Abstract
The suprabasin gene is a novel gene expressed in mouse and human differentiating keratinocytes. We identified a partial cDNA encoding suprabasin using a suppression subtractive hybridization method between the proliferative basal and differentiating suprabasal populations of the mouse epidermis. A 3′ gene-specific probe hybridized to transcripts of 0.7- and 2.2-kb pairs on Northern blots with specific detection in differentiated keratinocytes of stratified epithelia. The mouse gene was mapped to chromosome 7 by fluorescence in situ hybridization. This region is syntenic to human chromosome band 19q13.1, which contained the only region in the data bases with homology to the mouse suprabasin sequence. During embryonic mouse development, suprabasin mRNA was detected at day 15.5, coinciding with epidermal stratification. Suprabasin was detected in the suprabasal layers of the epithelia in the tongue, stomach, and epidermis. Differentiation of cultured primary epidermal keratinocytes with 0.12 mM Ca2+ or 12-O-tetradecanoylphorbol-13-acetate treatment resulted in the induction of suprabasin. The 2.2-kb cDNA transcript encodes a protein of 72 kDa with a predicted isoelectric point of 6.85. The translated sequence has an amino-terminal domain, a central domain composed of repeats rich in glycine and alanine, and a carboxyl-terminal domain. The alternatively spliced 0.7-kb transcript encodes a smaller protein that shares the NH2- and COOH-terminal regions but lacks the repeat domain region. Cross-linking experiments indicate that suprabasin is a substrate for transglutaminase 2 and 3 activity. Altogether, these results indicate that the suprabasin protein potentially plays a role in the process of epidermal differentiation.
Cells losing contact with the underlying basement membrane will undergo differentiation to form the mature epidermis. During mouse development, at day 14 the stratified epidermal layers begin to appear, and the keratinocytes follow a complex program of differentiation with the formation of the spinous and granular layers, leading to a highly stratified, water impermeable barrier at birth on day 19 (1, 2). The final phase of this complex differentiation program is the formation of cornified envelopes (CE).1 This process includes granular cell death, destruction of all organelles, ε-(γ-glutamyl)lysine isopeptide covalent cross-linking of cornified envelope precursors through the action of calcium-dependent transglutaminases (TGases), and attachment of lipid molecules to the cross-linked envelope (3, 4).
In the last two decades, a group of epidermal-specific genes that are components of the CE have been identified and characterized (reviewed in Refs. 5–7). In humans, many of these epidermal genes are closely linked in a 2.5-Mbp region, located in chromosome 1q21, that has been termed the epidermal differentiation complex (EDC) (8). Several of the reported mouse homolog genes have been mapped to the murine EDC in a syntenic region of chromosome 3. Three types of protein families that contribute to the cornification process are clustered in the EDC (9). The first, with proteins such as involucrin, loricrin, late envelope proteins, and small proline-rich proteins, is characterized by relatively small sizes with short tandem peptide repeats in the central region. The second family (fused-type), represented by profilaggrin and repetin, has EF-hand domains (Ca2+-binding domains) at the NH2-terminal region followed by multiple tandem repeats. The third family is represented by members of the S100 family and is characterized by presenting Ca2+-binding EF-hand domains. There are other epidermal genes that are incorporated into the CE that do not localize to the EDC, such as those for sciellin, envoplakin, and periplakin (10–12). It has been proposed that the cell envelope formation is a highly coordinated process in which cross-linking of specific groups of proteins occur in a sequential order (4). In recent reports from targeting experiments it has become clear that, in the absence of major envelope precursor proteins, other factor(s) are able to substitute or replace their function to culminate in the formation of a normal or “quasi-normal” cornified envelope (13–16). Recently, additional genes have been identified by different approaches: subtractive hybridization, rapid analysis of gene expression, positional cloning, and EST data base searches (17–21). It has been hypothesized that some of these newly characterized proteins would be able to compensate in function for other well known CE precursors (13, 19).
The novel suprabasin, which we have characterized in this study, is structurally similar to involucrin and loricrin; the open reading frame codes for a small protein rich in glycine, with short tandem repeats in the central region. The suprabasin gene is expressed in vivo in the differentiating layers of cornifying epithelia, and the expression can be induced in vitro by treatment of keratinocytes with Ca2+ or with TPA, dependent on PKC signaling. We propose that suprabasin is a new member of the epidermal-specific proteins that is a substrate for TGase activity and potentially plays a role in the epidermal differentiation process.
EXPERIMENTAL PROCEDURES
Suppression Subtractive Hybridization (SSH)
SSH (Clontech; Ref. 22) was performed following instructions of the manufacturer using basal cell RNA as a “driver” and suprabasal cell RNA as “tester.” The primary mouse basal and suprabasal keratinocytes were obtained from neonatal skins that were trypsinized overnight at 4 °C, and separated by a discontinuous Percoll gradient (23). From this screen, we identified a partial cDNA sequence (262 bp) that corresponded to part of the COOH-terminal end and 3′-untranslated region of suprabasin (nucleotides 2014 –2277). The complete cDNA sequence was obtained performing 5′-rapid amplification of cDNA ends (RACE) (Clontech). A canonical polyadenylation signal and poly(A)+ tail were identified at the 3′ end. The complete mRNA sequence was deposited in GenBankTM (accession no. AY115494).
Mapping of the Transcriptional Start Site
To determine the transcription start site, we performed the RACE method using the SMART RACE cDNA amplification kit (Clontech). The gene-specific oligonucleo-tide (5′-CTGCCCGGGCAGGCAGAGTCCC-3′) utilized was located at +100 bp from the putative translation initiation AUG in the coding sequence of suprabasin. The final PCR product was cloned and sequenced to determine the 5′ mRNA sequence.
Cloning and Analysis of Suprabasin Genomic Sequence
A genomic DNA region of ~110 kb was cloned through screening of a mouse VJ/129 BAC library (Genome Systems Inc.) using a 1.7-kb suprabasin cDNA as a probe. To analyze the genomic structure of the suprabasin gene, we performed PCR with the oligonucleotides corresponding to the 5′ end and 3′ end of cDNA and the BAC genomic DNA as a template. Comparison of the genomic and cDNA sequences determined the exon/intron boundaries and the sizes of intronic regions. To obtain further 5′ upstream and 3′ downstream sequence, we used a Chromosome Walking kit (Clontech) with gene-specific primers and genomic mouse DNA following the instructions specified by the manufacturer. We used BLAST analysis for sequence homology searches, made available through the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST/).
Chromosomal Localization by FISH
The genomic mouse supraba-sin BAC clone was labeled with digoxigenin dUTP by nick translation (carried out by Genome Systems Inc.). The labeled probe was combined with sheared mouse DNA and hybridized to normal metaphase chromosomes derived from mouse embryo fibroblast cells in a solution containing 50% formamide, 10% dextran sulfate, and 2× SSC. Specific hybridization signals were detected by incubating the hybridized slides in fluoresceinated antidigoxigenin antibodies followed by counterstain-ing with 4,6-diamidino-2-phenylindole.
Plasmids
Constructs were prepared by cloning a PCR-amplified coding region of suprabasin tagged with EcoRI and SalI sites into pGEMT-easy (Promega; pGEM/suprabasin) and pEGFP-C1 vector (Clontech; pGFP-suprabasin). The truncated sequences were generated by PCR using the following oligonucleotides: RI5′ suprabasin (5′-gcggaattctATGTATCTTGTCAGTTTGCTCAGCTCCTGC-3′) and 462Rsuprabasin (5′-gtggtcgacCCCCTGACCAAACTTCCCTGCTTCA-CTGCC-3′) for pGFP/suprabasin (aa 1–144); RI5′ suprabasin and 1304Rsuprabasin (5′-gtggtcgacCAGCCCCTTGCACCAGTCTGCCTC-3′) for pGFP/suprabasin (aa 1–425); RI5′ suprabasin and 1924Rsupra-basin (5′-gaggtcgacTCATTTCCTGGCCAGCAGCATGGTGGACAC-3′) for pGFP/suprabasin (aa 1–612); 427Fsuprabasin (5′-gaggaattctTCAG-GGGGGCAGTGAAGCAGGGAAG-3′) and 1924Rsuprabasin for pGFP/suprabasin (aa 133–612). Oligonucleotides 427Fsuprabasin and 1304Rsuprabasin were used to generate pGFP/suprabasin (aa 133–425) and RI5′ suprabasin and SalI3′ suprabasin for the full-length pGFP/suprabasin (aa 1–700). All PCR reactions were performed for 30 cycles of denaturation at 94 °C for 45 s, annealing at 55 °C for 45 s, and extension at 68 °C for 90 s.
Cell Culture and Transfection
Primary mouse keratinocytes were isolated from BALB/c trypsinized newborn mouse skins and grown in Eagle’s minimal essential medium lacking Ca2+ with 8% Chelex-treated fetal bovine serum (24 –26). Ca2+ concentrations were determined by analysis in an atomic absorption spectrophotometer. Unless otherwise indicated, the Ca2+ concentration of the medium was adjusted to 0.05 μM to maintain a basal cell-like population of undifferentiated cells. Cells were treated with different kinase inhibitors at 10 μM concentration for specific time periods. The inhibitors used were: the PKC inhibitor GF109203X (GF, bisindolylmaleimide I, Alexis), the PKA inhibitor H89 (N-[2–99p-bromocinnamyl-0-amino-0-ethyl]-5-isoquino-linesulfonamide, Alexis), or a CaM kinase II inhibitor (KN62, 1-[N,O-bis-(5-isoquinolinesulfonyl)-N-methyl-l-tyrosyl]-4-phenylpiperazine, Calbiochem). Primary mouse keratinocytes were transfected using the FuGENE6 reagent (Roche Molecular Biochemicals). Typically, 0.5 μg of GFP/suprabasin, GFP/suprabasin truncated constructs, or pEGFP-C1 control construct were transfected into cells plated and cultured in two-well chamber slides coated with rat tail collagen (0.1 mg/ml). After 4 h incubation, the cells were treated with 15% glycerol in KSF medium (Invitrogen) for 3.5 min and then maintained in medium with 0.05, 0.12, or 1.4 mM Ca2+.
Human keratinocytes (NHEK) were cultured in vitro in serum-free keratinocyte medium as described by Jang et al. (27), and were induced to differentiate by increasing the Ca2+ concentration in the media to 1.4 mM or by treatment with the PKC inducer TPA (100 nM). Cells were cultured for different time periods as indicated.
Northern Blots
Total RNA were isolated from mouse primary basal and suprabasal keratinocytes, and mouse and human keratinocytes differentiated in vitro using TRIzol (Invitrogen). The RNA samples (1–3 μg) were electrophoresed in 1.2% agarose/methymercuryhydroxide gels, electroblotted to nylon membranes, and hybridized according to Church and Gilbert (28). mRNA blots for human and mouse adult tissues were purchased from Clontech and used according to instructions. The probes used were 262-bp coding (mouse) and 500-bp coding (human) suprabasin cDNAs. After exposure, hybridized probes were removed by boiling filters in 0.1× SCC, 0.1% SDS. All blots were re-hybridized with a cDNA probe for glyceraldehyde-3-phosphate dehydrogenase (29) to control for RNA loading and integrity.
Radioactive in Situ Hybridization
RNA probes corresponding to the sense and antisense strands of mouse suprabasin partial cDNA (262 bp from 2014 to 2277) (pGEMT-easy/3′ suprabasin) were prepared using T7 and Sp6 RNA polymerase and 35S-labeled UTP. Sections of mouse embryos were subjected to in situ hybridization as described by Mackem and Mahon (Ref. 30 and molecular histology).
Suprabasin Expression, Purification, and Structure Predictions
The region coding for amino acids 1–612 was subcloned into a pET28b vector to generate a recombinant protein. The recombinant clone was overexpressed in BL21(DE3)RIL. The recombinant protein contains a His tag and a T7 tag at the NH2 terminus of the protein. The soluble and insoluble forms of the recombinant protein were purified over a nickel-nitrilotriacetic acid-agarose column (Qiagen) and Mono Q-FPLC. We analyzed the predicted protein sequence using the ISREC search tools (www.isrec.isb-sib.ch/), the Baylor College of Medicine Search launcher (searchlauncher.bcm.tmc.edu/), and MacVector 6.5.3.
TGase Assay and Western Analysis
To examine the cross-linking activity by TGase, 300 ng of recombinant suprabasin (aa 1–612) was incubated with 0.4 μg and 1.2 μg of TGase 2 (Sigma) or 3 μg and 6 μg of TGase 3 in buffer containing 20 mM CaCl2 and 5 mM dithiothreitol for 30 min at 37 °C. The reaction was stopped by addition of 6× SDS loading dye and heated at 90 °C for 3min. The products were separated on 4 –20% SDS-polyacrylamide gel. Involucrin was included as a positive control for the TGase activity. The gel was electroblotted to a polyvinylidene difluoride membrane, and an antibody against T7 (In-vitrogen) was used in Western analysis to determine the oligomerization of suprabasin by TGase.
RESULTS
Cloning and Characterization of Suprabasin cDNA
SSH, a PCR-based cDNA subtraction method (Ref. 31; Clontech) was used to compare the basal and suprabasal mRNA populations from neonatal epidermis. From a screen of 96 hybridization selected clones, the sequence of the candidate cDNAs was used to search the data bases, and to determine whether the isolated clones were novel and/or whether they contained motifs found in other factors. Several novel genes as well as known supra-basal-specific genes were identified. We used the partial cDNAs of a selected number of novel genes as probes in Northern blots containing poly(A)+ mRNAs from basal and supra-basal cells separated by Percoll gradients (23). Only the cDNAs that hybridized exclusively with the suprabasal mRNA fraction were further characterized. We utilized 5′-RACE methods to obtain the full-coding sequence of the novel suprabasal-specific gene that we termed suprabasin (Fig. 1A). A data base comparison with the full-length or repeat region sequence of suprabasin using the program BLAST did not reveal any homology to known protein sequences. The search of publicly available EST data bases revealed homology to a several overlapping mouse EST sequences (AA530183, AA727702, and 791703). By searching the data base of human genomic sequences, we identified a region in human chromosome 19 with homology to the mouse suprabasin cDNA sequence. To amplify the human homologue of suprabasin, we designed the oligonucleotides corresponding the NH2- and COOH-terminal region of suprabasin, which were conserved between the mouse cDNA sequence and the human genomic DNA sequence (forward, 5′-GAAGGGATCAACCGAGGGCTGAGCAATGCAG-3′; reverse, 5′-GCTCATAATGGGGTCAACCAAGCCAGCAAGG-3′). Using a RT-PCR method, we amplified a 500-bp DNA fragment from human skin RNA. Sequencing results showed that amplified fragment is identical to human EST cDNA BG742735. We performed Northern analysis of human suprabasin using the 0.5-kb amplified DNA fragment as a probe.
Fig. 1. Nucleotide and amino acid sequences of mouse suprabasin.

A, the nucleotide sequence of mouse suprabasin cDNA (2277 bp) was aligned with the predicted open reading frame amino acid sequence. Three open triangles indicate the sites of splicing (exon/intron boundaries), and the double-underlined nucleotides indicate the canonical poly(A) addition signal. B, the amino acid sequence of mouse suprabasin has been aligned to show the central repetitive domain region (boxed). The dotted line indicates the different repeat sequence region. The shaded residues highlight a predicted transmembrane region domain. The underlined sequences in the COOH-terminal domain indicate potential protein kinase C phosphorylation sites. Numbers in parentheses at the right side indicate number of residues.
The 2277-bp mouse suprabasin transcript included 30 bp of 5′-untranslated sequence and 144 bp of 3′-untranslated sequence. The transcript had a polyadenylation signal at 2251 bp, a poly (A)+ tail, and one putative open reading frame that translated into a 700-amino acid protein. Analysis of the open reading frame sequence by ISREC search tools (www.isrec.isb-sib.ch/), the Baylor College of Medicine Search launcher (searchlauncher.bcm.tmc.edu/), and MacVector 6.5.3. predicted a protein with a mass of 72 kDa and an isoelectric point for the unmodified protein of 6.85. The programs also identified a potential transmembrane domain in the amino-terminal region (shaded area in Fig. 1B) and a series of glycine- and histidine-rich short tandem repeats in the central domain (boxed area in Fig. 1B). Potential PKC phosphorylation sites were identified (underlined in Fig. 1B). Comparison of the open reading frame to the data bases also revealed 2 casein kinase II phosphorylation sites and 73 potential N-myristoylation sites. Analysis of the nucleotide sequence surrounding the AUG initiator codon for the suprabasin open reading frame (AACAACATGT) conformed to the consensus sequence for initiation of translation (GCCACCAUGG) in having a purine (A) at position –3, but lacked the consensus G found at position +4 (32). Further analysis must be done to determine whether there is a modulation of translation of suprabasin as a result of the primary sequence surrounding the AUG codon.
In Situ and Northern Blot Analysis of Suprabasin Expression
In situ hybridization of sagittal sections of E17.5 mouse embryos showed epidermal-specific expression (Fig. 2A). Higher magnification showed that the expression detected with the antisense mouse suprabasin probe was specific to the suprabasal-differentiated layers of interfollicular trunk and tail epidermis (Fig. 2, D and G). Expression was also detected in the stratified layers in the tongue and palate (Fig. 2J). In situ hybridization of an adult mouse mixed tissue block detected expression only in stomach tissue, with no detectable expression in kidney, liver, brain, spleen, or heart (data not shown). No expression was observed when using the control sense suprabasin probe (Fig. 2, B, E, H, and K).
Fig. 2. Expression of suprabasin.
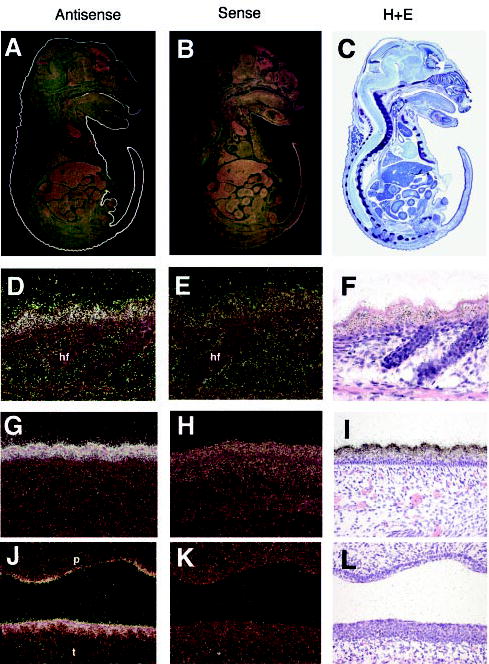
In situ hybridization was performed with antisense (A) and sense (B) suprabasin probes on sagittal sections of E17.5 mouse embryos. C, hematoxylin and eosin (H+E) staining of E17.5 sagittal sections. D, G, and J, 20× magnification of trunk skin (D), tail skin (G), and palate (J) with antisense suprabasin. Corresponding hybridization with sense suprabasin (E, H, and K) and hematoxylin and eosin stain (F, I, and L). hf, hair follicle; p, palate; t, tongue.
Analysis of suprabasin expression in mouse primary basal and suprabasal keratinocytes isolated by discontinuous Percoll gradient showed specific expression of the suprabasin 2.2-kb transcript in the differentiated epidermal cell layers (Fig. 3A). We identified a smaller 0.7-kb transcript that was also exclusively expressed in the differentiated layer.
Fig. 3. Northern blot analysis of mouse suprabasin.
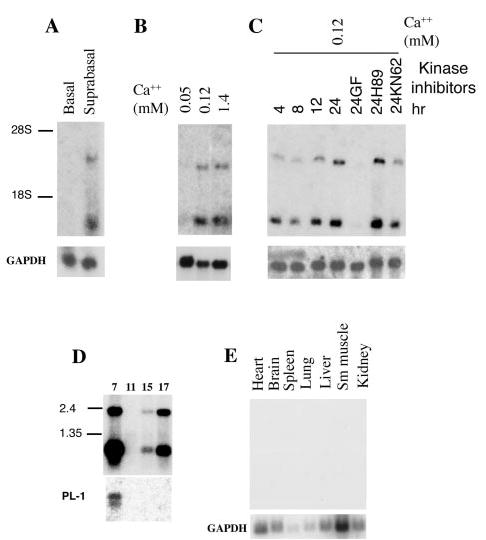
A, expression of suprabasin in the epidermis of newborn mice. Northern blot of poly(A)+ mRNA (2 μg) from basal and differentiated keratinocytes isolated by discontinuous Percoll gradient from neonatal epidermis. The position of the 18 and 28 S ribosomal RNAs are shown. B, expression of suprabasin in primary mouse keratinocytes cultured and differentiated in vitro by addition of Ca2+. C, time course of suprabasin expression in cultured mouse keratinocytes and after 24 h of treatment with different kinase inhibitors: GF (PKC inhibitor), H89 (PKA inhibitor), or KN62 (CaM kinase II inhibitor). D, mouse embryonic mRNA blot (Clontech). Panel shows 2 μg of poly(A)+ per lane from four embryonic developmental stages (7-, 11-, 15-, and 17-day embryos). RNA size marker is indicated on the left. E, adult multiple tissue mRNA Northern blot (Clontech). Tissue sample is indicated above each lane. All blots were hybridized with 262-bp suprabasin, glyceraldehyde-3-phosphate dehydrogenase, and PL-1 (placenta lactogen 1) probes, as indicated.
The 0.7-kb transcript detected by the suprabasin-specific probe in Northern analysis was amplified by RACE using a suprabasin-specific oligonucleotide corresponding to 3μ-untranslated region (bp 2133–2277). A 0.7-kb fragment was cloned into the pGEMT-easy vector, and by sequence analysis we determined that this shorter mRNA results from an alternative splicing that occurs between bp 384 and 1993, resulting in a transcript that lacks the coding region for the tandem repeats (Fig. 5B).
Fig. 5. Chromosomal localization and genomic structure of mouse suprabasin.
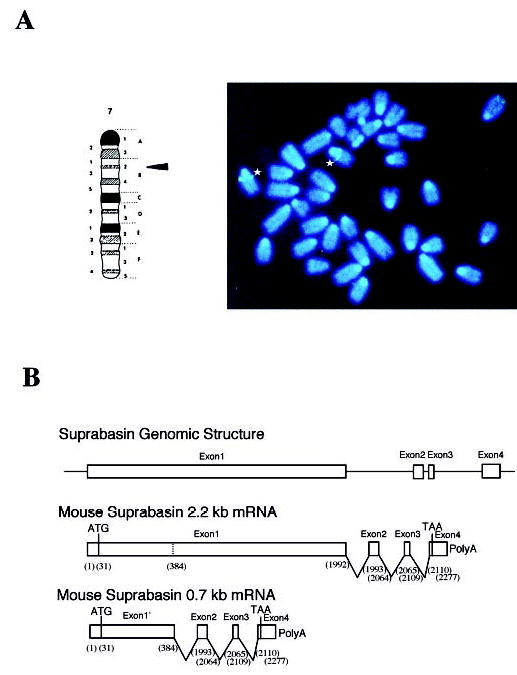
A, FISH localizes the mouse suprabasin gene to chromosome 7. Asterisks indicated the positive spots on the chromosomes 7 to band 7B2–7B3 (indicated by arrow in the schematic map of chromosome 7). B, sequence and genomic analysis of a BAC clone showed that mouse suprabasin consists of four exons and three introns. The small suprabasin transcript (0.7 kb) results from alternative splicing as indicated. Nucleotides (bp) are shown in parentheses.
The expression of suprabasin was also studied in keratinocytes cultured in vitro (Fig. 3, B and C). Suprabasin expression was detected only after the keratinocytes were induced to differentiate in vitro by incrementing Ca2+ concentration. A more detailed time course of induction showed that suprabasin expression was already detectable 4 h after the Ca2+ switch with high expression being reached after 24 h in culture. Treatment of cultured cells with different kinase (PKC, PKA, CaM kinase II) inhibitors showed a specific and complete repression of suprabasin expression with PKC inhibitor treatment (GF; Fig. 3C). This is reminiscent of results observed for other late epidermal differentiation factors, where expression is dependent on the PKC signaling pathway. No effect on expression was seen with treatment with a PKA inhibitor (H89) and a reproducible partial inhibition was observed with an inhibitor for the CaM kinase II (KN62) (Fig. 3C). Mouse suprabasin expression was assessed during embryonic development by Northern blot analysis (Fig. 3D). Expression was detected at E15 coinciding with stratification of the epidermis and dramatically increased by E17. The early detection at E7 was predicted to be caused by extra-embryonic tissue contribution that is present in the E7 sample. The extra-embryonic contribution in sample E7 was corroborated by re-hybridizing the Northern blot with PL-1, a placental specific marker. Other sites of expression detected from a dot blot analysis of different adult tissues were the uterus and thyroid (data not shown), whereas no expression was found in heart, brain, spleen, lung, liver, smooth muscle, or kidney (Fig. 3E).
As mentioned above, we also cloned a partial human supra-basin cDNA and analyzed the expression pattern by Northern blots. We found that human suprabasin is expressed as two transcripts of 2.2 and 1.1 kb . The expression was detected in the skin and in human keratinocytes (NHEK) cultured in vitro (Fig. 4A). A time course of induction for human suprabasin in NHEK showed that, as for the mouse primary keratinocytes, the expression was restricted to cells induced to differentiate in vitro (Fig. 4B). The expression could also be induced by treatment of the cells with TPA, an activator of the PKC signaling pathway. Northern blot analysis of different human tissues showed expression in the esophagus, uterus, and thymus (Fig. 4C).
Fig. 4. Northern blot analysis of human suprabasin.
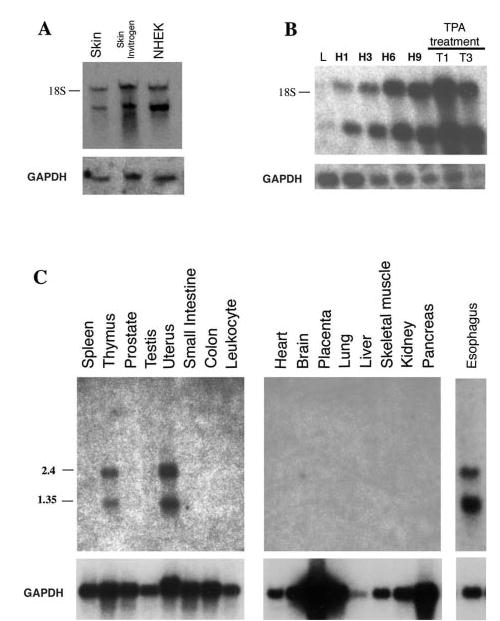
A, expression of suprabasin in NHEK. Northern blot of RNA (2 μg) from human skin, commercial preparation of human skin (Invitrogen), and human keratinocytes cultured in vitro in 1.4 mM Ca2+ (NHEK). The position of the 18 S ribosomal RNA is shown. B, time course of expression of suprabasin in human keratinocytes cultured and differentiated in vitro by addition of Ca2+. L, cultured in 0.05 mM Ca2+ concentration to maintain basal proliferative conditions. H, keratinocytes induced to differentiate in vitro by addition of Ca+ to 1.4 mM and cultured for different time periods (H1, 1 day; H3, 3 days; H6, 6 days; H9, 9 days. Keratinocytes were treated with TPA (100 nM) and cultured for 1 (T1) and 3 (T3) days. C, human multiple tissue mRNA blots (Clontech and Invitrogen); 2 μg of poly(A)+ per lane from tissues indicated above each lane. RNA size markers are indicated on the left. All blots were hybridized with a human 0.5-kb suprabasin probe and glyceraldehyde-3-phosphate dehydrogenase probe as indicated.
Chromosomal Localization and Structure of Mouse Suprabasin
An initial experiment resulted in specific labeling of the proximal region of which was believed to be chromosome 7 on the basis of 4,6-diamidino-2-phenylindole staining. A second experiment was conducted in which a probe specific for the telomeric region of chromosome 7 was co-hybridized with the genomic BAC suprabasin probe. This experiment resulted in the specific labeling of the telomere and the proximal portion of chromosome 7 (Fig. 5A). Measurements of 10 specifically labeled chromosomes 7 demonstrated that suprabasin is located at a position 19% of the distance from the heterochromatic-euchromatic boundary to the telomere of chromosome 7, an area that corresponds to band 7B2–7B3. A total of 80 metaphase cells were analyzed with 72 exhibiting specific labeling.
Search through the human genomic data bases localized only one site with high homology to the mouse suprabasin sequence to chromosome 19q13.1 (AC002389). This region in chromosome 19 is syntenic to the region in the mouse chromosome 7 where suprabasin was localized by FISH mapping. Analysis of the homologous genomic human region predicted exons of good and excellent quality.
As can be seen in the Northern blots with both human and mouse tissues, the suprabasin presents two alternatively spliced transcripts (2.2 and 0.7 kb in the mouse and 2.2 and 1.1 kb in the human). By comparison of the gene and cDNA sequences in the mouse, we determined the intronic and coding regions of suprabasin (Fig. 5B). The intron sizes are 1339 bp for intron 1, 121 bp for intron 2, and 918 bp for intron 3. Alternative splicing between bp 384 and 1993 generates the shorter 0.7-kb splice variant that lacks the coding region for the tandem repeats (Fig. 5B). Analysis of the proximal promoter region showed that it is AT-rich, lacks CpG islands, and contains a canonical TATA box, and revealed several potential transcription factor binding sites for AP1 and Sp1 (data not shown).
Expression of GFP/Suprabasin in Transfected Mouse Keratinocytes
To study the intracellular localization of suprabasin, we transfected GFP/suprabasin full-length and truncated fusion constructs into primary mouse keratinocytes (Fig. 6). In-tracellular localization of GFP fusion proteins was observed by direct fluorescence microscopy. The location of the nucleus was apparent by propidium iodide (PI) staining. The schematic representation of the GFP fusion constructs is in the left panel of Fig. 6 and indicates the suprabasin residues that are fused to GFP. Full-length suprabasin localizes in a highly patch-like expression pattern throughout the cytoplasm. To examine whether suprabasin co-localized with cellular organelles, we performed staining for mitochondria with MitoTracker (mitochondrion-selective stains, Molecular Probes) and immunocytochemistry with LAMP1 and -2 (lysosome-associated membrane glycoprotein, Santa Cruz). Neither co-localized with GFP/suprabasin expression in the transfected cells (data not shown). We also tested a series of truncated fusion constructs (Fig. 6). The expression of NH2-terminal region GFP/suprabasin-(1–144) resulted in expression throughout the cytoplasm that lacked the patchy phenotype of the COOH-terminal truncated (aa 1–612) or full-length (aa 1–700) fusion constructs. Expression of GFP/suprabasin-(133–425), which contains the first tandem repeat domain had effects on cell viability (data not shown), whereas the GFP/suprabasin-(133–612) that contained the complete tandem repeat region resulted in restricted expression around the nucleus. The expression of NH2 terminus with a partial tandem repeat domain of suprabasin-(1–425) caused a strong dense expression in the cytoplasm. This expression was distinct from the C-terminal truncated (aa 1–612) or the full-length (aa 1–700). GFP control was detected as bright green fluorescence throughout the cell (Fig. 6).
Fig. 6. Expression of GFP/suprabasin fusion proteins in primary mouse keratinocytes.
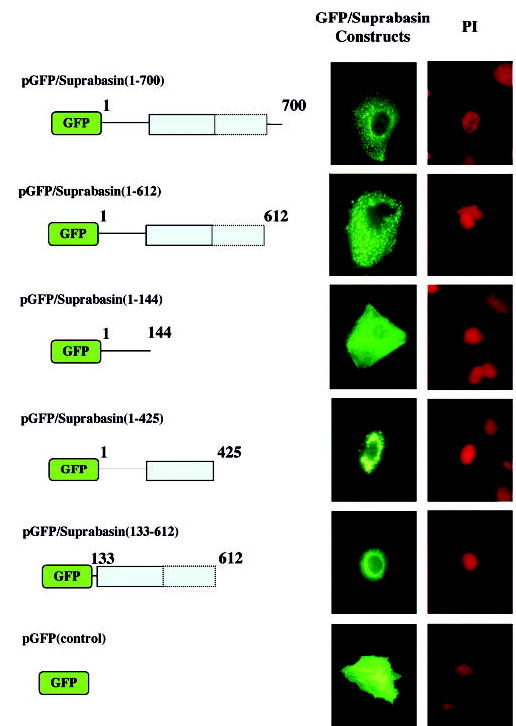
Figure is the schematic representation of full-length and truncated GFP/suprabasin fusion constructs transfected into mouse keratinocytes. The repeat domain region is represented as shaded box with the dotted line indicating the divergent repeat sequence and numbers indicating the amino acid residues in each construct. Middle panel, GFP, GFP/suprabasin, and GFP/suprabasin truncated proteins were expressed in keratinocytes differentiated in vitro with 0.12 mM Ca2+ and visualized by direct fluorescence microscopy with fluorescein isothiocyanate filter. The propidium iodide (PI) counterstain indicates the location of the nucleus within the cells (shown on right panel).
Transglutaminase Assays
We tested whether recombinant suprabasin (aa 1–612) was a substrate for TGase activity. Cross-linking assays were performed with TGase 2 and 3, with recombinant mouse suprabasin. TGase 3 is expressed in epithelia and is a proven participant in CE assembly. The monomeric recombinant suprabasin (aa 1–612) is shown in Fig. 7. After incubation at 37 °C with either TGase 2 (0.4 or 1.2 μg) or TGase 3 (3 or 9 μg), the samples were run in a SDS denaturing gel and transferred to a polyvinylidene difluoride membrane. Western blot analysis using an antibody against the T7 epitope (present in the amino terminus of suprabasin from the pET28 construct) showed that, after incubation with either TGase 2 or TGase 3, the monomeric suprabasin was cross-linked into a high molecular weight form (indicated by arrow in Fig. 7).
Fig. 7. Cross-linking assay of mouse suprabasin by TGase 2 and 3.
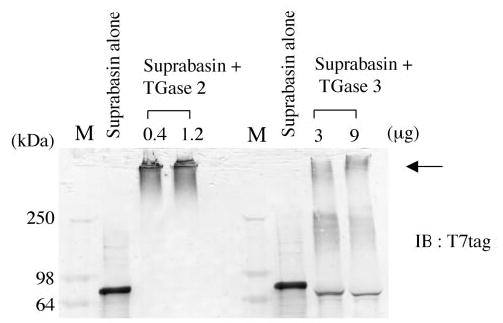
Recombinant suprabasin (aa 1–612) was incubated with 0.4 and 1.2 μg of TGase 2, or 3 and 9 μg of TGase 3 for 30 min at 37 °C (described under “Experimental Procedures”). A Western blot was performed with anti-T7 antibody. Arrow indicates the cross-linked supra-basin. M, lane for high molecular mass markers (SeeBlue, Invitrogen). Numbers on left side indicate molecular mass in kDa.
DISCUSSION
Identification of the diverse precursors that contribute to CE formation will help us elucidate the complex series of steps that are required for the formation of a functional biological barrier in the stratified epithelia. This highly cross-linked insoluble structure that protects individuals is essential for well being, and disruptions of its formation have been correlated with skin diseases (5).
Recently, through the use of different approaches and methodologies such as subtractive hybridization, rapid analysis of gene expression, positional cloning, and EST data base searches (17–21), several new epidermal-specific genes and CE precursors have been identified. In this report we present data on the identification and characterization of a novel gene with a potential role in epidermal stratification. The cDNA was initially identified by performing an SSH procedure. From the in situ hybridization data, we show that the suprabasin mRNA is restricted and highly expressed in the suprabasal layers of the neonatal epidermis. The suprabasin probe identifies transcripts of 2.2 and 0.7 kb in mouse RNA from stratified epithelial tissues and in extra-embryonic tissue (E7). The expression of the suprabasin transcripts in cultured keratinocytes was shown to be dependent on PKC signaling, because specific inhibition of PKC abolished the differentiation-induced expression. Alternatively, we also showed that the human suprabasin transcripts could be induced by treatment of NHEK cells with the PKC inducer, TPA. This response to TPA treatment has been shown for other epidermal late differentiation markers (33, 34), emphasizing the role of PKC in the Ca2+-mediated induction of suprabasin during differentiation.
Chromosomal FISH localized the mouse suprabasin gene to chromosome 7, at band 7B2–7B3. Data base searches identified only one homolog gene in the human genomic data bases in chromosome 19q13.1, in a region that is syntenic to the region where the mouse suprabasin gene was localized.
The 2.2-kb suprabasin transcript contains an open reading frame, which encodes a protein of 700 amino acids that is glycine-rich (~20%) and has a high content of basic residues such as lysine, histidine, and arginine. The deduced suprabasin protein sequence shares structural features with other differentiation markers such as involucrin and loricrin, and somewhat with sciellin, although suprabasin lacks a LIM domain sequence (10). Based on structural features, suprabasin can be divided into three domains: an amino-terminal region that contains a potential transmembrane sequence, a central domain with tandem repeats, and a carboxyl domain. 56% of the repetitive region in suprabasin is composed of glycine, glutamine, histidine, and alanine residues. The glutamine residues may be serving as acceptors of glutamyllysyl isopeptide bonds mediated by transglutaminases. TGases are Ca2+-dependent enzymes that catalyze the formation of ε-(γ-glutamyl)lysine isopep-tide cross-links and/or in the covalent incorporation of poly-amines and histamine. These covalent cross-links often result in the oligomerization of substrate proteins. Our results show that the recombinant suprabasin protein is a substrate for both, tissue TGase 2 and the epidermal TGase 3. Although we demonstrated the oligomerization of suprabasin in vitro by TGases, we did not detect the presence of previously determined transglutaminase target sequences in suprabasin (6, 7). It remains to be determined which are the specific residues that are reactive in supra-basin, whether these are also utilized in vivo, and whether the endogenous suprabasin is incorporated to the CE.
The relevance of each of the domains in the full-length suprabasin was shown with the intracellular localization of the GFP/suprabasin full-length and truncated proteins in transfected keratinocytes. The full-length GFP/suprabasin showed a highly patched phenotype that did not co-localized with mitochondria or lysosomes. This patched phenotype was absent when the protein was truncated to include only the amino-terminal domain, which presented a homogenous cytoplasmic distribution. The patched expression was shown to be caused at least in part by the repeat region, because the GFP/suprabasin- (1–425) had also a patched phenotype, although with a more perinuclear restricted expression. In addition, the mouse smaller transcript (0.7 kb) contains an open reading frame that shares the amino-terminal domain with the full-length supra-basin protein, but lacks the repeat domain. It will be of great interest to determine the role of the spliced transcript in the overall process of epidermal differentiation.
Acknowledgments
We thank Dr. E. Ralston and the members of the NIAMS Image Facility for help with the cell microscopy. We also thank Drs. P. Steinert, K. Boeshans, and B. Advazi for supplying active TGase 3. We are grateful to Noelia Rodriguez and Will Idler for technical assistance, Rick Dreyfuss for photographic assistance, and to Dr. Ulrike Lichti for Ca2+ concentration determinations and providing reagents.
Footnotes
The abbreviations used are: CE, cornified envelope; EDC, epidermal differentiation complex; E, embryonic day; PKA, cAMP-dependent protein kinase; PKC, protein kinase C; FISH, fluorescence in situ hybridization; SSH, suppression subtractive hybridization; NHEK, human keratinocyte; CaM, calmodulin; GFP, green fluorescent protein; RACE, rapid amplification of cDNA ends; TPA, 12-O-tetradecanoylphorbol-13-acetate; TGase, transglutaminase; EST, expressed sequence tag; aa, amino acid(s); GF, GF109203X (bisindolylmaleimide I); H89, N-[2–99p-bromocinnamyl-0-amino-0-ethyl]-5-isoquinolinesulfon-amide; KN62, 1-[N,O-bis-(5-isoquinolinesulfonyl)-N-methyl-l-tyrosyl]-4-phenylpiperazine.
References
- 1.Fuchs E, Byrne C. Curr Opin Genet Dev. 1994;4:725–736. doi: 10.1016/0959-437x(94)90140-x. [DOI] [PubMed] [Google Scholar]
- 2.Eckert RL, Welter JF. Mol Biol Rep. 1996;23:59 –70. doi: 10.1007/BF00357073. [DOI] [PubMed] [Google Scholar]
- 3.Steinert PM, Marekov LN, Parry DA. J Biol Chem. 1999;274:1657–1666. doi: 10.1074/jbc.274.3.1657. [DOI] [PubMed] [Google Scholar]
- 4.Kalinin A, Marekov LN, Steinert PM. J Cell Sci. 2001;114:3069 –3070. doi: 10.1242/jcs.114.17.3069. [DOI] [PubMed] [Google Scholar]
- 5.Presland RB, Dale BA. Crit Rev Oral Biol Med. 2000;11:383–408. doi: 10.1177/10454411000110040101. [DOI] [PubMed] [Google Scholar]
- 6.Robinson NA, Lapic S, Welter JF, Eckert RL. J Biol Chem. 1997;272:12035–12046. doi: 10.1074/jbc.272.18.12035. [DOI] [PubMed] [Google Scholar]
- 7.Steinert PM, Marekov LN. J Biol Chem. 1997;272:2021–2030. doi: 10.1074/jbc.272.3.2021. [DOI] [PubMed] [Google Scholar]
- 8.Volz A, Korge BP, Compton JG, Ziegler A, Steinert PM, Mischke D. Genomics. 1993;18:92–99. doi: 10.1006/geno.1993.1430. [DOI] [PubMed] [Google Scholar]
- 9.Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. J Invest Dermatol. 1996;106:989 –992. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- 10.Champliaud MF, Burgeson RE, Jin W, Baden HP, Olson PF. J Biol Chem. 1998;273:31547–31554. doi: 10.1074/jbc.273.47.31547. [DOI] [PubMed] [Google Scholar]
- 11.Maatta A, Ruhrberg C, Watt FM. J Biol Chem. 2000;275:19857–19865. doi: 10.1074/jbc.M001028200. [DOI] [PubMed] [Google Scholar]
- 12.Aho S, McLean WH, Li K, Uitto J. Genomics. 1998;48:242–247. doi: 10.1006/geno.1997.5188. [DOI] [PubMed] [Google Scholar]
- 13.Jarnik M, de Viragh PA, Scharer E, Bundman D, Simon MN, Roop DR, Steven AC. J Invest Dermatol. 2002;118:102–109. doi: 10.1046/j.0022-202x.2001.01661.x. [DOI] [PubMed] [Google Scholar]
- 14.Maatta A, DiColandrea T, Groot K, Watt FM. Mol Cell Biol. 2001;21:7047–7053. doi: 10.1128/MCB.21.20.7047-7053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djian P, Easley K, Green H. J Cell Biol. 2000;151:381–388. doi: 10.1083/jcb.151.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch PJ, de Viragh PA, Scharer E, Bundman D, Longley MA, Bickenbach J, Kawachi Y, Suga Y, Zhou Z, Huber M, Hohl D, Kartasova T, Jarnik M, Steven AC, Roop DR. J Cell Biol. 2000;151:389 –400. doi: 10.1083/jcb.151.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao XP, Elder JT. Genomics. 1997;45:250 –258. doi: 10.1006/geno.1997.4952. [DOI] [PubMed] [Google Scholar]
- 18.Marenholz I, Zirra M, Fischer DF, Backendorf C, Ziegler A, Mischke D. Genome Res. 2001;11:341–355. doi: 10.1101/gr.114801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall D, Hardman MJ, Nield KM, Byrne C. Proc Natl Acad Sci U S A. 2001;98:13031–13036. doi: 10.1073/pnas.231489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang A, Johnson DG, MacLeod MC. Genomics. 2001;73:284 –290. doi: 10.1006/geno.2001.6518. [DOI] [PubMed] [Google Scholar]
- 21.Makino T, Takaishi M, Morohashi M, Huh NH. J Biol Chem. 2001;276:47445–47452. doi: 10.1074/jbc.M107512200. [DOI] [PubMed] [Google Scholar]
- 22.Diatchenko L, Lukyanov S, Lau YF, Siebert PD. Methods Enzymol. 1999;303:349 –380. doi: 10.1016/s0076-6879(99)03022-0. [DOI] [PubMed] [Google Scholar]
- 23.Lichti U, Yuspa SH. Cancer Res. 1988;48:74 –81. [PubMed] [Google Scholar]
- 24.Hennings H, Michael D, Cheng C, Steinert P, Holbrook K, Yuspa SH. Cell. 1980;19:245–254. doi: 10.1016/0092-8674(80)90406-7. [DOI] [PubMed] [Google Scholar]
- 25.Yuspa SH, Kilkenny AE, Steinert PM, Roop DR. J Cell Biol. 1989;109:1207–1217. doi: 10.1083/jcb.109.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morasso MI, Jamrich M, Sargent TD. Dev Biol. 1994;162:267–276. doi: 10.1006/dbio.1994.1084. [DOI] [PubMed] [Google Scholar]
- 27.Jang SI, Steinert PM, Markova NG. J Biol Chem. 1996;271:24105–24114. doi: 10.1074/jbc.271.39.24105. [DOI] [PubMed] [Google Scholar]
- 28.Church GM, Gilbert W. Proc Natl Acad Sci U S A. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fort P, Marty L, Piechaczyk M, el Sabrouty S, Dani C, Jeanteur P, Blanchard JM. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackem S, Mahon KA. Development. 1991;112:791–806. doi: 10.1242/dev.112.3.791. [DOI] [PubMed] [Google Scholar]
- 31.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 33.Dlugosz AA, Yuspa SH. J Cell Biol. 1993;120:217–225. doi: 10.1083/jcb.120.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dlugosz AA, Yuspa SH. J Invest Dermatol. 1994;102:409 –414. doi: 10.1111/1523-1747.ep12372171. [DOI] [PubMed] [Google Scholar]


