 g), was noted (Fig. 7). Mice infected with SI sagB.KO.erm (106 CFU) maintained significant bacteremia over the 72-h trial. One mouse, euthanatized at 24 h postinoculation, harbored 105 CFU/ml in the blood, and 104 to 105 CFU/ml was recovered from the blood of the remaining six mice at 72 h. Similar levels of bacteria were recovered from mice infected with the wild-type strain 9117. SI sagB.KO.erm was capable of causing bacteremia (105 CFU/ml) from an inoculum as low as 102 CFU, as previously reported for strain 9117 (19). Bacteria isolated from the blood of SI sagB.KO.erm-infected mice remained NH under anaerobic culture conditions, confirming the integrity of the mutant phenotype.
FIG. 7.
Weight change observed in hairless female mice 72 h after subcutaneous inoculation with strains of S. iniae. Bars represent median weight gains ± ranges. Mice infected with the disease-associated S. iniae strain 9117 (shaded bar) or the sagB mutant (open bar) experienced significant weight loss, in contrast to a weight gain observed for mice inoculated with sterile Cytodex and no bacteria (striped bar) (0.75 g; range, 0.0 to 2.5 g) (P < 0.005 by the Wilcoxon signed-rank test).
Although the commensal strain 9066 does not induce invasive disease in mice, even at an inoculum as high as 108 CFU (19), it does give rise to a localized necrotic lesion at the site of inoculation, as does the disease-associated strain 9117. However, with the latter, mortality due to bacteremia occurs rapidly and progression of the necrotic lesion cannot be monitored. In order to examine the role of SLS in necrotic-lesion formation, we used the NH commensal mutant SI sagB.KO.Tn917, which contains a Tn917 insertion within the sagB ORF, in the murine model. Mice were inoculated either with 108 CFU of SI sagB.KO.Tn917 or with one of the wild-type strains 9174 and 9066. Strain 9174 caused necrotic lesions in 60% (3 of 5) of mice, compared to 100% (5 of 5) necrotic-lesion formation in 9066-infected mice. In contrast, SI sagB.KO.Tn917 did not cause necrotic lesions in mice (n = 5). Mice inoculated with 9066, 9174, or SI sagB.KO.Tn917 all experienced weight gains, with medians of 1.0 g (range, 0.0 to 2.5 g), 1.25 g (range, 1.0 to 3.0 g) and 0.25 g (range, 0.0 to 1.0 g), respectively, as did those inoculated with Cytodex and no bacteria (1.0 g; range, 0.0 to 2.5 g) (P = 0.95 by the Wilcoxon signed-rank test). None of these mice developed bacteremia. Culture of the inoculation site from SI sagB.KO.Tn917-inoculated mice yielded NH colonies on SBA with or without ERY, confirming the stability of the mutant in vivo.
Effect of SLS expression on S. iniae resistance to phagocytosis.
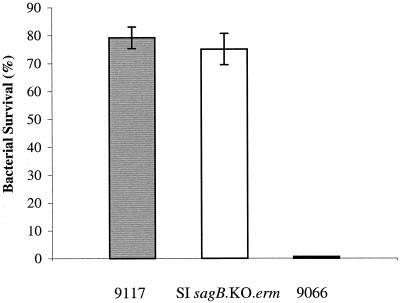
Disease-associated strains of S. iniae have been found to be highly resistant to killing by human blood, in contrast to the marked susceptibility of commensal isolates (19). Following exposure of the NH mutant SI sagB.KO.erm to whole human blood, the number of viable CFU recovered (percent survival) was comparable to that for the wild-type disease-associated strain 9117. As shown in Fig. 8, 75% of the SI sagB.KO.erm inoculum was recovered from blood, in comparison to 79% recovery of 9117. In contrast, the commensal isolate 9066 was completely susceptible to whole-blood killing, as previously described (19).
FIG. 8.
Resistance to phagocytosis of the disease-associated S. iniae strain 9117 (shaded bar), the sagB mutant (open bar), and the commensal strain 9066 (solid bar) in whole blood. Each bar represents the percentage of viable organisms relative to the initial inoculum (100%) remaining after 1.0 h of rotation in fresh human blood. Results are means ± standard deviations.
DISCUSSION
Disease-associated strains of S. iniae, represented by a unique genetic profile, cause soft-tissue infection in humans and are able to sustain invasive disease in a murine model of soft-tissue infection (19). However, the precise bacterial factors contributing to disease have not been elucidated. We hypothesized that the S. iniae cytolysin, responsible for its hemolytic phenotype, may represent one such virulence determinant. Insertional mutagenesis of a disease-associated strain of S. iniae and subsequent sequencing steps led to the discovery that S. iniae cytolytic activity is encoded by a gene cluster that shares significant genetic similarities with the 9-gene GAS sag operon for SLS biosynthesis (38). The first gene of this operon, sagA, encodes a putative bacteriocin-like prepropeptide with sequence similarity to the microcin B17 precursor (McbA) from E. coli. Immunological studies have recently provided convincing evidence that sagA is the structural gene encoding SLS. Antibodies raised against a synthetic peptide of SagA effectively abolished the cytolytic activity of SLS (10). The remaining gene products of the operon are contiguously aligned (sagB to sagI) and are considered important for the processing and transport of SagA. In addition, while the GAS sag operon is both necessary and sufficient for SLS production, there is evidence that sagA may also possess a regulatory function affecting the expression of SLS and other GAS virulence factors through a complex mechanism (6, 32, 34).
The discovery of an SLS homologue in S. iniae adds to a small list of streptococcal pathogens harboring a version of this potent exotoxin. Human large-colony, beta-hemolytic group C and group G streptococci, belonging to Streptococcus equisimilis subsp. dysgalactiae, were recently shown to possess sag-like genetic loci for SLS biosynthesis (25). Streptococcus equi, the cause of strangles in horses, produces a beta-hemolysin with SLS-like characteristics (15). SLS is distinct from the other GAS hemolysin, SLO, a cholesterol-dependent, oxygen-labile cytolysin encoded by a single gene (21, 27), and the beta-hemolysin/cytolysin of GBS, encoded by the cylE gene (43, 45).
Comparative analysis between the GAS and S. iniae sag clusters revealed significant amino acid similarity and an identical gene arrangement. Additional features shared with GAS were a putative promoter region upstream of the sagA ORF, ribosomal binding sites and translational stop codons for each gene, and two noncoding regions that harbor putative rho-independent terminators, located between the sagA and sagB ORFs and downstream of the sagI ORF (38). Residues that are considered important for SagA maturation in GAS are conserved in S. iniae. The Gly-Gly sequence is thought to precede a cleavage site that would remove the putative leader sequence and yield a product equivalent to the predicted size of SLS in GAS (38). The propeptide sequence of S. iniae also contains a high number of residues (8 cysteines, 3 serines, and 5 glycines) that are considered precursors for posttranslational modification in GAS SagA (38). Complementation and trypan blue inhibition studies demonstrated the functional relatedness of the sagA gene products. When the sagA locus of S. iniae was cloned and transformed into a sagA allelic replacement mutant of GAS, cytolytic activity was restored. Therefore, these findings demonstrate that, in addition to conserved amino acid sequence, the biological activities of the SagA peptides are similar and that the additional proteins required for processing and/or transport of SLS do not discriminate between these two SagA peptides.
The SagB product of S. iniae contains a protein domain conserved among a group of bacterial flavoproteins, most notably the NR of E. cloacae and the NO of T. thermophilus (7, 40). Furthermore, the FMN-binding regions of T. thermophilus NO (24) are potentially conserved in other flavoproteins (47) as well as SagB. The amino acid sequences of the SagB products also share weak homology with McbC of E. coli (20). In conjunction with McbB and McbD, McbC forms the microcin B17 synthetase that directs the chemical modification of McbA prepropeptide amino acids (31). Interestingly, preliminary studies have suggested that McbC is a flavoprotein in that recombinant protein is associated with stoichiometric quantities of FMN (36). The bacteriocin epidermin, produced by Staphylococcus epidermidis, also requires the enzymatic action of a flavoprotein, EpiD, for posttranslational modification (28). In addition, at least one of the conserved FMN-binding motifs mentioned above is present in both McbC and EpiD. These observations, in combination with the predicted cytoplasmic localization of SagB (38), support the hypothesis that these conserved regions in SagB are involved in FMN binding and that SagB contributes to the chemical modification of SagA. Liu et al. reported elimination of SLS activity in a transposon Tn916 mutant of GAS deficient in riboflavin biosynthesis (34). Considering that FMN is a direct product of riboflavin metabolism, a requirement for FMN would provide the link between SLS activity and riboflavin biosynthesis.
Although the S. iniae cytolysin confers a beta-hemolytic phenotype on SBA, the hemolytic profile of disease-associated strains is less profound than that of commensal strains when cultures are grown aerobically. In vitro growth exhibits weak beta-hemolysis encircling a zone of alpha-hemolysis and can easily be confused with the alpha-hemolysis of viridans streptococcal species. Yet the extent of cytolysis is equivalent when disease-associated and commensal strains are cultured anaerobically. The nucleotide sequence of sagA, including the promoter region and >600 bp upstream of the sagA ORF, is identical in disease-associated and commensal isolates (data not shown). Following complementation of an NH strain of GAS with the S. iniae sagA gene from a disease-associated strain, a beta-hemolytic phenotype resulted, regardless of oxygen tension. These findings suggest that a regulatory mechanism, rather than structural differences in SagA, affects the oxygen-dependent expression of SLS in disease-associated strains. In GAS, the CovR-CovS two-component regulatory system and the negative regulators Nra and RofA negatively influence sagA transcription (2, 14, 37), and rofA is also affected by oxygen conditions (16, 17). A small number of strains expressing hemolytic phenotypes opposite to those described for their genotypes (disease associated versus commensal) have been identified. These include two commensal strains and the ATCC type strain 29178, expressing the alpha/beta-hemolytic phenotype, as well as two constitutively beta-hemolytic disease-associated strains, all grown under aerobic and anaerobic conditions (data not shown). Although the selective advantage that this oxygen-dependent SLS phenotype confers on S. iniae is not immediately apparent, these observations lend further support to the hypothesis that SLS expression is under some degree of regulation.
By use of a murine model of subcutaneous infection, SLS activity in GAS has been linked to tissue necrosis (4). Recent work showed that targeted mutagenesis of each of the sag genes generated an NH mutant, while mutagenesis immediately downstream of the operon did not abolish hemolysis, demonstrating a direct link between the hemolytic phenotype and the defined sag locus (38). When a GAS sagA allelic replacement mutant (NZ131 sagAΔcat) was examined in a murine model of subcutaneous infection, a complete attenuation of necrotic-lesion formation, similar to that in previous studies (4), was observed (unpublished data), emphasizing the importance of sagA expression in GAS virulence. By using a disease-associated strain of S. iniae, a sagB insertion-deletion mutant was generated to examine the role of SLS in the virulence of this pathogen. Previous work with S. iniae demonstrated that disease-associated strains, in stark contrast to commensal isolates, established bacteremic infection in the murine model following subcutaneous inoculation and were highly resistant to phagocytic killing in whole human blood (19). Interestingly, the SLS-deficient SI sagB.KO.erm mutant was also able to cause invasive disease in this model, even at low inocula, with no evident signs of attenuated virulence in comparison to that with the wild type. Furthermore, this mutant remained resistant to whole-blood killing in vitro. From these studies, it is apparent that the contribution of S. iniae SLS to bacteremia and resistance to phagocytosis is negligible. It was previously reported that disease-associated and commensal strains of S. iniae can elicit localized necrosis in the murine model, similar to that elicited by GAS, if a higher inoculum of 108 CFU is used (19). However, mice infected with disease-associated strains at this dose are at risk of terminal disease, likely due to the invasiveness of these strains. Thus, we examined necrotic-lesion formation caused by a commensal isolate and its SLS-deficient counterpart. In contrast to the commensal wild-type isolate, which caused necrotic lesions in 60% of animals, the SLS-deficient mutant, SI sagB.KO.Tn917, did not cause localized cell damage in mice. These results suggest that SLS of S. iniae contributes to tissue damage but not to bacteremia.
In summary, we have identified an SLS-associated gene cluster within the chromosome of S. iniae with strong homology to that of GAS and demonstrated the requirement for SLS in causing tissue damage in a mammalian model. While SLS does seem to contribute to virulence by promoting local tissue necrosis, it does not play a critical role in the establishment of bacteremia in mice. However, S. iniae infection in fish is characterized by meningoencephalitis, and SLS may be required for brain endothelial-cell injury. Future studies will test this hypothesis in a fish infection model of meningoencephalitis.
Acknowledgments
This work was supported by a grant from the Canadian Bacterial Diseases Network.
Editor: E. I. Tuomanen
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Beckert, S., B. Kreikemeyer, and A. Podbielski. 2001. Group A streptococcal rofA gene is involved in the control of several virulence genes and eukaryotic cell attachment and internalization. Infect. Immun. 69:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, A. M., J. Yother, D. E. Briles, D. Hansman, and J. C. Paton. 1989. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect. Immun. 57:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betschel, S. D., S. M. Borgia, N. L. Barg, D. E. Low, and J. C. S. de Azavedo. 1998. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect. Immun. 66:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, I., P. Germon, K. McDade, and J. R. Scott. 2001. Generation and surface localization of intact M protein in Streptococcus pyogenes are dependent on sagA. Infect. Immun. 69:7029-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant, C., L. Hubbard, and W. D. McElroy. 1991. Cloning, nucleotide sequence, and expression of the nitroreductase gene from Enterobacter cloacae. J. Biol. Chem. 266:4126-4130. [PubMed] [Google Scholar]
- 8.Chaffin, D. O., and C. E. Rubens. 1998. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219:91-99. [DOI] [PubMed] [Google Scholar]
- 9.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 10.Dale, J. B., E. Y. Chiang, D. L. Hasty, and H. S. Courtney. 2002. Antibodies against a synthetic peptide of SagA neutralize the cytolytic activity of streptolysin S from group A streptococci. Infect. Immun. 70:2166-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eldar, A., Y. Bejerano, and H. Bercovier. 1994. Streptococcus shiloi and Streptococcus difficile: two new streptococcal species causing a meningoencephalitis in fish. Curr. Microbiol. 28:139-143. [Google Scholar]
- 13.Eldar, A., P. F. Frelier, L. Assenta, P. W. Varner, S. Lawhon, and H. Bercovier. 1995. Streptococcus shiloi, the name for an agent causing septicemic infection in fish, is a junior synonym of Streptococcus iniae. Int. J. Syst. Bacteriol. 45:840-842. [DOI] [PubMed] [Google Scholar]
- 14.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flanagan, J., N. Collin, J. Timoney, T. Mitchell, J. A. Mumford, and N. Chanter. 1998. Characterization of the haemolytic activity of Streptococcus equi. Microb. Pathog. 24:211-221. [DOI] [PubMed] [Google Scholar]
- 16.Fogg, G. C., and M. G. Caparon. 1997. Constitutive expression of fibronectin binding in Streptococcus pyogenes as a result of anaerobic activation of rofA. J. Bacteriol. 179:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogg, G. C., C. M. Gibson, and M. G. Caparon. 1994. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an m gamma delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol. Microbiol. 11:671-684. [DOI] [PubMed] [Google Scholar]
- 18.Framson, P., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller, J. D., D. J. Bast, V. Nizet, D. E. Low, and J. C. S. de Azavedo. 2001. Streptococcus iniae virulence is associated with a distinct genetic profile. Infect. Immun. 69:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genilloud, O., F. Moreno, and R. Kolter. 1989. DNA sequence, products, and transcriptional pattern of the genes involved in production of the DNA replication inhibitor microcin B17. J. Bacteriol. 171:1126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginsberg, I. 1970. Streptolysin S, p. 99-171. In T. C. Montie, S. Kadis, and S. J. Ajl (ed.), Microbial toxins, vol. 3. Bacterial protein toxins. Academic Press, New York, N.Y.
- 22.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillman, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hecht, H. J., H. Erdmann, H. J. Park, M. Sprinzl, and R. D. Schmid. 1995. Crystal structure of NADH oxidase from Thermus thermophilus. Nat. Struct. Biol. 2:1109-1114. [DOI] [PubMed] [Google Scholar]
- 25.Humar, D., V. Datta, D. J. Bast, B. Beall, J. C. S. De Azavedo, and V. Nizet. 2002. Streptolysin S and necrotising infections produced by group G streptococcus. Lancet 359:124-129. [DOI] [PubMed] [Google Scholar]
- 26.Karlyshev, A. V., M. J. Pallen, and B. W. Wren. 2000. Single-primer PCR procedure for rapid identification of transposon insertion sites. BioTechniques 28:1078, 1080, 1082. [DOI] [PubMed] [Google Scholar]
- 27.Kehoe, M., and K. N. Timmis. 1984. Cloning and expression in Escherichia coli of the streptolysin O determinant from Streptococcus pyogenes: characterization of the cloned streptolysin O determinant and demonstration of the absence of substantial homology with determinants of other thiol-activated toxins. Infect. Immun. 43:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kupke, T., M. Uebele, D. Schmid, G. Jung, M. Blaesse, and S. Steinbacher. 2000. Molecular characterization of lantibiotic-synthesizing enzyme EpiD reveals a function for bacterial Dfp proteins in coenzyme A biosynthesis. J. Biol. Chem. 275:31838-31846. [DOI] [PubMed] [Google Scholar]
- 29.Lancefield, R. C. 1962. Current knowledge of the type-specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 30.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y. M., J. C. Milne, L. L. Madison, R. Kolter, and C. T. Walsh. 1996. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science 274:1188-1193. [DOI] [PubMed] [Google Scholar]
- 32.Li, Z., D. D. Sledjeski, B. Kreikemeyer, A. Podbielski, and M. D. Boyle. 1999. Identification of pel, a Streptococcus pyogenes locus that affects both surface and secreted proteins. J. Bacteriol. 181:6019-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Limbago, B., V. Penumalli, B. Weinrick, and J. R. Scott. 2000. Role of streptolysin O in a mouse model of invasive group A streptococcal disease. Infect. Immun. 68:6384-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, S., S. Sela, G. Cohen, J. Jadoun, A. Cheung, and I. Ofek. 1997. Insertional inactivation of streptolysin S expression is associated with altered riboflavin metabolism in Streptococcus pyogenes. Microb. Pathog. 22:227-234. [DOI] [PubMed] [Google Scholar]
- 35.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milne, J. C., R. S. Roy, A. C. Eliot, N. L. Kelleher, A. Wokhlu, B. Nickels, and C. T. Walsh. 1999. Cofactor requirements and reconstitution of microcin B17 synthetase: a multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17. Biochemistry 38:4768-4781. [DOI] [PubMed] [Google Scholar]
- 37.Molinari, G., M. Rohde, S. R. Talay, G. S. Chhatwal, S. Beckert, and A. Podbielski. 2001. The role played by the group A streptococcal negative regulator Nra on bacterial interactions with epithelial cells. Mol. Microbiol. 40:99-114. [DOI] [PubMed] [Google Scholar]
- 38.Nizet, V., B. Beall, D. J. Bast, V. Datta, L. Kilburn, D. E. Low, and J. C. S. de Azavedo. 2000. Genetic locus for streptolysin S production by group A streptococcus. Infect. Immun. 68:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, H. J., R. Kreutzer, C. O. Reiser, and M. Sprinzl. 1993. Molecular cloning and nucleotide sequence of the gene encoding a H2O2-forming NADH oxidase from the extreme thermophilic Thermus thermophilus HB8 and its expression in Escherichia coli. Eur. J. Biochem. 211:909.. [PubMed] [Google Scholar]
- 41.Perera, R. P., S. K. Johnson, M. D. Collins, and D. H. Lewis. 1994. Streptococcus iniae associated with mortality of Tilapia nilotica × T. aurea hybrids. J. Aquat. Anim. Health 6:335-340. [Google Scholar]
- 42.Pier, G. B., and S. H. Madin. 1976. Streptococcus iniae sp. nov., a beta-hemolytic streptococcus isolated from an Amazon freshwater dolphin, Inia geoffrensis. Int. J. Syst. Bacteriol. 26:545-553. [Google Scholar]
- 43.Pritzlaff, C. A., J. C. Chang, S. P. Kuo, G. S. Tamura, C. E. Rubens, and V. Nizet. 2001. Genetic basis for the beta-haemolytic/cytolytic activity of group B Streptococcus. Mol. Microbiol. 39:236-247. [DOI] [PubMed] [Google Scholar]
- 44.Simon, D., and J. J. Ferretti. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 66:219-224. [DOI] [PubMed] [Google Scholar]
- 45.Spellerberg, B., B. Pohl, G. Haase, S. Martin, J. Weber-Heynemann, and R. Lutticken. 1999. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J. Bacteriol. 181:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vriesema, A. J. M., S. A. J. Zaat, and J. Dankert. 1996. A simple procedure for isolation of cloning vectors and endogenous plasmids from viridans group streptococci and Staphylococcus aureus. Appl. Environ. Microbiol. 62:3527-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe, M., T. Nishino, K. Takio, T. Sofuni, and T. Nohmi. 1998. Purification and characterization of wild-type and mutant “classical” nitroreductases of Salmonella typhimurium. L33R mutation greatly diminishes binding of FMN to the nitroreductase of S. typhimurium. J. Biol. Chem. 273:23922-23928. [DOI] [PubMed] [Google Scholar]
- 48.Weinstein, M. R., M. Litt, D. A. Kertesz, P. Wyper, D. Rose, M. Coulter, A. McGeer, R. Facklam, C. Ostach, B. M. Willey, A. Borczyk, D. E. Low, et al. 1997. Invasive infections due to a fish pathogen, Streptococcus iniae. N. Engl. J. Med. 337:589-594. [DOI] [PubMed] [Google Scholar]
- 49.Wennerstrom, D. E., J. C. Tsaihong, and J. T. Crawford. 1985. Evaluation of the role of hemolysin and pigment in the pathogenesis of early onset group B streptococcal infection, p. 155-156. In Y. Kimura, S. Kotami, and Y. Shiokawa (ed.), Recent advances in streptococci and streptococcal diseases. Reedbooks, Bracknell, United Kingdom.
- 50.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]