Abstract
The human T-cell recognition of the low-molecular-mass culture filtrate antigen TB10.4 was evaluated in detail. The molecule was strongly recognized by T cells isolated from tuberculosis (TB) patients and from BCG-vaccinated donors. The epitopes on TB10.4 were mapped with overlapping peptides and found to be distributed throughout the molecule. The broadest response was found in TB patients, whereas the response in BCG-vaccinated donors was focused mainly toward a dominant epitope located in the N terminus (amino acids 1 to 18). The gene encoding TB10.4 was found to belong to a subfamily within the esat-6 family that consists of the three highly homologous proteins TB10.4, TB10.3, and TB12.9 (Rv0288, Rv3019c, and Rv3017c, respectively). Southern blot analysis combined with database searches revealed that the three members of the TB10.4 family were present only in strains of the Mycobacterium tuberculosis complex, including BCG, and M. kansasii, whereas other atypical mycobacteria had either one (M. avium, M. intracellulare, and M. marinum) or none (M. scrofulaceum, M. fortuitum, and M. szulgai) of the genes. The fine specificity of the T-cell response to the three closely related esat-6 family members was markedly different, with only a few epitopes shared between the molecules. Minimal differences in the amino acid sequence translated into large differences in recognition by T cells and secretion of gamma interferon. In general, the peptides from TB10.4 stimulated the largest responses, but epitopes unique to both TB10.3 and TB12.9 were found. The relevance of the findings for TB vaccine development and as a potential mechanism for immune evasion is discussed.
The bacterium Mycobacterium tuberculosis was isolated and identified as the causative agent of tuberculosis (TB) by Robert Koch in 1882. Despite very significant research efforts in recent years, major gaps still remain in our knowledge of the bacterium. Designing an effective vaccine against the disease has been an international research priority since human trials demonstrated a highly variable protective efficacy of the current vaccine BCG (8). The recent sequencing of the M. tuberculosis genome (6) has opened the way to many new antigen discovery approaches; among these are the definition and screening of immunodominant gene families identified in silico. One such family, the esat-6 gene family, has attracted significant interest and has been demonstrated to encode several immunodominant molecules that are strongly recognized by the immune systems in different animal models of TB, as well as by T cells from human beings exposed to M. tuberculosis (1, 15, 17, 18).
The esat-6 family consists of 14 to 23 low-mass proteins, depending on the definition criteria used (18). Of the 23 proteins in this family at least 13 can be divided further into subfamilies due to high sequence relatedness of the molecules in the individual gene-protein subfamilies. One esat-6 subfamily is the Mtb9.9 family, consisting of five open reading frames (ORFs) (Rv1037c, Rv1198, Rv1793, Rv2346c, and Rv3619c), which was recently described by Alderson et al. (1). A second subfamily (the QILSS family [7]) consists of the five neighboring ORFs (Rv1038c, Rv1197, Rv1792, Rv2347c, and Rv3620c) that share individual identity on the protein level of >98%. Both of these families probably result from relatively recent gene duplications, as indicated by the high homology of the proteins (96 to 99% identity).
Here we analyze the T-cell epitopes on the TB10.4 antigen and describe the identification and characterization of two highly homologous proteins, TB10.3 and TB12.9, which together with TB10.4 constitute another subfamily within the esat-6 gene family. Although these molecules are highly homologous, our data indicate that the fine specificity of the T-cell responses to these molecules differ markedly, and very few T-cell epitopes are shared among the three family members. The possible role of these findings for antigenic variation in M. tuberculosis and as a potential mechanism for immune evasion is discussed.
MATERIALS AND METHODS
Southern blotting.
Genomic DNA from the different mycobacterial species listed in Table 2 was prepared as described previously (2). Southern blotting was carried out as described elsewhere (17).
TABLE 3.
Biochemical properties of the TB10.4 family membersa
| Gene | Antigen | No. of aa/ no. of bases | Molecular mass (kDa) | pI | % Homology to TB10.4b | Homologous protein in:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CSU#93c
|
M. bovisd
|
M. aviume,f
|
|||||||||
| Status TIGR ID | % Homology | Status | % Homology | Status | % Homology | ||||||
| Rv 0288 | TB10.4 | 96/291 | 10.391 | 4.43 | 100 (96/96) | Yes, MT0301 | 100 (96/96) | Yes | 100 (96/96) | Yesf | 79 (76/96) |
| Rv3019c | TB10.3 | 96/291 | 10.313 | 4.2 | 84 (81/96) | Yes, MT3104 | 100 (96/96) | Yes | 100 (96/96) | Yesf | 80 (77/96) |
| Rv3017c | TB12.9 | 120/363 | 12.923 | 7.92 | 65 (45/69) | Yes, MT3097 | 100 (120/120) | Yes | 89 (107/120) | No | 62 (44/70) |
aa, amino acids. Numbers in parentheses refer to the number of homologous amino acids/total number of amino acids.
Determined by database BLASTP searches at http://www.sanger.ac.uk/Projects/M_tuberculosis/blast_server.shtml.
Determined by database BLASTP searches at http://198.76.161.137/cmr-blast/Index.cgi?database=GMT.pep. TIGR ID refers to the TIGR database identification number.
Determined by database BLASTP searches at http://www.sanger.ac.uk/Projects/M_bovis/blast_server.shtml.
Determined by database BLASTP searches at http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?organism=m_avium.
Only one copy of the TB10.4 family is present in M. avium (Table 2). It is not possible to determine if this protein is the homologue of TB10.4 or TB10.3.
Recombinant proteins and synthetic peptides.
Recombinant TB10.4 was produced as previously described (17).
Preliminary experiments have shown that T-cell responses to TB10.4 and a mixture of all overlapping peptides are almost identical. The same has previously been demonstrated for two other esat-6 family members: ESAT-6 and CFP-10 (3). Therefore, we used a mixture of overlapping synthetic peptides of TB10.3 and TB12.9 as a substitute for whole protein.
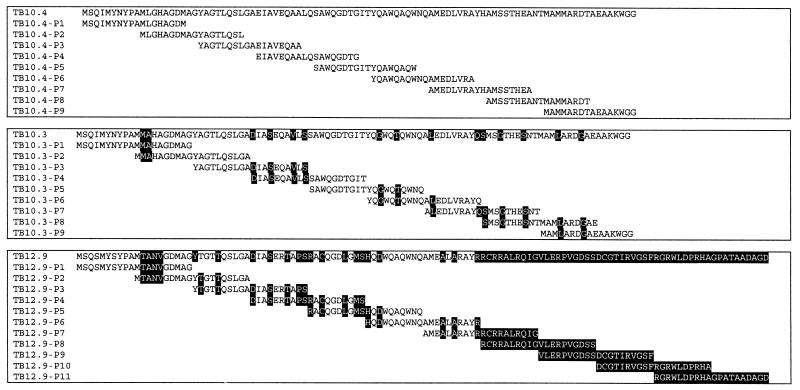
Synthetic overlapping peptides (18- or 20-mer) covering the complete primary structure of the three proteins (see Fig. 4) were synthesized by standard solid-phase methods on an ABIMED peptide synthesizer (ABIMED, Langenfeld, Germany) at the Department of Infectious Diseases and Immunohematology/Bloodbank C5-P, Leiden University Medical Centre, Leiden, The Netherlands (TB10.4), or at Schafer-N, Copenhagen, Denmark. The peptides were purified by reverse-phase high-pressure liquid chromatography. Purified peptides were lyophilized and stored dry until reconstitution in phosphate-buffered saline (PBS).
FIG. 4.
Amino acid sequences of the TB10.4 family members (TB10.4, TB10.3, and TB12.9). Sequences of overlapping peptides are given below. Residues shown in black boxes indicate differences from the sequence of TB10.4.
Human T-cell lines.
T-cell lines were generated as described previously (12). Briefly, peripheral blood mononuclear cells (PBMC) were obtained from HLA-DRB1*-typed TB patients (the patients were HLA-DRB1 typed by PCR amplification of DNA obtained from peripheral blood cells or phytohemagglutinin (PHA)-stimulated blast cells with sequence-specific primers, identifying polymorphisms corresponding to the serologically defined series DR1-DR18). PMBC were incubated at 1 × 106 to 2 × 106 cells/well in 24-well plates (Nunc, Roskilde, Denmark) in the presence of antigen at 5 μg/ml for 6 days and then expanded with recombinant interleukin-2. The T-cell lines were then frozen and stored in liquid nitrogen. Five cell lines were generated with short-term culture filtrate (TCL2-6) and one with purified protein derivative (PPD; TCL1). Only T-cell lines that were M. tuberculosis reactive, i.e., responded to M. tuberculosis sonicate or PPD (tuberculin RT23; Statens Serum Institute, Copenhagen, Denmark) but not to tetanus toxoid, were used in the present study. For the analysis of antigen-specific responses, T-cell lines (1.5 × 104/well) were incubated with irradiated autologous or HLA-matched PBMC (5 × 104/well), with or without antigen (PPD at 5 μg/ml; recombinant antigen at 0.2, 1, or 5 μg/ml; peptide mixtures at 0.2, 1, or 5 μg of total peptides/ml; or individual peptides at 1 or 5 μg/ml), in a total volume of 200 μl/well in triplicate in 96-well flat-bottom microtiter plates. The proliferation was measured by determining the [3H]thymidine incorporation at day 4 and is expressed as the stimulation index (SI), which is defined as the mean counts per minute (cpm) in stimulated cultures divided by the mean cpm in unstimulated cultures.
For this part of the study, the protocol (P136/97) was approved by the Ethics Committee of the Leiden University Medical Center.
Human donors.
PBMC were obtained from 3 healthy nonvaccinated donors, 10 healthy BCG-vaccinated donors with a positive in vitro response to PPD, and 10 TB patients with microscopy- or culture-proven infection. Blood samples were drawn from TB patients 0 to 6 months after diagnosis. This part of the study was approved by the Local Ethical Committee for Copenhagen and Frederiksberg (RH 01-282/96 and KF 01-369/98).
Lymphocyte preparations and cell culture.
PBMC were freshly isolated by gradient centrifugation of heparinized blood on Lymphoprep (Nycomed, Oslo, Norway) and stored in liquid nitrogen until use. The cells were resuspended in complete RPMI 1640 medium (Gibco-BRL/Life Technologies) supplemented with 1% penicillin-streptomycin (Gibco-BRL/Life Technologies), 1% nonessential amino acids (FLOW; ICN Biomedicals), and 10% heat-inactivated normal human AB serum. The viability and number of the cells were determined by Nigrosin staining. Cell cultures were established with 1.25 × 105 PBMC in 100 μl in microtiter plates (Nunc) and stimulated with 5 μg of PPD/ml, 5 μg of recombinant TB10.4 or single peptides/ml at final concentrations of 10, 5, or 0.5 μg/ml, or peptide pools corresponding to the full-length proteins in which each peptide was included at concentrations of 0.5, 0.1, or 0.05 μg/ml.
“No antigen” was included as a negative control, and PHA was used as a positive control (results not shown). Cell cultures were incubated for 5 days at 37°C (5% CO2, 95% air), and supernatants were harvested for cytokine analysis.
Cytokine analysis.
Gamma interferon (IFN-γ) was detected with a standard sandwich enzyme-linked immunosorbent assay with a commercially available pair of monoclonal antibodies (Endogen) and used according to the manufacturer's instruction. Recombinant IFN-γ (also from Endogen) was used as a standard.
Enzyme-linked immunospot (ELISPOT) assay for IFN-γ.
We blocked 96-well polyvinylidene difluoride-packed plates (MAIPS 4510; Millipore) precoated with anti-human IFN-γ (Endogen) with 1% bovine serum albumin in PBS for 2 h at 37°C. Afterward, cells were added in different known concentrations, together with antigen (10 μg/ml), and then incubated 18 h at 37°C (5% CO2, 95% air). After being washed with 0.05% Tween 20 in PBS, anti-human IFN-γ-Biotin (Endogen) secondary antibody was added, and the mixture was incubated for 2 h. Alkaline phosphatase-conjugated streptavidin (1:1,000 in PBS-1% BSA) was added and left for 1 h at 37°C. Plates were developed with nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate; Sigma) and incubated for 15 min at 37°C in the dark.
Spots were counted under microscope and expressed as spot-forming units per 105 cells.
RESULTS
T-cell responses to the TB10.4 antigen.
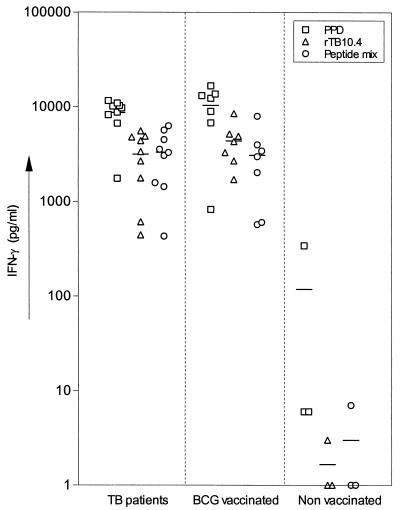
PBMC from human TB patients, healthy BCG-vaccinated patients, and nonvaccinated controls were stimulated in vitro with recombinant TB10.4 (rTB10.4), a mixture of peptides covering the complete TB10.4 sequence (18-mer peptides overlapping with eight amino acids), or PPD (Fig. 1). TB10.4 was not recognized in healthy nonvaccinated donors, and the response induced in the group of TB patients and BCG-vaccinated donors was at the same level, a finding in agreement with the presence of the gene for TB10.4 in different strains of BCG (reference 17 and unpublished results). The response induced by the recombinant preparation was at the same level as responses to the mixture of peptides and typically constituted 25 to 50% of the response to PPD at the individual donor level. To investigate the magnitude of the response to TB10.4 in more detail, we evaluated the number of cells responding to PPD, rTB10.4, and the mixture of TB10.4 peptides by ELISPOT assay. In the two BCG-vaccinated healthy donors investigated, the number of responding cells correlated closely with the IFN-γ response, and the number of TB10.4-specific cells was 40 to 45% of the frequency of cells responding to PPD (Table 1), thereby confirming the immuno-dominant nature of this antigen.
FIG. 1.
Individual IFN-γ responses of 10 TB patients, 7 BCG-vaccinated donors, and 3 nonvaccinated healthy donors stimulated with PPD, recombinant TB10.4 (rTB10.4), or TB10.4 peptide mixture (nine peptides covering the full-length TB10.4 protein). Each symbol represents one donor. Mean values (solid bars) are indicated.
TABLE 1.
Frequency of IFN-γ-producing cells responding to rTB10.4 and TB10.4 peptide mix in comparison to PPD as determined by ELISPOT on two PPD-positive BCG-vaccinated donorsa
| Antigen | Donor 1
|
Donor 2
|
||
|---|---|---|---|---|
| IFN-γ concn (pg/ml) | Spots/105 cells | IFN-γ concn (pg/ml) | Spots/105 cells | |
| PPD | 113,000 | 87 | 18,000 | 14 |
| rTB10.4 | 51,000 | 39 | 7,000 | 5 |
| TB10.4 mix | 41,000 | 32 | 6,000 | 5 |
No antigen is included as a negative control. Antigens were tested in concentrations of 10 μg/ml.
Epitope mapping of TB10.4.
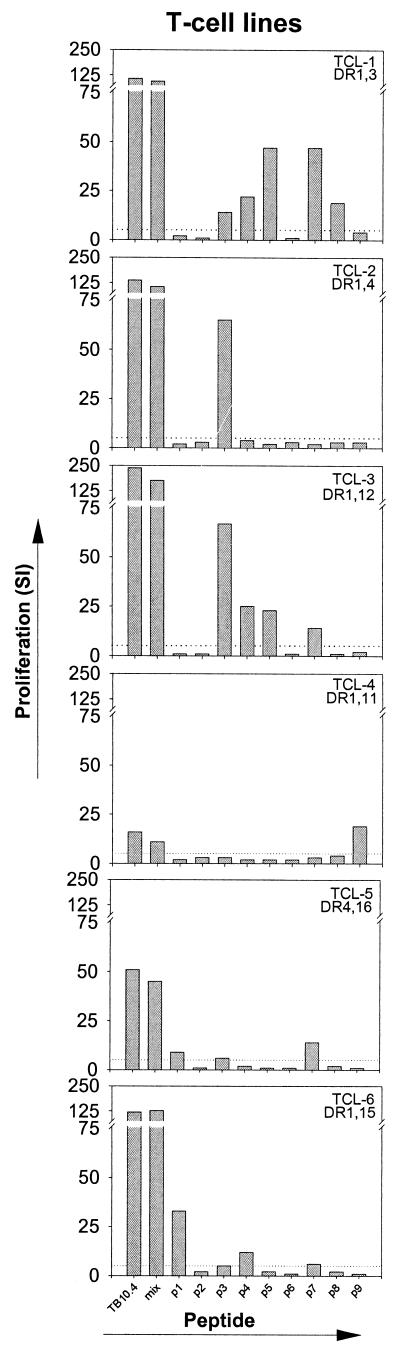
The T-cell response against TB10.4 was characterized in more detail by mapping the epitopes recognized. For this analysis the panel of nine overlapping peptides (18 amino acids) covering the complete sequence of TB10.4 was used to stimulate T-cell lines derived from human TB patients. The peptides were used either as single peptides or in pools covering the full-length protein. The pattern of the T-cell proliferation after stimulation with TB10.4-derived peptides was heterogeneous (Fig. 2) in the sense that almost all peptides were recognized by at least one T-cell line (SI > 5), and in some cases the T-cell lines recognized multiple peptides throughout the sequence.
FIG.2.
Individual proliferative responses of six T-cell lines (generated from blood from TB patients) stimulated with single TB10.4 peptides, TB10.4 peptide mixture, or recombinant TB10.4 (rTB10.4). T-cell lines were driven with either PPD (TCL1) or STCF (TCL2-6). Results are given as SI values (i.e., mean cpm in the presence of antigen/mean cpm without antigen). The maximum stimulation in the presence of either 1 or 5 μg of antigen/ml is given. Responses that are >5 SI (dotted line) are regarded as positive.
The specificity of the T-cell responses to TB10.4 peptides was compared for TB patients and BCG-vaccinated donors. PBMC from nine TB patients and nine healthy BCG-vaccinated donors were stimulated with the individual peptides and a mixture of all overlapping peptides covering the complete sequence of TB10.4.
The response induced in PBMC by the TB10.4-derived peptides were, as with the T-cell lines, highly heterogeneous (Fig. 3). The profile of T-cell responses to the TB10.4-derived peptides further demonstrated the presence of interindividual differences in the epitope recognition (Fig. 3).
FIG. 3.
Individual IFN-γ responses of nine TB patients and nine BCG-vaccinated healthy donors stimulated with single TB10.4 peptides or TB10.4 peptide mixture. Each symbol represents one donor. The maximum stimulation in the presence of either 1 or 5 μg of antigen/ml is given. Mean values (solid bars) are indicated. Responses of >200 pg of IFN-γ/ml (dotted line) are regarded as positive.
Almost all peptides (with peptide 8 as an exception) induced high IFN-γ responses in at least one donor, and most donors recognized multiple peptides throughout the sequence (Fig. 3). The magnitude of the IFN-γ response to the dominant peptides was in many cases comparable to or even greater than the level induced by the peptide mixture and thus the recombinant protein.
The tendency found was that TB patients recognized a broader panel of epitopes than the BCG-vaccinated healthy donors. Donors in the BCG-vaccinated group had responses focused to one or two peptides (Fig. 3), whereas the TB patients often recognized several peptides and, for some donors, as many as six peptides. The overall dominant epitope for both groups was located at the N-terminal end of the protein (amino acids 1 to 18), and this region gave significant PBMC responses in the majority of the TB patients (eight of nine) and BCG-vaccinated donors (five of nine) (Fig. 3), whereas the recognition of this peptide was less pronounced when we analyzed T-cell lines (Fig. 2). In TB patients, p2 (amino acids 11 to 29) and p9 (amino acids 78 to 96) were also strongly recognized by several donors, with responses of >1,000 pg of IFN-γ/ml.
tb10.4 subfamily: homology and distribution.
To identify other genes closely related to tb10.4 on the genome of M. tuberculosis and other mycobacterial species, a Southern blot analysis was performed. Genomic DNA from a number of mycobacterial strains was digested with PvuII and probed with tb10.4. This analysis resulted in three cross-hybridization bands in lanes corresponding to M. tuberculosis H37Rv, M. bovis, M. bovis BCG Danish, M. bovis BCG Pasteur, and M. kansasii (Table 2). In the more distantly related strains M. avium and M. intracellulare, we only observed one band, and in M. scrofulaceum, M. fortuitum, and M. szulgai no hybridizing bands were detected (Table 2).
TABLE 2.
Interspecies distribution of the tb10.4 family members
| Strain | No. of tb10.4 family membersa |
|---|---|
| M. tuberculosis H37Rv | 3b |
| M. tuberculosis clinical isolate CSU#93 | 3c |
| M. bovis | 3b |
| BCG Danish | 3 |
| BCG Pasteur | 3 |
| M. kansasii | 3 |
| M. avium | 1b |
| M. intracellulare | 1 |
| M. marinum | 1 |
| M. scrofulaceum | 0 |
| M. fortuitum | 0 |
| M. szulgai | 0 |
Determined by Southern blot analysis.
Verified by database searches (http://www.sanger.ac.uk/Projects/M_tuberculosis/blast_server.shtml, http://www.sanger.ac.uk/Projects/M_bovis/blast_server.shtml, http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?organism=m_avium).
Determined by database searches (http://198.76.161.137/cmr-blast/index.cgi?database=GMT.pep).
Since no internal PvuII sites are present in the tb10.4 gene, this result suggested that tb10.4 was present in more than one copy on the genome of the species in the M. tuberculosis complex. This finding was confirmed by BLAST searches in the M. tuberculosis H37Rv sequence database Tuberculist (http://genolist.pasteur.fr/TubercuList/), which identified two ORFs that were highly homologous to tb10.4 (on the DNA and amino acid levels). One ORF (Rv3019c) was 84% identical to tb10.4 (247 of 291 bp), and the other ORF (Rv3017c) shared 74% sequence identity to tb10.4 (156 of 206 bp at the 5′ end). Together, these molecules constitute the TB10.4 subfamily (Fig. 4 and Table 3).
Furthermore, in agreement with the Southern blot analysis, a BLAST search of tb10.4 in the incomplete sequence databases of M. bovis (http://www.sanger.ac.uk) and the M. tuberculosis clinical isolate CSU#93 (http://www.tigr.org/tdb/) identified the presence of three TB10.4 family members (Tables 2 and 3). The corresponding protein sequences of TB10.4, TB10.3, and TB12.9 in CSU#93 and M. bovis were 100% identical to the H37Rv sequences except for the TB12.9 sequence of M. bovis, which was only 88.4% identical (Table 3).
Additionally, a search in the incomplete sequence database of the M. avium genome (http://www.tigr.org/tdb) identified only one ORF with high homology to tb10.4 (Table 2). This ORF was not identical to any of the three family members but had high homology to especially tb10.3 and tb10.4 (Table 3).
The results from the two approaches thus correlate well, and it is apparent that the TB10.4 family is conserved in the species belonging to the M. tuberculosis complex and species closely related to this one (e.g., M. kansasii), whereas only one candidate is present in the M. avium complex, and the rest of the strains lack this family completely.
T-cell recognition of TB10.4 family members.
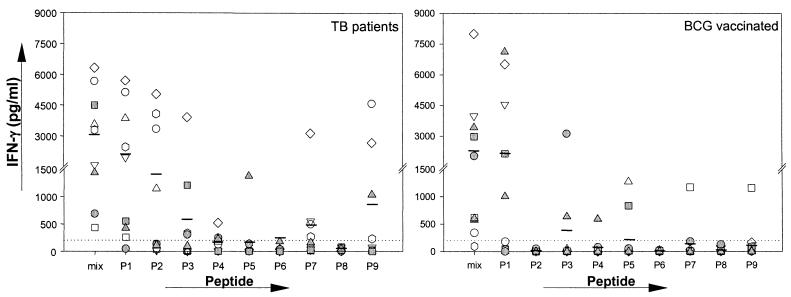
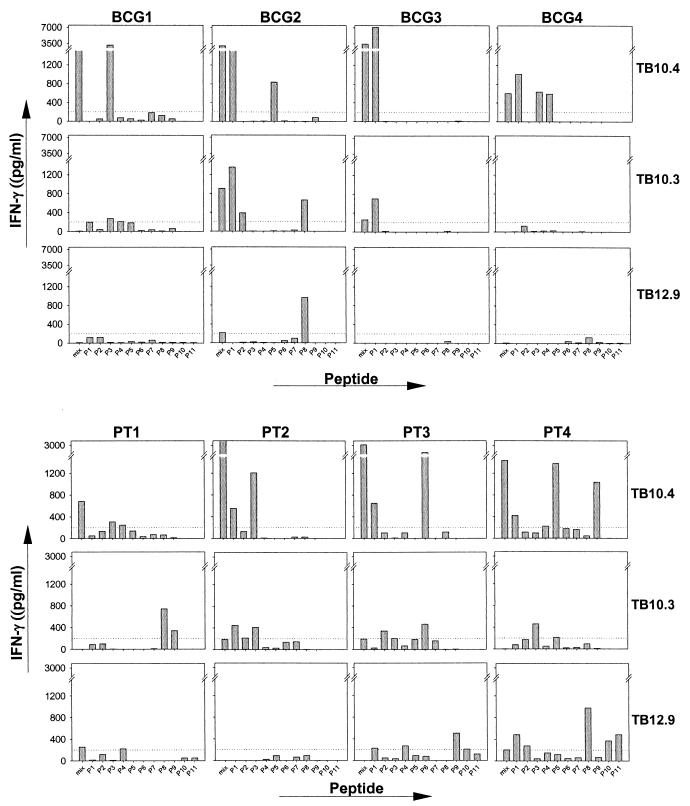
The fine specificity of the human T-cell recognition of the three closely related TB10.4 family members was analyzed by screening of a panel of overlapping peptides covering the complete sequences of TB10.3, TB10.4, and TB12.9 (Fig. 4).
PBMC from eight donors (four TB patients and four BCG-vaccinated donors) were used to compare the responses to the three sets of peptides (Fig. 5). Although the molecules exhibit a significant sequence homology, the recognition patterns found in the eight donors were quite different. The peptides from TB10.4 stimulated the largest responses, with IFN-γ levels to a single peptide in the range of 3,500 to 7,000 pg/ml (e.g., p1 [amino acids 1 to 18] in BCG3 and p3 [amino acids 21 to 38] in BCG1). For these donors with responses focused to one particular region, some recognition of the corresponding region in the other family members was found but, in general, minimal changes in the sequence translated into large differences at the stimulation level found with these peptides. In the highly conserved N-terminal region, the substitution of two amino acids—i.e., amino acids 12 and 13 (Leu-Gly substituted with Met-Ala)—in peptide 1 in TB10.3 reduced the IFN-γ release from BCG3 almost 10-fold, and changing four amino acids in the corresponding peptide from TB12.9 (Leu-Gly-His-Ala substituted with Trp-Ala-Asn-Val) completely ablated the recognition. Similarly, a substitution of three amino acids in peptide 6 (amino acids 54, 57, and 63) almost completely ablated the strong recognition of this epitope by PT3. The epitope mapping also resulted in the identification of a number of epitopes unique for TB10.3 such as peptide 8 (residues 71 to 90) in PT1 and BCG2. TB12.9 is the least similar among the three molecules, in particular due to the presence of an extra 51 C-terminal amino acids. This part of the molecule, which is lacking completely from TB10.4 and TB10.3, was also found to contain unique epitopes, especially peptide 8, recognized by two donors (BCG2 and PT4).
FIG. 5.
Individual IFN-γ responses of four TB patients and four BCG-vaccinated healthy donors stimulated with the single peptides TB10.4, TB10.3, and TB12.9 or with mixtures of these peptides. The maximum stimulation in the presence of either 1 or 5 μg of antigen/ml is given. Each column represents one donor, and each row represents one antigen. Responses of >200 pg of IFN-γ/ml (dottedt line) are regarded as positive.
An analysis of the three molecules on four T-cell lines indicated that epitopes shared between all three molecules existed (results not shown). One T-cell line recognized peptide 3 of all three family members. The response was comparable when stimulated with peptide 3 of TB10.4 and TB10.3 but was lower when stimulated with the corresponding peptide of TB12.9.
DISCUSSION
Data from several laboratories have demonstrated the vaccine potential of antigens secreted by M. tuberculosis, and high-resolution separation techniques have identified molecules in the low-molecular-mass region of culture filtrate (masses of <15 kDa) to be of particular relevance (2). Antigen discovery efforts from several laboratories have in recent years resulted in the identification of a number of these low-molecular-mass antigens (Mtb8.4 [1]; Mtb12 [20]; GroES, CFP10 [4]; TB10.4 [17]; Mtb9.9 [1]; alpha chrystallin, CFP6 [5]; TB7.3 [17]; and TB9.8, TB9.4, TB11.0, and TB11.2 [14]). In analyses of these molecules, it is striking that a significant number of them were found to be located in one mycobacterial protein-gene family-the esat-6 family. This family consists of at least 14 low-mass proteins with some homology to ESAT-6 and a defined genomic organization. With the proteins described here, a total of 10 proteins (ESAT-6 [15], CFP10 [17], MTB9.9A-E [1], TB10.4 [17], TB10.3, and TB12.9) from this family have now been published as strongly recognized T-cell antigens.
In the present study we describe the identification and immunological evaluation of the TB10.4 family, consisting of TB10.3, TB10.4, and TB12.9, as a new subfamily of the esat-6 family. We used synthetic peptides to identify and compare the epitopes recognized in these proteins and found that especially TB10.4 contained a number of strongly recognized T-cell epitopes distributed throughout the protein sequence. The major epitopes recognized at the PBMC level were located in the N-terminal part of the molecule, but this region was less frequently recognized by T-cell lines, which may suggest that the skewing of responses occurs during in vitro propagation of a T-cell line. TB10.3 and TB12.9 were in general less antigenic than TB10.4 but contained T-cell epitopes, several of which were unique to these proteins. Sharing of the major T-cell epitopes by these proteins would be expected based on their highly homologous amino acid sequence, but the present study, together with recent observations from Alderson et al. (1), clearly demonstrate that the highly homologous proteins from the esat-6 family (Mtb9.9 and TB10.4 subfamilies) contain predominantly protein-specific epitopes and that shared epitopes are an exception. This finding underlines the highly specific binding of peptides to HLA-class II molecules and demonstrates that a difference of just one amino acid at critical positions may have dramatic consequences for T-cell recognition.
Expression of TB10.4 has been verified by N-terminal sequencing of the corresponding protein spot in a two-dimensional gel of a M. tuberculosis short-term culture filtrate fraction enriched in monoclonal antibody PV-2 (a monoclonal antibody that recognizes TB10.4) reactive protein (17). Although not formally demonstrated, the recognition of a number of unique epitopes on TB10.3 and TB12.9 also strongly suggests that these proteins are expressed by the bacteria during TB infection.
Pathogenic organisms will often use sequence polymorphism of highly immunogenic proteins to escape the host's immune defenses (9, 10, 16). This is not the case for M. tuberculosis as recently demonstrated by Musser et al. (11) and Streevatsan et al. (19), who found negligible sequence diversity in a large number of M. tuberculosis genes encoding important antigens recognized by the host immune system. This finding is in agreement with the recent finding of a lack of sequence diversity in the TB10.4 sequence originating from 13 clinical isolates of M. tuberculosis obtained from different geographical locations (unpublished observations).
An interesting possibility is that M. tuberculosis may compensate for this lack of sequence polymorphism by having duplicated the genes encoding major T-cell antigens, leading to several copies (homologues) of proteins that can substitute each other functionally but which differ in their immunodominant epitopes. Tightly controlled expression of these homologues may result in antigen variation and immune evasion. This hypothesis may be relevant for the whole esat-6 family, since the genomic organization of these proteins suggests that these proteins have similar functions. Apart from the esat-6 family, the immunodominant molecules within the antigen 85 complex in which the three molecules presumably have the same function in living bacteria could be examples of this phenomenon as well. In this regard, the TIGR database defines the three tb10.4 family genes plus esat-6 as one paralogous gene family (http://www.tigr.org), meaning that these genes possibly have been duplicated in M. tuberculosis during evolution. These duplication events may even involve whole clusters of genes, including regions surrounding the esat-6 family members such as PE and PPE genes as suggested by Cole et al. (7)
The lack of sequence homology to proteins and genes from other organisms illustrates that esat-6 family members have Mycobacterium-specific functions, and the need for several copies of the same gene may indicate that the genes encode function(s) of crucial importance for bacterial survival in the host. Using differential display for comparison of gene expression in M. tuberculosis H37Rv and the avirulent H37Ra, Rindi et al. (13) showed that TB10.4 was produced in the virulent strain but not in the avirulent strain, suggesting that this protein or gene may be involved in functions important for M. tuberculosis virulence. The presence of this gene in BCG (which is deleted of the esat-6-cfp10 operon) illustrates that attenuation can be obtained through the inactivation of various combinations of these different genes.
The availability of the M. tuberculosis genome and the current efforts to sequence a large number of additional mycobacterial genomes has set the stage for postgenomic approaches to the identification of novel antigens. The identification and characterization of protein-gene families is a very important outcome of these initiatives. If the gene families contain components that can substitute for each other functionally and represent mechanisms for immune evasion, the finding of markedly different epitope recognition of the individual components, as observed in the present study, may have important consequences for vaccine design. The extreme consequence may be that a successful TB vaccine will have to promote an immune response not only to a selected target antigen such as TB10.4 but also to epitopes characteristic for the rest of the closely related gene family. We are currently conducting experiments to address this important question.
Acknowledgments
This study received financial support from the Danish National Association against Lung Diseases, The Royal Netherland Academy of Arts and Sciences, and The European Community (project no. QLK2-1999-01093)
We are grateful to Axel Kok-Jensen, Department of Pulmonary Diseases, Gentofte Hospital, Copenhagen, Denmark, for recruiting the patients and to Vita Skov for excellent technical assistance.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Alderson, M. R., T. Bement, C. H. Day, L. Zhu, D. Molesh, Y. A. Skeiky, R. Coler, D. M. Lewinsohn, S. G. Reed, and D. C. Dillon. 2000. Expression cloning of an immunodominant family of Mycobacterium tuberculosis antigens using human CD4+ T cells. J. Exp. Med. 191:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, P., D. Askgaard, A. Gottschau, J. Bennedsen, S. Nagai, and I. Heron. 1992. Identification of immunodominant antigens during infection with Mycobacterium tuberculosis. Scand. J. Immunol. 36:823-831. [DOI] [PubMed] [Google Scholar]
- 3.Arend, S. M., A. Geluk, K. E. van Meijgaarden, J. T. van Dissel, M. Theisen, P. Andersen, and T. H. Ottenhoff. 2000. Antigenic equivalence of human T-cell responses to Mycobacterium tuberculosis-specific RD1-encoded protein antigens ESAT-6 and culture filtrate protein 10 and to mixtures of synthetic peptides. Infect. Immun. 68:3314-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthet, F. X., P. B. Rasmussen, I. Rosenkrands, P. Andersen, and B. Gicquel. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144:3195-3203. [DOI] [PubMed] [Google Scholar]
- 5.Bhaskar, S., S. P. Khanna, and R. Mukherjee. 2000. Isolation, purification and immunological characterization of novel low molecular weight protein antigen CFP 6 from culture filtrate of Mycobacterium tuberculosis. Vaccine 18:2856-2866. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T. 1996. Why sequence the genome of Mycobacterium tuberculosis? Tuberc. Lung Dis. 77:486-490. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Fine, P. E. M. 1995. Variation in protection by BCG: implications of and for heterologous immunity. Lancet 346:1339-1345. [DOI] [PubMed] [Google Scholar]
- 9.Fitch, W. M., J. M. Leiter, X. Q. Li, and P. Palese. 1991. Positive Darwinian evolution in human influenza A viruses. Proc. Natl. Acad. Sci. USA 88:4270-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes, M. K., and A. L. Hughes. 1995. Natural selection on Plasmodium surface proteins. Mol. Biochem. Parasitol. 71:99-113. [DOI] [PubMed] [Google Scholar]
- 11.Musser, J. M., A. Amin, and S. Ramaswamy. 2000. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics 155:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ottenhoff, T. H., D. G. Elferink, J. Hermans, and R. R. de Vries. 1985. HLA class II restriction repertoire of antigen-specific T cells. I. The main restriction determinants for antigen presentation are associated with HLA-D/DR and not with DP and DQ. Hum. Immunol. 13:105-116. [DOI] [PubMed] [Google Scholar]
- 13.Rindi, L., N. Lari, and C. Garzelli. 1999. Search for genes potentially involved in Mycobacterium tuberculosis virulence by mRNA differential display. Biochem. Biophys. Res. Commun. 258:94-101. [DOI] [PubMed] [Google Scholar]
- 14.Rosenkrands, I., K. Weldingh, S. Jacobsen, C. V. Hansen, W. Florio, I. Gianetri, and P. Andersen. 2000. Mapping and identification of Mycobacterium tuberculosis proteins by two-dimensional gel electrophoresis, microsequencing and immunodetection. Electrophoresis 21:935-948. [DOI] [PubMed] [Google Scholar]
- 15.Sørensen, A. L., S. Nagai, G. Houen, P. Andersen, and Å. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 63:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seibert, S. A., C. Y. Howell, M. K. Hughes, and A. L. Hughes. 1995. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol. Biol. Evol. 12:803-813. [DOI] [PubMed] [Google Scholar]
- 17.Skjøt, R. L., T. Oettinger, I. Rosenkrands, P. Ravn, I. Brock, S. Jacobsen, and P. Andersen. 2000. Comparative evaluation of low-molecular-mass proteins from Mycobacterium tuberculosis identifies members of the ESAT-6 family as immunodominant T-cell antigens. Infect. Immun. 68:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skjøt, R. L. V., E. M. Agger, and P. Andersen. 2001. Antigen discovery and tuberculosis vaccine development in the post-genomic era. Scand. J. Infect. Dis. 33:643-647. [DOI] [PubMed] [Google Scholar]
- 19.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webb, J. R., T. S. Vedvick, M. R. Alderson, J. A. Guderian, S. S. Jen, P. J. Ovendale, S. M. Johnson, S. G. Reed, and Y. A. Skeiky. 1998. Molecular cloning, expression, and immunogenicity of MTB12, a novel low-molecular-weight antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 66:4208-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]