Abstract
Porphyromonas gingivalis is an important pathogen associated with destructive periodontal disease and is able to invade the epithelial cell barrier. Its cysteine proteases are recognized as major virulence factors, and in this study, we examined the interaction of the arginine-specific protease with epithelial cells in culture. Three cell lines (KB, HeLa, and SCC4) were incubated with strain W50 culture supernatant; stained with monoclonal antibody 1A1, which recognizes an epitope on the adhesin (β) component of the cysteine protease-adhesin (α/β) heterodimer; and viewed using immunofluorescence microscopy. Within 1 h, the protease traversed the plasma membrane and was localized around the nucleus before becoming concentrated in the cytoplasm after 24 to 48 h. In contrast, the purified arginine-specific heterodimeric protease (HRgpA) rapidly entered the nucleus within 15 to 30 min. This nuclear targeting (i) was seen with active and Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK)-inactivated HRgpA, indicating it was independent of the proteolytic activity; (ii) occurred at both 4 and 37°C; and (iii) failed to occur with the monomeric protease (RgpAcat), indicating the importance of the adhesin chain of the HRgpA protease to this process. Rapid cell entry was also observed with recombinant catalytic (α) and adhesin (β) chains, with the latter again targeting the nuclear area. After 48 h of incubation with HRgpA, significant dose-dependent stimulation of metabolic activity was observed (measured by reduction of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide), and a doubling of mitotic activity combined with the presence of apoptotic cells indicated that HRgpA may interfere with cell cycle control mechanisms. These effects were seen with both active and TLCK-inactivated protease, confirming that they were not dependent on proteolytic activity, and thus provide new insights into the functioning of this P. gingivalis protease.
Periodontal diseases are chronic inflammatory conditions resulting from the host response to microbial challenge and are characterized by the formation of deepening periodontal pockets and the destruction of both hard and soft tissues supporting the teeth. An essential step in initiating any infection involves adherence to, and/or invasion of, host cells by pathogens or their products. As epithelial cells form the first barrier to invasion by bacteria which colonize the gingival crevice, the nature of their interaction with potential pathogens is likely to have a major influence on the progression of disease.
Porphyromonas gingivalis is an anaerobic gram-negative bacterium important in destructive periodontal disease whose main ecological niche is the subgingival crevice adjacent to the tooth surface and the epithelial attachment apparatus of the soft tissues. The organism can invade gingival tissue in advanced periodontitis (18) and can be taken up by gingival and pocket epithelial cells, oral epithelial cell lines, and endothelial cells in vitro (13, 23, 33, 40). The nature of the eukaryotic surface receptor molecules and the signal transduction pathways required for internalization of P. gingivalis by oral epithelial cells remain to be clarified. However both fimbriae and bacterial proteases have been suggested to be important mediators in this interaction.
The cysteine proteases of P. gingivalis with specificity for arginyl- and lysyl-peptide bonds are encoded by a family of three genes, now designated rgpA, rgpB, and kgp (8). Evidence that they are expressed by the organism in vivo and are the targets of specific immune response has been provided by comparison of antibody levels in sera from periodontitis patients and healthy controls (10). These proteases are recognized as major virulence factors which are able to deregulate plasma-based immune and inflammatory systems and homeostatic mechanisms operating in periodontal tissues (6, 24, 38). They are also considered important colonization determinants of the organism through the development of attachment epitopes and provision of metabolizable peptides and micronutrients.
We previously reported the damaging effects of P. gingivalis proteases on fibronectin-integrin interactions in human gingival fibroblasts using HRgpA, a heterodimer derived from rgpA consisting of an arginine-specific catalytic α chain in noncovalent association with a β component which functions as an adhesin. The specificity of action of the heterodimer is achieved through binding of its adhesin chain to fibronectin deposited by the fibroblasts and consequent targeting of the catalytic chain to sites of extracellular matrix. This results in proteolytic destruction of the fibronectin and the loss of the main fibronectin integrin receptor, α5β1 (35). The adhesin component of HRgpA is also known to bind to laminin, an important component of basement membrane which interacts with the integrins α6β4 and α3β1 on epithelial cells. This suggested that a similar mechanism of adhesin-mediated, targeted destruction of laminin-integrin interactions may operate in epithelium. Contrary to expectations, however, we demonstrated that the HRgpA protease rapidly enters epithelial cells and localizes in and around the nucleus. To our knowledge, this is the first report of a bacterial product which is able both to direct its entry across the cytoplasmic membrane and to localize in the nuclear compartment of target cells. Here, we describe these observations and discuss the potential mechanisms involved and their implications for the functioning of important barrier cells of the periodontium.
MATERIALS AND METHODS
Bacterial culture and supernatant preparation.
P. gingivalis strain W50 was grown in brain heart infusion broth supplemented with hemin (5 mg liter−1) in an atmosphere of 80% N2, 10% H2, and 10% CO2 at 37°C for 6 days. The culture supernatant was obtained by centrifugation (11,000 × g for 20 min at 5°C), sterilized by passage through a 0.2-μm-pore-size cellulose acetate membrane, and stored at −70°C.
Arginine-specific protease activity was measured by hydrolysis of Nα-benzoyl-dl-arginine p-nitroanilide (BApNA) as previously described (31). One unit of protease activity is defined as the amount of enzyme causing an increase in the A405 of 1.0 min−1.
Purification of arginine-specific proteases.
The arginine-specific proteases HRgpA and RgpAcat were purified from strain W50 culture supernatant as previously described (31). Before being applied to cultured epithelial cells, they were dialyzed using 25 mM sodium acetate buffer (pH 5.3) and stored at 4°C. Where indicated, the enzyme activity was irreversibly inactivated by incubation for 30 min at 4°C with 1 mM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) in the presence of 10 mM β-mercaptoethanol and CaCl2. Excess inhibitor was removed by dialysis using the same sodium acetate buffer. The enzyme activities of protease samples before and after TLCK treatment were measured by BApNA hydrolysis.
Recombinant protein synthesis; recombinant RgpAα (rRgpAα) and rRgpAβ.
The individual coding regions for the α and β domains of RgpA were amplified from P. gingivalis W50 chromosomal DNA by PCR and then cloned into His6 expression vectors. Full details are given in Slaney et al. (37). In brief, the incorporation of appropriate restriction enzyme sites within the primers permitted directional cloning of the products into pJFQ3010 (a derivative of pQE30 [11]) for the α domain and pJF119EH (14) for the β domain. The recombinant plasmids were then propagated in Escherichia coli XL-1 Blue (Stratagene) with selection for ampicillin resistance. These strategies resulted in constructs encoding the RgpA catalytic domain, R227 to R719 (rRgpAα), and the adhesin domain, G721 to R1262 (rRgpAβ), each fused to a His6 purification tag at the N terminus.
E. coli XL-1 Blue transformed with the appropriate expression construct was grown overnight at 37°C in Luria-Bertani medium (32) incorporating ampicillin (50 μg/ml). Cells expressing rRgpAα and rRgpAβ were harvested by centrifugation, solubilized in 6 M guanidine HCl (pH 8), and purified by column chromatography on Ni-nitrilotriacetic acid resin (Qiagen). Recombinant proteins were eluted with 8 M urea in 0.5 M saline plus 0.05 M Tris buffer (pH 5.9) and dialyzed at 4°C using progressively decreasing concentrations of urea in the saline-Tris buffer followed by progressively decreasing concentrations of saline from 0.5 to 0.15 M over 3 days before filter sterilization and storage at −20°C. The identities of the recombinant proteins were confirmed by Western blotting using specific antibody to P. gingivalis proteins and by automated N-terminal sequencing.
rRagB.
The primer pair, HRAGBF1 (5′-ATATATGAGCTCCGCGACCCCGAAGGAAAAG-3′) and HRAGBRI (5′-TATATAGTCGACGAAAAGATAGGGGCTGCGAC-3′) (the italicized sequences represent SacI and SalI sites, respectively) were designed to amplify the region spanning R24 to I501 of the outer membrane protein RagB using either P. gingivalis W50 DNA or pPM1 (17) as the template in a PCR. The amplification was for 25 cycles, and the parameters were 94°C, 1 min; 60°C, 1 min; and 72°C, 4 min. The 1,508-bp partial SstI-SalI amplicon was subsequently cloned into the SstI-SalI sites of pJFQ3010 (11) as pNY34. This scheme ensured that RagB, minus the first 23-amino-acid putative signal peptide sequence, was synthesized as an N-terminal His6 fusion protein under the control of the tac promoter and IPTG (isopropyl-β-d-thiogalactopyranoside) induction in E. coli XL-1 Blue. The predicted size of the recombinant protein was 55.7 kDa.
E. coli XL-1 Blue expressing rRagB was solubilized in 0.01 M imidazole lysis buffer. Supernatant from the lysate was incubated for 1 h at room temperature with Ni-nitrilotriacetic acid agarose (Qiagen) and packed into a column which was first washed with lysis buffer containing 0.02 M imidazole and eluted with 0.25 M imidazole in lysis buffer. The eluted protein was dialyzed against 0.15 M saline, filter sterilized, and stored at −20°C.
Epithelial cell culture and treatment with P. gingivalis preparations.
Epithelial cell cultures (HeLa, European Collection of Cell Cultures reference number 93021013, and KB cells, reference number 94050408) were maintained in Dulbecco's modified Eagle's medium (DMEM; Life Technologies Ltd., Paisley, Scotland) supplemented with 10% (vol/vol) fetal bovine serum (Globepharm Ltd., Surrey, England) in 5% CO2 in air. SCC4 cells, (ATCC reference number CRL 1624) were maintained in DMEM-Hams F12 medium (Life Technologies Ltd.) supplemented with fetal bovine serum as described above.
For immunofluorescence studies, suspensions containing 104 cells were inoculated into each well of 24-well plates containing sterilized 13-mm-diameter glass coverslips and incubated at 37°C for approximately 3 days to establish adherent monolayers. The cells were then treated with P. gingivalis culture products, diluted in DMEM, for intervals from 15 min to 48 h. The bacterial products tested included culture supernatant from strain W50, HRgpA, TLCK-treated HRgpA, RgpAcat, recombinant RgpAα (rRgpAα), rRgpAβ, and rRagB. Following incubation, the cells were fixed for 5 min at 4°C in 3.7% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.4) and permeabilized in 1% Triton X-100 in PBS for 5 min before antibody staining to demonstrate the localization of the bacterial products. As a control, in some experiments, the permeabilization step was omitted to confirm an intracellular location of the bacterial product.
Immunofluorescence studies.
Details of the primary and secondary antibodies are given in Table 1. All dilutions were made in PBS (pH 7.4) containing 0.05% (wt/vol) sodium azide and 5% (vol/vol) human serum. Cells to be stained with mouse monoclonal primary antibodies (MAbs) were preincubated for 30 min in a blocking solution of 10% (vol/vol) normal goat serum (G9023; Sigma Chemical Company Ltd., Poole, England) in PBS containing 0.05% sodium azide to reduce nonspecific staining. Coverslips with adherent cells were incubated overnight with primary antibody at room temperature and washed three times with PBS, followed by incubation for 60 min with a 1/200 concentration of the appropriate biotin-conjugated anti-mouse or anti-rabbit antibody. After further washing, a 1/100 concentration of streptavidin Texas Red complex (RPN 1233; Amersham International, Little Chalfont, England) was added for 60 min. Nuclei were then stained with DAPI (4,6-diamidino-2-phenylindole [D9542]; Sigma Chemical Company Ltd.) (10 ng ml−1 in PBS) for 5 min before the coverslips were mounted on glass slides using Immumount (Thermo Shandon, Runcorn, Cheshire, England) containing 2.5 mg of 1,4-diazabicyclo [2,2,2] octane/ml to reduce fading. All experiments included negative controls in which incubation with primary antibody was omitted. Nonspecific primary antibodies were also included in certain experiments where indicated in the text. Cells were viewed with a Nikon Microphot FXA microscope equipped for epifluorescence with an excitation filter of 510 to 560 nm for Texas Red and a UV filter of 330 to 380 nm for DAPI. Photographs of treated and control cells were taken with standardized exposure times using Fujichrome Provia 1600 film, ASA 3200, and E6 processing pushed two stops.
TABLE 1.
Antibodies used in immunofluorescence studies
| Primary antibody (reference) | Species and isotypea | Specificityb | Optimum working concn | Secondary antibody (no. and source) |
|---|---|---|---|---|
| 1A1 (4) | Mouse monoclonal IgG1 | Adhesin component of HRgpA, rRgpAβ | 1/50 | Biotin-conjugated goat anti-mouse antibody (B0529; Sigma Chemical Company Ltd.) |
| 1B5 (11) | Mouse monoclonal IgG2a | LPS and polysaccharide components on RgpAcat, mt-RgpAc | 1/20 | Biotin-conjugated goat anti-mouse antibody (B0529; Sigma Chemical Company Ltd.) |
| B15 (28) | Mouse monoclonal IgG2a | RagB | 1/20 | Biotin-conjugated goat anti-mouse antibody (B0529; Sigma Chemical Company Ltd.) |
| PgWC (9) | Rabbit polyclonal | P. gingivalis cells and secreted products; rRgpAα, rRgpAβ | 1/5,000 | Biotin-conjugated swine anti-rabbit antibody (E0353; Dako Ltd.) |
IgG1, immunoglobulin G1.
LPS, lipopolysaccharide.
mt, membrane type.
Confocal analysis was conducted with a Zeiss laser scanning microscope (model LSM510; 63× objective). Optical sectioning was used to collect sequential images on the x and y axes, starting at the basal surface of the cell adherent to the coverslip and rising at 0.5-μm intervals to the apical surface. Images were manipulated using Zeiss LSM image-browsing software.
Measurement of mitotic and metabolic activities of epithelial cells treated with HRgpA.
Mitotic activity was measured using KB cells grown as described above for immunofluorescence studies. The cells were incubated for 48 h with HRgpA (optical density at 280 nm, 0.27; specific activity, 30 U of BApNA mg−1 with doubling dilutions in DMEM from 1/8 to 1/1,024; BApNA activities, 1.0 to 0.0078 U ml−1), fixed, permeabilized, and stained with DAPI. The percentage of dividing cells at each protease concentration was estimated by examining at least 1,500 cells across the maximum diameter of each coverslip.
Metabolic activity was assessed using 96-well plates containing 3 × 103 KB cells per well in 100 μl of DMEM. After overnight incubation to allow the cells to settle and attach, 100 μl of HRgpA or TLCK-treated HRgpA in DMEM was added to columns of wells to give final dilutions of 1/8 to 1/1,024, and the plates were incubated for 48 h. For the final 4 h, each well was inoculated with 20 μl of a solution of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT; Sigma Chemical Co. Ltd.) at 5 μg ml−1 in PBS. The ability of mitochondrial dehydrogenase enzymes in metabolically active cells to reduce MTT to an insoluble formazan (29) was determined at the end of the incubation period by solubilizing the reduced formazan using 100 μl of dimethyl sulfoxide and measuring the absorbance at 570 nm. The mean absorbance of blank wells containing dimethyl sulfoxide alone was subtracted from the values for treated wells before further data analysis.
Each experiment was conducted in duplicate, and the mean mitotic activities or absorbance values of treated cells were compared with the corresponding DMEM control values using Student's t test.
RESULTS
Interaction of P. gingivalis culture supernatant with epithelial cells.
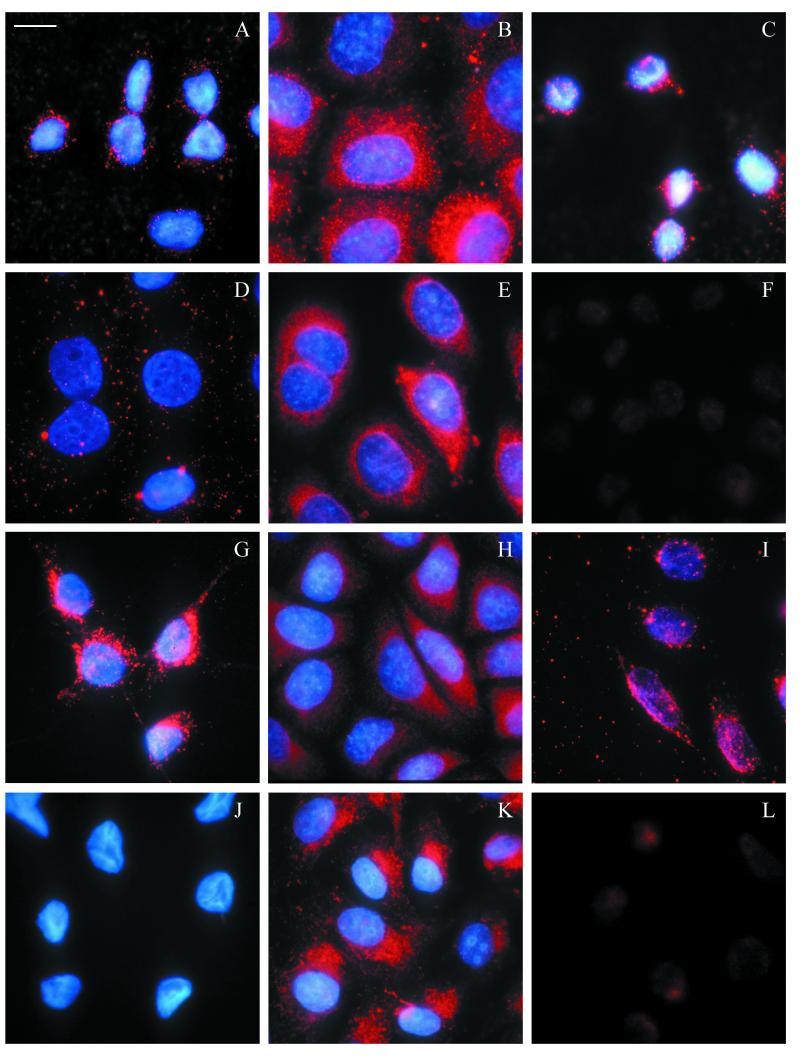
The time course of interaction of P. gingivalis extracellular products with epithelial cells was first examined by incubating KB cells with strain W50 culture supernatant diluted in DMEM culture medium. The mean BApNA activity of three batches of supernatant used in this work was 3.6 U ml−1, and doubling dilutions were made from 1/2 to 1/128 (BApNA activity, 1.8 to 0.028 U ml−1). Localization of the bacterial exoproducts was followed by staining with MAb 1A1, which recognizes an epitope on the adhesin component of HRgpA, Kgp, and other members of the hemagglutinin family (7). This showed that after 15 to 30 min the immunoreactive material began to be concentrated in small, discrete loci around the nuclei. Careful focusing indicated that these loci were close to the undulations in the nuclear membrane, which were particularly prominent in KB cells. The intensity of staining around the nuclei was greatest after 1 h (Fig. 1A) and became more diffuse and randomly distributed by 4 to 8 h (Fig. 1D). After 16 h, the staining began to concentrate in granular structures within the cytoplasm, and the maximum cytoplasmic staining intensity was reached after 24 h (Fig. 1G) and was maintained beyond 48 h. The most intense staining was observed using supernatant dilutions from 1/16 to 1/64; higher concentrations caused cell rounding and detachment after prolonged incubation periods.
FIG.1.
Time course of localization of MAb 1A1-immunoreactive material from P. gingivalis strain W50 culture supernatant in epithelial cells. The left-hand column shows KB cells incubated for 1 (A), 4 (D), and 24 h (G) with a 1/32 dilution of P. gingivalis culture supernatant and stained with MAb 1A1, which recognizes an adhesin component of the bacterial cysteine protease, streptavidin Texas Red, and nuclei stained with DAPI. Control KB cells (J) incubated in DMEM without bacterial culture supernatant and stained as described above show negligible Texas Red staining, but the nuclei are clearly visible. The center column shows HeLa cells incubated for 30 min (B) and 1 (E), 4 (H), and 24 h (K) with a 1/4 dilution of W50 culture supernatant and stained with MAb 1A1 as for KB cells. Immunoreactive material was initially localized around the nuclei, although the time course varied between KB and HeLa cells. After 24 h, immunoreactive material became concentrated in cytoplasmic vesicles in both cell types. SCC4 cells (not shown) closely resembled KB cells in their optimum concentration of culture supernatant and time course of response. The right-hand column shows KB cells incubated for 1 h with a 1/32 dilution of W50 culture supernatant stained with a panel of antibodies. MAb 1B5, which recognizes carbohydrate modifications to the catalytic chain of RgpAcat and mt-RgpA (C), shows bright focal staining around the nuclear membrane similar to that with MAb 1A1 (compare Fig. 1A), and MAb B15 against the 55-kDa RagB outer membrane protein of W50 (F) shows no specific staining, whereas PgWC rabbit polyclonal antibody, raised against P. gingivalis whole cells and secreted products (I), shows both cytoplasmic and perinuclear staining compared with control cells incubated with DMEM culture medium stained with PgWC (L). Magnification, ×980; bar = 10 μm.
A similar pattern of nuclear targeting by MAb 1A1-reactive proteins was observed when SCC4 cells (data not shown) and HeLa cells (Fig. 1, center column) were exposed to W50 culture supernatant. In the case of HeLa cells, a higher concentration of bacterial supernatant was required to observe the nuclear localization of immunoreactive material, but the process occurred more rapidly than with KB cells (Fig. 1B). Further experiments were conducted using KB cells and confirmed, where indicated, with other cell types.
To explore the role of protease activity in the phenomenon of nuclear targeting and subsequent cytoplasmic localization, KB cells were treated with native, unheated supernatant from strain W50 and supernatant heated to 80°C for 30 min in order to abolish BApNA activity (36). The treatments resulted in similar staining patterns, indicating that the translocation and localization phenomenon was not dependent on active protease (data for the heated supernatant is not shown).
Cells exposed to W50 supernatant for 1 h were also stained with a panel of primary antibodies against different P. gingivalis components (Table 1). In the case of MAb 1B5 (Fig. 1C), which recognizes a carbohydrate determinant on a polysaccharide of P. gingivalis found on the cell surface and in culture supernatants, as well as glycan modifications to some of the Arg-gingipains, the staining resembled the pattern described above with antibody 1A1 (Fig. 1A). In contrast, MAb B15, which recognizes the outer membrane protein RagB, gave no specific staining (Fig. 1F). Generalized cellular staining was seen with the polyclonal antibody PgWC, raised against P. gingivalis whole cells, indicating that, in addition to targeting the nucleus, bacterial factors were scattered throughout the cytoplasm (Fig. 1I).
The observations using MAbs 1A1 and 1B5 implicated members of the cysteine protease/hemagglutinin family in the nuclear targeting phenomenon. Further experiments were conducted using the purified arginine-specific protease (HRgpA), a 110-kDa heterodimer consisting of an α chain with protease activity and a β chain with adhesin-like properties, with RgpAcat, which lacks the adhesin domain, and finally with recombinant RgpAα and RgpAβ polypeptides.
Interaction of active HRgpA with epithelial cells.
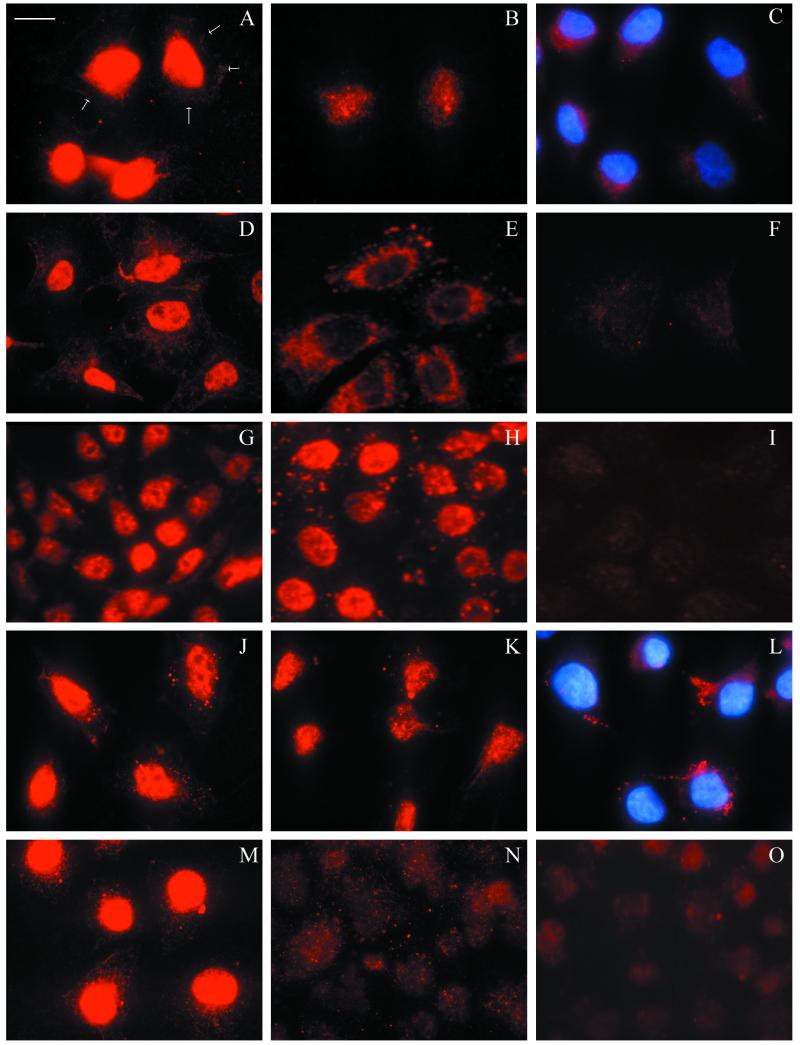
KB cells were treated with HRgpA, diluted in DMEM from 1/4 to 1/16, and were then stained with MAb 1A1. The resultant pattern resembled that of cells exposed to W50 culture supernatant except that, from 15 min, staining appeared to be localized within the nucleus rather than around the nuclear membrane. The nuclear staining intensity increased by 1 h (Fig. 2A), faded and became diffuse after 4 h (Fig. 2B), and was seen in the cytoplasm by 24 to 48 h (Fig. 2C). Patterns of strong initial intranuclear staining, similar to those seen with KB cells, were also observed with HeLa cells (Fig. 2D to F) and SCC4 cells (Fig. 2G to I) incubated with HRgpA. As we had observed previously with the W50 culture supernatant, the nuclear localization occurred more rapidly when HeLa cells were incubated with HRgpA than it did with the other cell lines. Both KB and SCC4 cells precooled to 4°C and treated with HRgpA for 1 h at 4°C showed nuclear staining with MAb 1A1 similar to that observed at 37°C, indicating that cell metabolic activity is not required for nuclear localization.
FIG.2.
Time course of localization of HRgpA in epithelial cells. Cells were treated with active HRgpA diluted in DMEM to a BApNA activity of 1 U ml−1 (A to E, G, H, and M) or TLCK-inactivated HRgpA at a comparable concentration (J to L). The primary antibodies were MAb 1A1 (A to L) and PgWC (M to O), with DAPI staining of nuclei (C and L). After treatment of KB cells for 1 h (A), the protease showed strong staining localized in the nucleus with faint cytoplasmic staining (the arrows indicate the cell periphery); by 4 h (B), nuclear staining had become less intense, and after 24 h (C), some staining was visible in the cytoplasm. HeLa cells showed similar strong nuclear localization of the protease after a shorter time of only 15 min (D) and diffuse staining by 1 h (E) compared with background staining in two control cells without protease treatment (F). The response of SCC4 cells to HRgpA after 1 and 24 h (G and H) was similar to that of KB cells, although SCC4 cells showed more obvious granular cytoplasmic staining after 24 h. Control cells are also shown (I). KB cells treated with TLCK-inactivated HRgpA for 1, 4, and 24 h (J to L) showed similar responses to the active HRgpA (A to C), except that occasional small specks of positive staining were seen in the cytoplasm. KB cells incubated with HRgpA and stained with PgWC antibody (M) showed nuclear localization similar to that with MAb 1A1 (A), whereas RgpAcat, which lacks the adhesin component, showed diffuse cytoplasmic staining but failed to demonstrate nuclear localization (N). Control cells stained with PgWC are also shown (O). Magnification, ×980; bar = 10 μm.
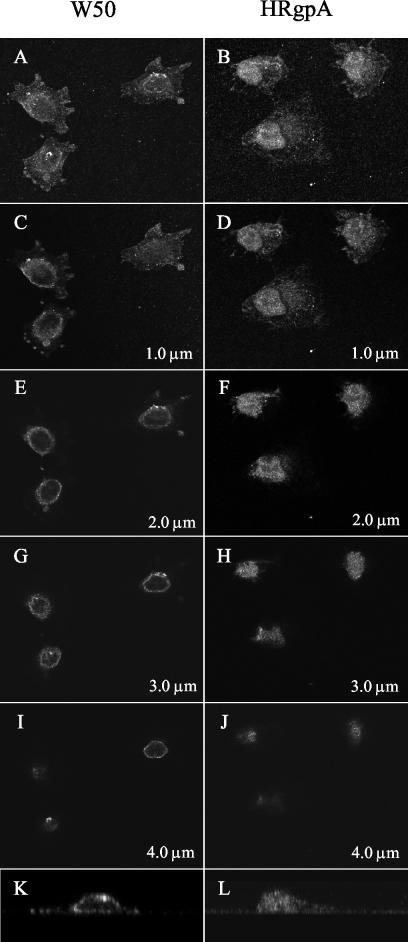
Confocal microscopy was used to confirm the different patterns of initial localization of MAb 1A1-reactive material around the nuclear membrane in W50-treated KB cells (Fig. 3, left-hand column) and within the nucleus in HRgpA-treated cells (Fig. 3, right-hand column). Enlarged lateral views (z plane optical sections) of typical cells demonstrated MAb 1A1-stained material around the nuclear periphery after W50 supernatant treatment (Fig. 3K), in contrast with staining throughout the nucleus after HRgpA treatment (Fig. 3L).
FIG. 3.
Confocal laser scanning microscope analysis of KB cells incubated for 30 min with 1/32 dilution of W50 culture supernatant (left) or 1 U of HRgpA ml−1 (right), fixed, and stained for immunofluorescence using MAb 1A1 as described in the text. Optical 0.5-μm-thick horizontal sections were taken from the basal to the apical surface of the cells. The combined stack of images (A and B) and representative x-y sections selected at 1-μm intervals (C to J) show localization of the MAb 1A1-reactive material in a ring around the nuclei with culture supernatant (C, E, G, and I), contrasting with localization within the nuclei with HRgpA (D, F, H, and J). Enlarged lateral views (z plane optical section) of typical cells are shown after treatment with W50 supernatant (K) and HRgpA (L).
Combined interaction of P. gingivalis culture supernatant and HRgpA with epithelial cells.
The observation that purified HRgpA enters the nuclei of epithelial cells whereas MAb 1A1-immunoreactive material in the culture supernatant of W50 remains in the perinuclear region led us to examine whether the culture supernatant contains moieties which are inhibitory to the nuclear localization of HRgpA. KB cells were incubated for 1 h with W50 culture supernatant alone, HRgpA alone, HRgpA combined with W50 culture supernatant, or HRgpA combined with brain heart infusion broth as a control for the culture supernatant. In each case, concentrations previously shown to give optimum staining were selected. The typical discrete loci of MAb 1A1-reactive material were seen around the nuclei of cells incubated with W50 culture supernatant, and staining was present within the nuclei of HRgpA-treated cells (as previously shown [Fig. 1A and 2A, respectively]). However, when culture supernatant was combined with HRgpA, no specific MAb 1A1 staining within the nuclei was observed. Instead, perinuclear positive staining characteristic of the supernatant was seen, with an increase in diffuse cytoplasmic staining in some cells (data not shown). Cells incubated with HRgpA combined with brain heart infusion broth showed nuclear staining comparable with that seen with HRgpA alone, indicating that components of the growth medium were not responsible for preventing nuclear entry.
Role of the catalytic component of HRgpA in interaction with epithelial cells.
The role of the catalytic component of HRgpA in cell entry and nuclear localization was investigated in two ways. First, cells exposed to active HRgpA were compared with those exposed to HRgpA which had been irreversibly inactivated by TLCK treatment. Staining patterns in cells incubated with inactive HRgpA (Fig. 2J to L) were comparable with those for the active HRgpA (compare Fig. 2A to C), confirming that the localization phenomenon was not dependent on proteolytic activity. Second, cells treated with active HRgpA were compared with those exposed to RgpAcat, the catalytic component of the protease, which lacks the adhesin chain present in HRgpA. After being stained with PgWC antibody, HRgpA-treated cells (Fig. 2M) showed nuclear localization typical of that observed using MAb 1A1, whereas treatment with RgpAcat resulted in diffuse staining throughout the cytoplasm and the absence of nuclear localization (Fig. 2N). Therefore, the adhesin component of HRgpA appears to be important in nuclear localization. To extend these observations, recombinant RgpAα and -β chain polypeptides were tested for the ability to enter epithelial cells and localize to the nucleus.
Interaction of recombinant proteins with epithelial cells.
KB cells were incubated for 1 h with rRgpAα and rRgpAβ (optical densities at 280 nm, 0.2 and 0.3, respectively) diluted up to 1/4 in DMEM. rRagB at a comparable concentration was used as a negative control, and HRgpA (BApNA activity, 1 U ml−1) was used as a positive control. The localization of bacterial products was determined by staining cells following all treatments with the polyclonal PgWC antibody and the use of the specific Mabs 1A1 and B15 for rRgpAβ and rRagB, respectively. rRgpAα, detected with PgWC antibody (Fig. 4A), was able to enter cells and was scattered diffusely within the cytoplasm but did not show the nuclear localization typical of HRgpA (compare Fig. 2M), whereas rRgpAβ localized around the nuclei in a pattern that closely resembled that seen with W50 supernatant when stained with either PgWC antibody (Fig. 4B) or MAb 1A1 (not shown). In contrast, no cell entry or localization was seen with cells incubated with rRagB using either PgWC (Fig. 4C) or MAb B15 (not shown). These observations using rRgpAβ support data from the experiments with the heterodimeric versus monomeric proteases and indicate that the catalytic component only translocates into the cytoplasmic compartment of epithelial cells whereas the adhesin component is able to both enter and localize at the nuclei of these cells.
FIG. 4.
Localization of recombinant RgpAα, RgpAβ, and Rag B proteins in KB cells after 1 h of incubation and staining with PgWC antibody. rRgpAα (A) was present throughout the cytoplasm, whereas rRgpAβ (B) localized predominantly around the nuclei (the arrows indicate the cell periphery). No specific staining was seen with rRagB (C) or control cells (not shown). All photographs were taken using constant exposure times, so direct comparisons can be made. Magnification, ×980; bar = 10 μm.
Effect of HRgpA on cell morphology, proliferation, and metabolic activity.
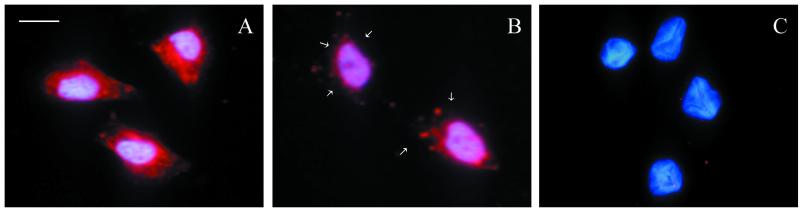
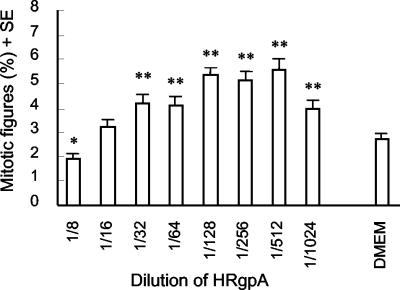
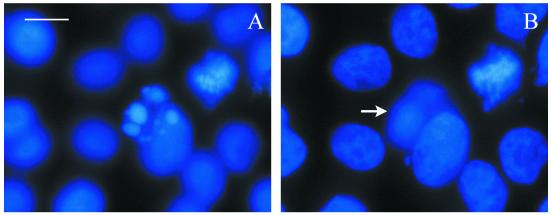
Microscopical examination of cells treated with HRgpA indicated transient rounding at higher concentrations after approximately 2 h, followed by flattening and apparent alterations in cell growth compared with untreated cells after prolonged incubation. In order to quantitate these changes, mitotic activity was examined in KB cells after 48 h of exposure to a range of concentrations of HRgpA (Fig. 5). The highest protease concentration tested (1/8 dilution; BApNA activity, 1 U ml−1) significantly suppressed mitotic activity (P < 0.05), whereas lower concentrations (≤1/32 dilution; BApNA activities, ≤0.25 U ml−1) stimulated mitotic activity, with a typical dose-response curve showing a doubling of mitotic activity at 1/128 to 1/512 (BApNA activities, 0.0625 to 0.0156 U ml−1; P < 0.005) compared with DMEM-treated cells. Cells displaying nuclear condensation characteristic of apoptosis (Fig. 6) were observed at all protease concentrations with both active and TLCK-inactivated HRgpA but were more prominent around the middle of both dilution series. Apoptotic cells were usually in a plane above that of the main cell monolayer, indicating that they were less adherent and therefore liable to be detached during processing. Thus, the true numbers of apoptotic cells are likely to exceed those observed using stained coverslips.
FIG. 5.
Mitotic activities of KB cells incubated with HRgpA for 48 h. Cells were grown on coverslips, fixed, permeabilized, and stained with DAPI as described in Materials and Methods. At least 1,500 cells were examined on each coverslip, and the mitotic figures are expressed as the percentage (+ standard error [SE]) of the total cell number at each protease dilution. Typical results from two experiments are shown. ∗, P < 0.05; ∗∗, P < 0.005.
FIG. 6.
DAPI staining of KB cells incubated for 48 h with 1/128 dilution of HRgpA showing an apoptotic cell (A) and the same field focusing on the cell monolayer showing mitotic figures (B), with the position of the apoptotic cell marked with an arrow. Magnification, ×980; bar = 10 μm.
All protease treatments stimulated metabolic activity, measured by reduction of MTT, above control values (Table 2). The two highest protease concentrations produced the maximum increases of approximately 25% (P < 0.05), with progressively reduced stimulation as the protease concentration decreased. Concentration-dependent patterns were seen with both active and TLCK-treated HRgpA, but although stimulation of metabolic activity expressed as a percentage of the untreated control value was consistently greater for active HRgpA than for TLCK-treated HRgpA at all except the highest protease concentration, these differences were not statistically significant.
TABLE 2.
Metabolic activity of KB cells incubated with HRgpA or TLCK-HRgpA for 48 h
| Dilution of HRgpA | BApNA activity (U ml−1) | MTT absorbance (% of control)a
|
|||
|---|---|---|---|---|---|
| HRgpA | P | TLCK-HRgpA | P | ||
| 1/8 | 1.0 | 124.7 | <0.05 | 125.3 | <0.05 |
| 1/16 | 0.5 | 127.3 | <0.05 | 125.5 | <0.05 |
| 1/32 | 0.25 | 120.7 | <0.05 | 110.1 | NS |
| 1/64 | 0.125 | 116.0 | NS | 109.9 | NS |
| 1/128 | 0.0625 | 115.6 | NS | 108.0 | NS |
| 1/256 | 0.0312 | 114.1 | NS | 102.8 | NS |
| 1/512 | 0.0156 | 106.3 | NS | 103.4 | NS |
| 1/1,024 | 0.0078 | 106.2 | NS | 105.6 | NS |
Mean MTT absorbance values (570 nm) from groups of eight wells are expressed as percentages of the DMEM control values. NS, not significant. The results shown are representative of two experiments.
DISCUSSION
It is well recognized that the cysteine proteases of P. gingivalis are able to proteolytically degrade a wide range of host macromolecules in plasma as well as structural components of the extracellular matrix. However the molecular mechanisms by which P. gingivalis interacts with the resident host cells of the periodontal tissues is an area of growing interest. In particular, the cysteine proteases of this organism have been shown to exert deregulatory effects on host cells via a number of mechanisms, including proteolytic removal or activation of cell surface receptors (27), disruption of cell-cell and extracellular matrix-integrin interactions (3, 21, 35), and up-regulation of matrix metalloprotease and proinflammatory cytokine release (12, 27). These cellular effects have reinforced the view that the cysteine proteases of this organism play a significant role in the perturbation of immune, inflammatory, and tissue homeostatic mechanisms through proteolysis of host proteins in the extracellular environment. However the data in the present report are the first to suggest that proteases of this bacterium may also have a role to play in the intracellular environment of host cells.
Upon exposure to a range of human epithelial cell types, HRgpA is rapidly internalized and localizes within the nucleus. The distribution of a ring of more brightly stained areas within the nuclear membrane of some HRgpA-treated cells within 1 h of treatment could reflect entry at nuclear membrane pores and controlled transport to specialized nuclear sites. This contrasts with the generalized cellular distribution of RgpAcat, which lacks the adhesin component. The reduction in nuclear staining intensity of HRgpA seen after 4 h may indicate export from the nucleus, degradation, or modification of the antibody binding epitope. The molecular mechanisms responsible for cell entry and nuclear localization of HRgpA are not known, although interrogation of motif databases has failed to demonstrate the presence of a known nuclear localization signal within the HRgpA amino acid sequence. Furthermore, our preliminary studies using inhibitors of receptor-mediated endocytosis and intracellular trafficking pathways failed to prevent nuclear localization of this protease, and similar data (not shown) were obtained at both 37 and 4°C, indicating that this process does not require cellular metabolism.
The similar patterns of localization observed with unheated and heat-treated culture supernatants from strain W50, and the comparable nuclear localization obtained using active and TLCK-treated HRgpA, indicate that both cell entry and nuclear localization were independent of proteolysis. Together with the nuclear targeting seen with the recombinant β chain, these data suggest that the β component of the bacterial protease may possess hitherto-unrecognized properties. There is good evidence that the β component is involved in adhesion and binding processes, possibly via its hydrophilic repeat sequences, characteristic of bacterial proteins which bind to mammalian extracellular matrix (5). Our previous observations that the adhesin component of HRgpA mediates targeted destruction of fibronectin in fibroblast cultures (35) supports this property. Binding to fibronectin, laminin, and fibrinogen, with degradation by the functional protease domain, has also been demonstrated in cell-free assays (30). Furthermore, the importance of the β component has been shown in vivo by suppression of recolonization by P. gingivalis in patients passively immunized via subgingival application of a MAb to a determinant on this polypeptide (2).
The phenomenon of internalization and intracellular targeting has been described previously with single polypeptides of viral origin and with a component of the Antennapedia homeodomain, a Drosophila transcription factor (34). All these molecules contain domains which have the ability to cross biological membranes without involving classical receptor-, transporter-, or endosome-mediated mechanisms. Experimentally produced chimeric proteins capable of intranuclear delivery have also been synthesized. For example EtXB-Pol consists of the nontoxic B subunit of E. coli heat-labile enterotoxin fused to a peptide derived from the DNA polymerase of herpes simplex virus type 1 (26). In contrast with the peptides mentioned above, EtXB-Pol appears to enter cells via endocytosis. Cleavage of the two components occurs in acidic organelles, and the Pol peptide, which contains a sequence resembling a nuclear localization signal, enters the nucleus while EtXB remains in the cytoplasm.
The basic subunit organization of the heterodimeric HRgpA protease is reminiscent of some bacterial protein toxins whose cell-damaging mechanisms have received much attention, e.g., Pseudomonas aeruginosa exotoxin, diphtheria toxin, E. coli enterotoxin, cholera toxin, and Shiga-like toxins. All these bacterial toxins possess an enzymatic A subunit linked to one or more B subunits which mediate binding to cell surface receptors (25). They accumulate in early endosomes before following different pathways of intracellular membrane trafficking in which the catalytically active A fragment is cleaved from the B subunit(s) and enters the cytosol, leading to ADP ribosylation of specific cytoplasmic target proteins and ultimately cell death. Consequently, in these instances the B subunit plays an important role in intracellular delivery. The fate of the internalized HRgpA heterodimer will require further study to determine whether the catalytic α component dissociates from the adhesin component and where this occurs.
The rapid nuclear entry of HRgpA contrasted with the peripheral nuclear targeting seen both with MAb 1A1-reactive material in W50 culture supernatant and with rRgpAβ. The absence of MAb 1A1-reactive material localized in the nuclei of KB cells incubated with a combination of culture supernatant and HRgpA indicates that material present in the culture supernatant may have physically interfered with nuclear entry of HRgpA or influenced the nuclear translocation pathway. Vesicles or membrane-bound enzyme, which constitute approximately 60 to 70% of the total BApNA activity of the culture supernatant used in this study, could contribute to such interference. Alternatively, interference with nuclear entry of HRgpA could be due to the formation of higher-molecular-mass complexes of the soluble enzyme. As the BApNA activity of the culture supernatant which produced optimum responses in KB cells was only approximately 20% of that of HRgpA, it could be argued that the use of supernatant with a comparable protease activity would result in nuclear entry rather than localization around the nuclear membrane. However, higher concentrations of the supernatant could not be tested with KB cells, as prolonged incubation resulted in morphological changes, including rounding and detachment of adherent cells, possibly because of the action of other bacterial components present in the culture supernatant. Nevertheless, HeLa cells, which were more resistant than KB cells to detachment at high concentrations of supernatant, still retained differences between their staining patterns with supernatant and HRgpA when these cells were exposed to comparable BApNA levels.
Internalization of viable P. gingivalis organisms by epithelial cells in vitro has been described previously by a number of groups (1, 13, 23), and intracellular bacteria have been shown to release outer membrane vesicles both in monolayer cultures of KB cells (19) and in multilayered cultures of human pocket epithelium (33). Interestingly, the perinuclear localization of the viable whole organisms described in gingival epithelial cells and KB cells (1, 19) resembled the nuclear targeting we observed with MAb 1A1-reactive material in W50 culture supernatant. Belton et al. (1) stated that the perinuclear region of keratinocytes contains many organelles, but the functional significance of the association remained to be determined. The presence of adhesin chain proteins, or other proteins with homologous domains, on the surfaces of whole P. gingivalis bacteria may have contributed to this phenomenon.
KB cells have been widely used in studies of P. gingivalis invasion of epithelial cells and contributed to the choice of this cell line for the majority of the present studies. While it is recognized that expression of KB cell surface receptors for P. gingivalis may differ from that of nontransformed cells (20), significant similarities have also been reported with human gingival epithelial cells (16). With the exception of some variation in the protease treatment time and dose to achieve optimum results, we consistently observed the phenomenon of nuclear entry of HRgpA with cell lines of both oral and nonoral origin.
The stimulation of metabolic and mitotic activity, accompanied by the presence of apoptosis in epithelial cells following exposure to HRgpA, indicates that cells are responding to a balance of proliferative and cytotoxic signals and is likely to reflect perturbation in the control of proteins regulating cell cycle progression. Apoptosis in host cells is a well-recognized response to a number of bacterial pathogens (41), although the nature of the apoptotic mediators produced by P. gingivalis appears to be variable. Butyric acid, as well as other volatile fatty acids released as byproducts of anaerobic metabolism by P. gingivalis, are known to induce apoptosis (22) and are likely candidates for the heat-stable factors in culture supernatant reported to cause lymphocyte apoptosis (15). Significantly increased apoptosis has also been observed in both epithelial cells and human gingival fibroblasts exposed to active P. gingivalis protease preparations (3, 39), although in the latter case, this did not occur when the protease was inactivated. The similar responses with both active and TLCK-treated HRpgA in the present study indicate that the action of this molecule is independent of its proteolytic activity, and it is thus unlikely to be operating via activation of protease-activated receptors on epithelial cells (27).
As epithelial cells form the first barrier to bacterial invasion, the outcome of their interaction with pathogenic organisms will inevitably influence the progression of disease. It is tempting to speculate that the stimulation of cell division and apoptosis by P. gingivalis proteases in vivo could contribute not only to pocket deepening and the overall remodeling of gingival tissue in periodontal disease but also possibly to more widespread effects in susceptible individuals. Further clarification of the changes in the gene expression and concomitant alterations to the epithelial cell phenotype following exposure to HRgpA are required to substantiate these possibilities.
Acknowledgments
This work was supported by grants from the Special Trustees of the Royal London Hospital to M.A.S. and the Medical Research Council (grant no. PG 9318173) to M.A.C.
We thank Ali Alsam for his assistance with preparation of the Photoshop images.
Editor: D. L. Burns
REFERENCES
- 1.Belton, C. M., K. T. Izutsu, P. C. Goodwin, Y. Park, and R. J. Lamont. 1999. Fluorescent image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell. Microbiol. 1:215-223. [DOI] [PubMed] [Google Scholar]
- 2.Booth, V., F. P. Ashley, and T. Lehner. 1996. Passive immunization with monoclonal antibodies against Porphyromonas gingivalis in patients with periodontitis. Infect. Immun. 64:422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Z., C. A. Casiano, and H. M. Fletcher. 2001. Protease-active extracellular protein preparations from Porphyromonas gingivalis W83 induce N-cadherin proteolysis, loss of cell adhesion, and apoptosis in human epithelial cells. J. Periodontol. 72:641-650. [DOI] [PubMed] [Google Scholar]
- 4.Cridland, J. C., V. Booth, F. P. Ashley, M. A. Curtis, R. F. Wilson, and P. Shepherd. 1994. Preliminary characterisation of antigens recognised by monoclonal antibodies raised to Porphyromonas gingivalis and by sera from patients with periodontitis. J. Periodontal Res. 29:339-347. [DOI] [PubMed] [Google Scholar]
- 5.Curtis, M. A. 1997. Analysis of the protease and adhesin domains of the PrpR1 of Porphyromonas gingivalis. J. Periodontal Res. 32:133-139. [DOI] [PubMed] [Google Scholar]
- 6.Curtis, M. A., J. Aduse-Opoku, and M. Rangarajan. 2001. Cysteine proteases of Porphyromonas gingivalis. Crit. Rev. Oral Biol. Med. 12:192-216. [DOI] [PubMed] [Google Scholar]
- 7.Curtis, M. A., J. Aduse-Opoku, J. M. Slaney, M. Rangarajan, V. Booth, J. Cridland, and P. Shepherd. 1996. Characterization of an adherence and antigenic determinant of the ArgI protease of Porphyromonas gingivalis which is present on multiple gene products. Infect. Immun. 64:2532-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis, M. A., H. K. Kuramitsu, M. Lantz, F. L. Macrina, K. Nakayama, J. Potempa, E. C. Reynolds, and J. Aduse-Opoku. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J. Periodontal Res. 34:464-472. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, M. A., M. Ramakrishnan, and J. M. Slaney. 1993. Characterization of the trypsin-like enzymes of Porphyromonas gingivalis W83 using a radiolabelled active-site-directed inhibitor. J. Gen. Microbiol. 139:949-955. [DOI] [PubMed] [Google Scholar]
- 10.Curtis, M. A., J. M. Slaney, R. J. Carman, and N. W. Johnson. 1991. Identification of the major surface protein antigens of Porphyromonas gingivalis using IgG antibody reactivity of periodontal case-control serum. Oral Microbiol. Immunol. 6:321-326. [DOI] [PubMed] [Google Scholar]
- 11.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shepherd, N. Paramanov, and E. F. Hounsell. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeCarlo, A. A., H. E. Grenett, G. J. Harber, L. J. Windsor, M. K. Bodden, B. Birkedal-Hansen, and H. Birkedal-Hansen. 1998. Induction of matrix metalloproteinases and a collagen-degrading phenotype in fibroblasts and epithelial cells by secreted Porphyromonas gingivalis proteinase. J. Periodontal Res. 33:408-420. [DOI] [PubMed] [Google Scholar]
- 13.Dorn, B. R., W. A. Dunn, Jr., and A. Progulske-Fox. 1999. Invasion of human coronary artery cells by periodontal pathogens. Infect. Immun. 67:5792-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 15.Geatch, D. R., J. I. Harris, P. A. Heasman, and J. J. Taylor. 1999. In vitro studies of lymphocyte apoptosis induced by the periodontal pathogen Porphyromonas gingivalis. J. Periodontal Res. 34:70-78. [DOI] [PubMed] [Google Scholar]
- 16.Han, Y. W., W. Shi, G. T.-J. Huang, S. K. Haake, N.-H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanley, S. A., J. Aduse-Opoku, and M. A. Curtis. 1999. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect. Immun. 67:1157-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillmann, G., S. Dogan, and W. Geurtsen. 1998. Histopathological investigation of gingival tissue from patients with rapidly progressive periodontitis. J. Periodontol. 69:195-208. [DOI] [PubMed] [Google Scholar]
- 19.Houalet-Jeanne, S., P. Pellen-Mussi, S. Tricot-Doleux, J. Apiou, and M. Bonnaure-Mallet. 2001. Assessment of internalization and viability of Porphyromonas gingivalis in KB epithelial cells by confocal microscopy. Infect. Immun. 69:7146-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huard-Delcourt, A., L. Du, P. Pellen-Mussi, S. Tricot-Doleux, and M. Bonnaure-Mallet. 1998. Adherence of Porphyromonas gingivalis to epithelial cells: analysis by flow cytometry. Eur. J. Oral Sci. 106:938-944. [DOI] [PubMed] [Google Scholar]
- 21.Katz, J., V. Sambandam, J. H. Wu, S. M. Michalek, and D. F. Balkovetz. 2000. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect. Immun. 68:1441-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurita-Ochiai, T., K. Ochiai, and K. Fukushima. 1998. Volatile fatty acid, metabolic by-product of periodontopathic bacteria, induces apoptosis in WEHI 231 and RAJI B lymphoma cells and splenic B cells. Infect. Immun. 66:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lord, J. M., D. C. Smith, and L. M. Roberts. 1999. Toxin entry: how bacterial proteins get into mammalian cells. Cell. Microbiol. 1:85-91. [DOI] [PubMed] [Google Scholar]
- 26.Loregian, A., E. Papini, B. Satin, H. S. Marsden, T. R. Hirst, and G. Palu. 1999. Intranuclear delivery of an antiviral peptide mediated by the B subunit of Escherichia coli heat-labile enterotoxin. Proc. Natl. Acad. Sci. USA 96:5221-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lourbakos, A., J. Potempa, J. Travis, M. R. D'Andrea, P. Andrade-Gordon, R. Santulli, E. J. Mackie, and R. N. Pike. 2001. Arginine-specific protease from Porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion. Infect. Immun. 69:5121-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millar, D. J., E. E. Scott, J. M. Slaney, S. U, P. Benjamin, and M. A. Curtis. 1993. Production and characterisation of monoclonal antibodies to the principal sonicate antigens of Porphyromonas gingivalis W50. FEMS Immunol. Med. Microbiol. 7:211-222. [DOI] [PubMed] [Google Scholar]
- 29.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 30.Pike, R. N., J. Potempa, W. McGraw, T. H. T. Coetzer, and J. Travis. 1996. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J. Bacteriol. 178:2876-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangarajan, M., S. J. M. Smith, S. U, and M. A. Curtis. 1997. Biochemical characterization of the arginine-specific proteases of Porphyromonas gingivalis W50 suggests a common precursor. Biochem. J. 323:701-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sandros, J., P. N. Papapanou, U. Nannmark, and G. Dahlen. 1994. Porphyromonas gingivalis invades human pocket epithelium in vitro. J. Periodontal Res. 29:62-69. [DOI] [PubMed] [Google Scholar]
- 34.Schwarze, S. R., and S. F. Dowdy. 2000. In vivo protein transduction: intracellular delivery of biologically active proteins, compounds and DNA. Trends Pharmacol. Sci. 21:45-48. [DOI] [PubMed] [Google Scholar]
- 35.Scragg, M. A., S. J. Cannon, M. Rangarajan, D. M. Williams, and M. A. Curtis. 1999. Targeted disruption of fibronectin-integrin interactions in human gingival fibroblasts by the R1 protease of Porphyromonas gingivalis W50. Infect. Immun. 67:1837-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scragg, M. A., S. J. Cannon, and D. M. Williams. 1996. The secreted products of Porphyromonas gingivalis alter human gingival fibroblast morphology by selective damage to integrin-substrate interactions. Microb. Ecol. Health Dis. 9:167-179. [Google Scholar]
- 37.Slaney, J. M., M. Rangarajan, J. Aduse-Opoku, S. Fawell, I. Darby, D. Kinane, and M. A. Curtis. 2002. Recognition of the carbohydrate modifications to the RgpA protease of Porphyromonas gingivalis by periodontal patient serum IgG. J. Periodontal Res. 37:215-222. [DOI] [PubMed] [Google Scholar]
- 38.Travis, J., R. Pike, T. Imamura, and J. Potempa. 1997. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J. Periodontal Res. 32:120-125. [DOI] [PubMed] [Google Scholar]
- 39.Wang, P.-L., S. Shirasu, M. Shinohara, M. Daito, M. Oido, Y. Kowashi, and K. Ohura. 1999. Induction of apoptosis in human gingival fibroblasts by a Porphyromonas gingivalis protease preparation. Arch. Oral Biol. 44:337-342. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zychlinsky, A., and P. Sansonetti. 1997. Apoptosis in bacterial pathogenesis. J. Clin. Investig. 100:493-496. [DOI] [PMC free article] [PubMed] [Google Scholar]