Abstract
During the process of epidermal differentiation, intracellular and extracellular calcium (Ca++ ) concentrations induce an array of signaling pathways [Berridge, M.J., Lipp, P., Bootman, M.D., 2000. The versatility and universality of calcium signaling. Nature Rev. Mol. Cell. Biol. 1, 11–21]. Keratinocytes follow a complex Ca++ -dependent program of differentiation moving from the basal proliferative layer, through the spinous and granular differentiated layers to ultimately culminate in the formation of the cornified layer of the epidermis. Members of the Ca++ -binding proteins play a central role in the transduction of Ca++ signals. Utilizing a suppressive subtractive hybridization screen comparing basal and differentiated keratinocytes, we identified the novel Ca++ -binding protein genes, Scarf (skin Calmodulin-related factor) and Scarf2, which have homology to calmodulin (CaM). In this study, we present a comprehensive analysis of the expression pattern for Scarf and Scarf2 transcripts and proteins in the developing mouse. To examine Scarf2 expression during embryogenesis, we performed in situ hybridization, and detected expression in the hair follicle, skin and nasal epithelium. These results showed substantial overlap with the previously reported Scarf gene expression [Hwang, M., Morasso, M.I., 2003. The novel murine Ca2+-binding protein, Scarf, is differentially expressed during epidermal differentiation. J. Biol. Chem. 278, 47827–47833]. Comparing the expression patterns of Scarf and Scarf2 proteins in neonatal and adult mouse skin with several structural epidermal proteins, i.e. keratin 14 (K14), keratin 1 (K1), loricrin (LOR) and filaggrin (FIL) showed that their expression overlaps K1, an early marker of keratinocyte differentiation. Interestingly, Scarf and Scarf2 were also detected in the tongue and oral epithelia, rib bone undergoing ossification and in the medullar region of thymus.
Keywords: Calcium-binding protein, Skin, Differentiation, Scarf, Scarf2, Epidermal development
1. Results and discussion
Ca++ is known as a secondary messenger in many cellular processes: proliferation, differentiation and apoptosis. Many aspects of these processes are mediated by Ca++ - binding proteins (Nelson and Chazin, 1998). CaM, the most characterized Ca++ -binding protein, is ubiquitously expressed and binds several dozen cellular proteins, from enzymatic to structural proteins, in response the Ca++ signals (Crivici and Ikura, 1995). CaM and CaM-like Ca++ -binding proteins are characterized by the presence of four EF-hand domains, the putative Ca++ -binding domains (Haeseleer et al., 2000; Haeseleer and Palczewski, 2002; Mehul et al., 2000; Rogers et al., 2001). It is through the binding of Ca++ that these proteins undergo conformational changes that allow for the association and regulation of their specific target proteins.
In contrast to CaM, some Ca++ -binding proteins such as the GCAPs show a cell-specific expression pattern (Dizhoor, 2000), suggesting that this family of genes plays a crucial function in the retina. Likewise, here we present evidence that Scarf and Scarf2 are CaM-like proteins that are expressed in a specific temporal and spatial pattern during epidermal development and are differentially expressed during the Ca++ -dependent differentiation process of the epidermis (Hennings et al., 1980; Yuspa et al., 1989).
1.1. Scarf2 expression during mouse embryo development
The initial characterization of the novel CaM-like protein Scarf was previously reported (Hwang and Morasso, 2003). The intronless Scarf gene was localized on chromosome 13, with its expression initiating at day 15 of mouse development and highly restricted to the differentiating epidermis. Scarf2 was reported as a gene with 83% nucleotide sequence homology to Scarf, that localized approximately 15 kb apart on chromosome 13. The high degree of homology extended to 1.6 kb upstream and 1.2 kb downstream sequence of the coding regions, indicating homology in the potential regulatory sequences including the putative proximal promoter region. To examine the level of similarity in the expression pattern between Scarf and Scarf2, the expression of Scarf2 was studied by radioactive in situ hybridization during mouse embryogenesis. Hybridization of sagittal sections of 13-day (Fig. 1A,B), 15-day (Fig. 1C,D) and 16-day (Fig. 1E,F) mouse embryos with an antisense Scarf2 probe, showed no expression at 13-day and strong epidermal expression at 15-day and 16-day developmental stages. Scarf2 expression was also detected in the nasal epithelial cells, the dorsum of the tongue and in the vibrissae (indicated by arrows in Fig. 1C,E, and presented in magnified view in Fig. 1G–J). In neonatal skin, the expression of Scarf2 was restricted to the suprabasal (differentiated) layers of the epidermis (Fig. 1M,N). The expression pattern of Scarf2 transcript was very similar to that of Scarf (Hwang and Morasso, 2003). No expression was detected with the use of Scarf2 sense probe (data not shown).
Fig. 1.
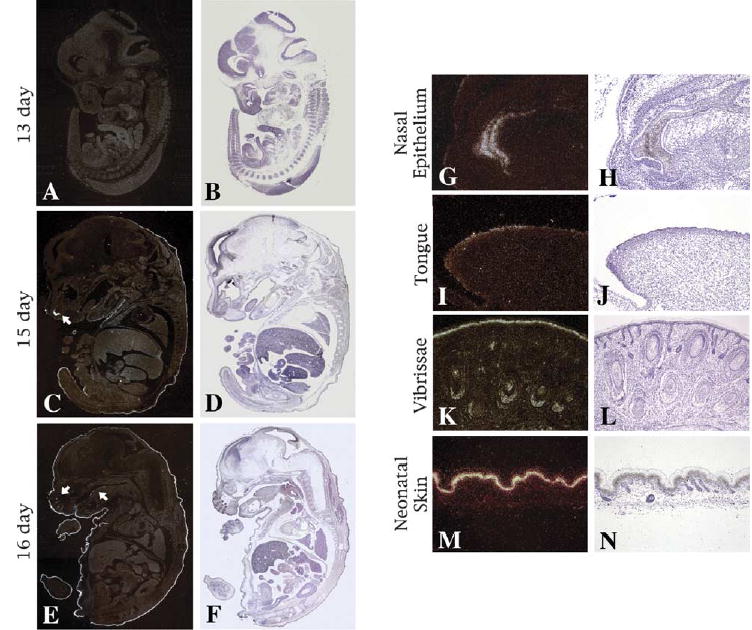
Expression of Scarf2 during mouse development. In situ hybridization was performed with an antisense Scarf2 probe on sagittal sections of day 13 (A), day 15 (C) and day 16 (E) mouse embryos. Corresponding hematoxylin–eosin staining on panels B, D and F, respectively. Magnification 1.25×. Arrows point to sites of Scarf2 expression at 15-day, the nasal epithelium (C) and at day 16, in the vibrissae and dorsum of the tongue (E). (G–L) Magnified view of Scarf2 expression in the nasal epithelium (G) and tongue (I). Scarf2 expression in vibrissae (K) and in the differentiated layers of neonatal skin (M). Corresponding hematoxylin–eosin staining (H–N, respectively). Magnification 10× (G–L) and 20× (M–N).
Using a commercial Northern blot of total embryo mRNA at different developmental stages, Scarf2 was detected at 15 days of embryogenesis and the level of expression increased by day 17, corroborating the results obtained by in situ hybridization (Fig. 2A). Scarf2 expression was not detected in heart, brain, spleen, lung, liver, skeletal muscle, kidney or testis (data not shown), whereas Scarf was weakly detected in skeletal muscle (Hwang and Morasso, 2003).
Fig. 2.
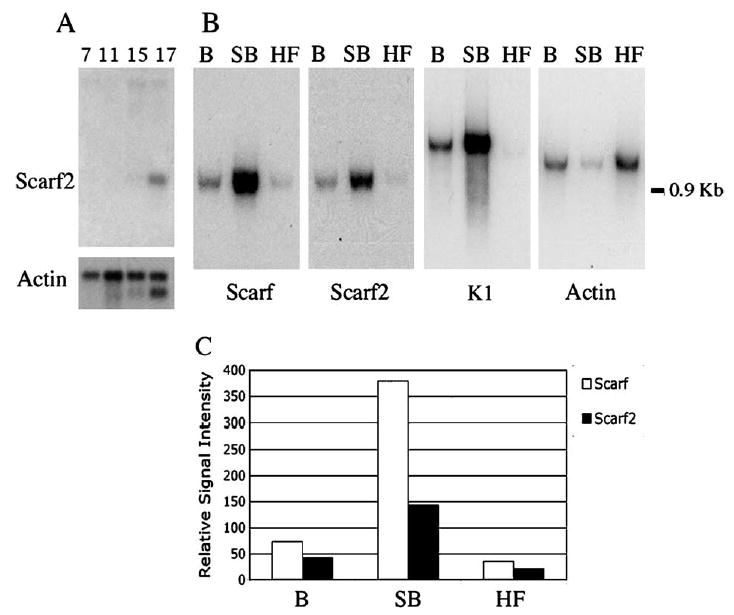
Expression of Scarf and Scarf2 in the stratified epidermis and adult tissues. (A) Expression of Scarf2 during mouse embryonic development (7, 11, 15 and 17 day embryos). (B) Northern blots of 0.5 μg poly (A) RNA from basal (B) and suprabasal (SB) epidermal cells, and hair follicle (HF) cells were hybridized with Scarf, Scarf 2, K1 and β-actin cDNA probes. Molecular weight of RNA is indicated on the right. (C) Graph summarizes the densitometric analysis of (B) with normalization using β-actin, allowing for quantification of the level of expression of Scarf and Scarf2 in (B) and (SB) keratinocytes and in (HF) cells. Normalized densitometric values; Scarf (white bars) and Scarf2 (black bars).
Percoll gradients are utilized to separate subpopulations of cells in the epidermis by density centrifugation (Fisher et al., 1982). Scarf2 expression was detected in the differentiated (suprabasal) epidermal cells obtained by Percoll gradient preparations from neonatal skins, corroborating the results obtained by in situ hybridization (Fig. 2B). During the gradient formation in the centrifugation, some suprabasal cells may remain and be detected as contaminant in the basal cell fraction (Fisher et al., 1982; Lichti and Yuspa, 1988). The differentiation-specific K1 marker was utilized to validate the Percoll separation method for basal (B) and suprabasal (SB) cells. Scarf, Scarf2 and K1 are suprabasal-specific transcripts primarily detected in the mRNA from the SB cell fraction. Both Scarf and Scarf2 transcripts were also detected in mRNA from isolated hair follicles, conforming the results obtained by in situ hybridization (Fig. 1K). Measurements of signal intensity and normalization using β-actin allowed for quantification of the levels of expression for Scarf and Scarf2 in the differentiated epidermal cells; Scarf expression was 2.7 times higher than Scarf2 in the suprabasal cells (Fig. 2C).
1.2. The expression pattern of Scarf and Scarf2 proteins during mouse development
Polyclonal antibody against the full-length Scarf protein was generated in hens (Hwang and Morasso, 2003). Due to the high homology and sequence identity present between Scarf and Scarf2, and in order to develop an antibody that would recognize specifically Scarf2, the antibody was generated against a peptide that corresponds to the most C-terminal sequence of Scarf2 protein (Fig. 3A). Anti-Scarf2 antibody recognized the Scarf2 recombinant protein and there was lack of cross-reactivity to Scarf recombinant protein, and no cross-reactivity to human CaM, Clp or Clsp recombinant proteins (Fig. 3B). Anti-CaM antibody did not detect either Scarf or Scarf2 recombinant proteins (data not shown). The Scarf2-specific antibody also detected a band of correct size in whole cell extract from keratinocytes obtained from neonatal skins (Fig. 3C). Scarf and Scarf2 are homologous to the conserved and ubiquitously expressed CaM protein, 56 and 50%, respectively. They also share homology with the human Ca++ -binding proteins Clp and Clsp (Rogers et al., 2001; Mehul et al., 2000, 2001). All these proteins are members of the CaM-like protein family and are characterized as small, acidic, Ca++ -binding proteins that contain EF-hand motifs.
Fig. 3.
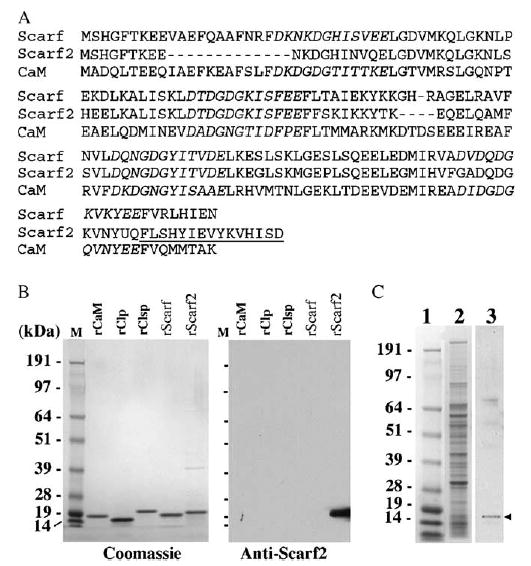
Specificity of Scarf2 antibody. (A) Alignment of the predicted amino acid sequence of Scarf and Scarf2 highlighting the EF-hand domains (italics) and the amino acid sequence of the synthetic peptide utilized for antibody generation (underlined). (B) Recombinant CaM, Clp, Clsp, Scarf, Scarf2 proteins (1 μg) were electrophoresed and transferred to a nitrocellulose membrane. Western blot analysis was performed using anti-Scarf2 antibody, an HRP conjugated secondary antibody and visualized using chemiluminescence. A replicate gel was also stained with Coomassie blue (left panel). Molecular weights are indicated on the left. (C) Keratinocyte whole cell extract (40 μg) were electrophoresed (lane 2) and transferred to nitrocellulose membrane. Western blot analysis was performed using anti-Scarf2 antibody (lane 3), an HRP conjugated secondary antibody and visualized using chemiluminescence. The molecular weights are indicated in lane 1. The specific band detected by western blot is indicated with an arrowhead, and corresponds to the molecular weight of Scarf2 (16 kDa).
Analysis of the expression of Scarf and Scarf2 proteins was performed by immunohistochemistry on sagittal sections of mouse embryos and in sections of neonatal and adult mouse skin. Scarf and Scarf2 proteins were detected in the differentiated layers of the stratified epidermis and in dorsal aspect of the ossifying rib of 16-day mouse embryos (Fig. 4A,B). To confirm the identity of the Scarf and Scarf2-positive cells in the ossifying rib, colocalization experiments were performed utilizing anti-osteocalcin (OC) antibody and trichrome staining on serial sections (Fig. 4C,D). OC is a bone-specific marker that is expressed in the differentiating osteoblasts undergoing mineralization (Marks and Herney, 1996) and trichrome staining is used for histological stain of connective tissue and collagen fibers. During murine embryogenesis, OC expression is detectable in a subset of osteoblasts of the ribs undergoing endochondral ossification but not in chondrocytes (Bidder et al., 1998). By embryonic day 16, as shown for OC, Scarf and Scarf2 are detected specifically in perichondral osteoblasts, with no expression in chondrocytes (Fig. 4E–G). At this stage of development the ventral portion of the ribs are still not ossifying and there is absence of OC expression (Bidder et al., 1998; Fig 4J). Likewise, no expression of either Scarf or Scarf2 was detected in the ventral portion of the ribs at this stage. (Fig. 4H–J). Scarf2 was also detected in the non-stratified squamous epithelial cells of the bladder (Fig. 4L). On cross-sections of 16-day embryo heads, Scarf and Scarf2 were detected in the tongue and oral epithelia (Fig. 4M,N). Scarf2 positive cells were also present in the primodium of the incisor tooth (Fig. 4P).
Fig. 4.
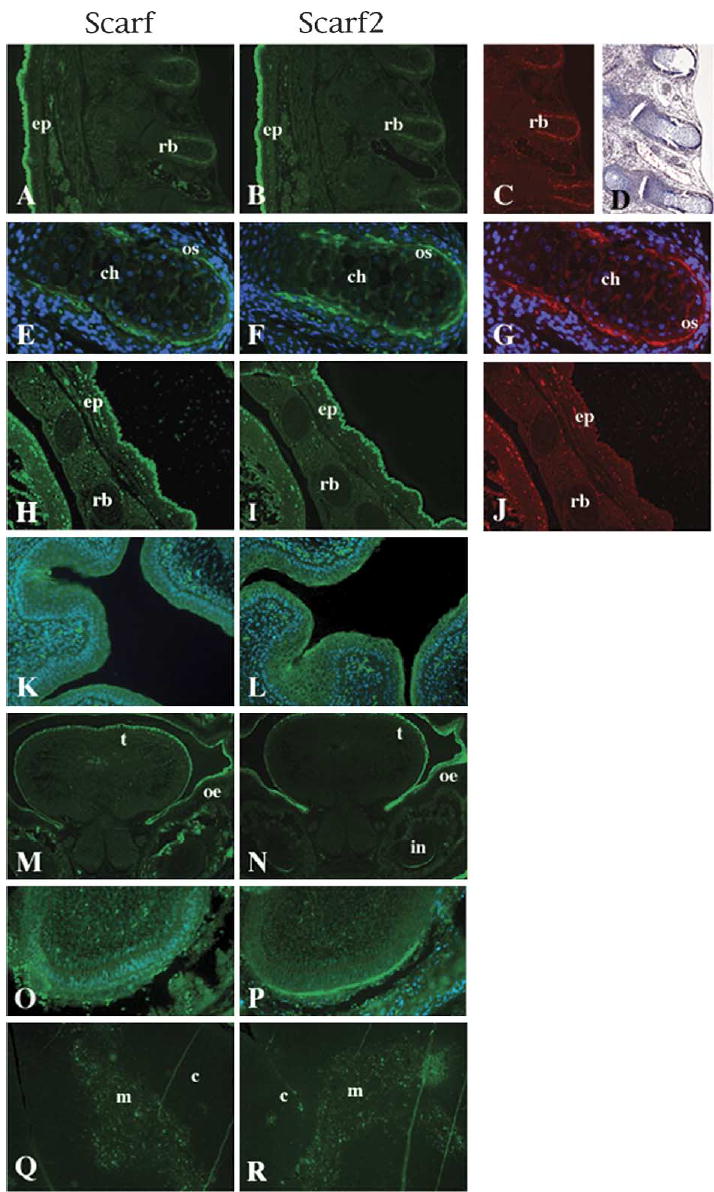
Scarf and Scarf2 protein expression. Dorsal view of sagittal sections of 16-day embryos treated with anti-Scarf (A), anti-Scarf2 (B), anti-OC (C) and corresponding Trichrome (Masson) staining (D). Treatments for OC and Trichrome staining were performed on serial sections. (E–G) 20× magnification of the dorsal aspect of rib bone, anti-Scarf (E), anti-Scarf2 (F) and anti-OC (G), with DAPI used for nuclear counterstain. The ventral aspects of sagittal section of corresponding (A)–(C): anti-Scarf (H), anti-Scarf2 (I) and anti-OC (J). (K–L) Scarf and Scarf2 staining in the epithelial cells of the bladder counterstained with DAPI. (M–N) Cross-sections of the head of 16-day embryo stained with anti-Scarf (M) and anti-Scarf2 (N). (O–P) 20× magnification of the incisor region from panels M and N, respectively. (Q–R) Adult thymus sections were stained with anti-Scarf (Q) and anti-Scarf2 (R), showing detection in the medullar region of thymus. Ep, epidermis; rb, rib bone; ch, chrondrocyte; os, osteoblast; t, tongue; oe, oral epithelia; in, incisor tooth; c, cortex; m, medulla.
We examined the expression of Scarf and Scarf2 utilizing mouse adult tissues microarray containing brain, heart, kidney, liver, lung, muscle, pancreas and spleen (data not shown). Neither Scarf nor Scarf2 proteins were detected in any of these tissues, results that are consistent with the results obtained for mRNA by northern blot analysis (data not shown). However, expression of Scarf and Scarf2 was detected in the thymus (Fig. 4Q,R). It has been reported that K13, K14 and K17 are expressed in the medullar epithelial cells (Shezen et al., 1995). Detection of expression of Scarf and Scarf2 in the thymus showed localization in the medullar region of thymus, but not in the cortex region (Fig. 4Q,R).
The expression of the epidermal keratins, K5 and K14, starts at day 9.5 and is generalized at day 10.5 of mouse embryonic development (McGowan and Coulombe, 1998). Using whole mount staining, Byrne et al. (1994) showed that the differentiated keratins, K1 and keratin 10 (K10), are initially detected by day 13.5 only in the nasal epithelia. Until day 15, the mouse embryo is covered by a two-layer epithelium, where K1 is not detected. At this stage of embryogenesis, the stratification of the epidermis commences, and the expression of K1 and K10 is then readily detectable in the differentiated layers of the epidermis (McGowan and Coulombe, 1998; Byrne et al., 1994).
To examine Scarf and Scarf2 protein expression in neonatal and adult mouse skin, we performed co-localization studies of Scarf and Scarf2 with well-characterized keratinocyte differentiation markers: K14, K1, LOR and FIL. K14 is expressed in the proliferative basal layer of the epidermis (Fig. 5A,B). K1 is an early differentiation marker and LOR and FIL are late differentiation markers (Yuspa et al., 1989; Steven et al., 1990). In neonatal skin, Scarf and Scarf2 proteins were found in the spinous and granular (differentiated) layers, overlapping with K1, LOR and FIL expression (Fig. 5C–H). Scarf and Scarf2 were also detected in the hair follicles (Fig. 5I,B, respectively). Although adult epidermis is relatively thin when compared to neonatal murine epidermis, expression of Scarf and Scarf2 was detected only in the suprabasal layers (Fig. 5I,J), and co-localized with K1 expression (Fig. 5K,L).
Fig. 5.
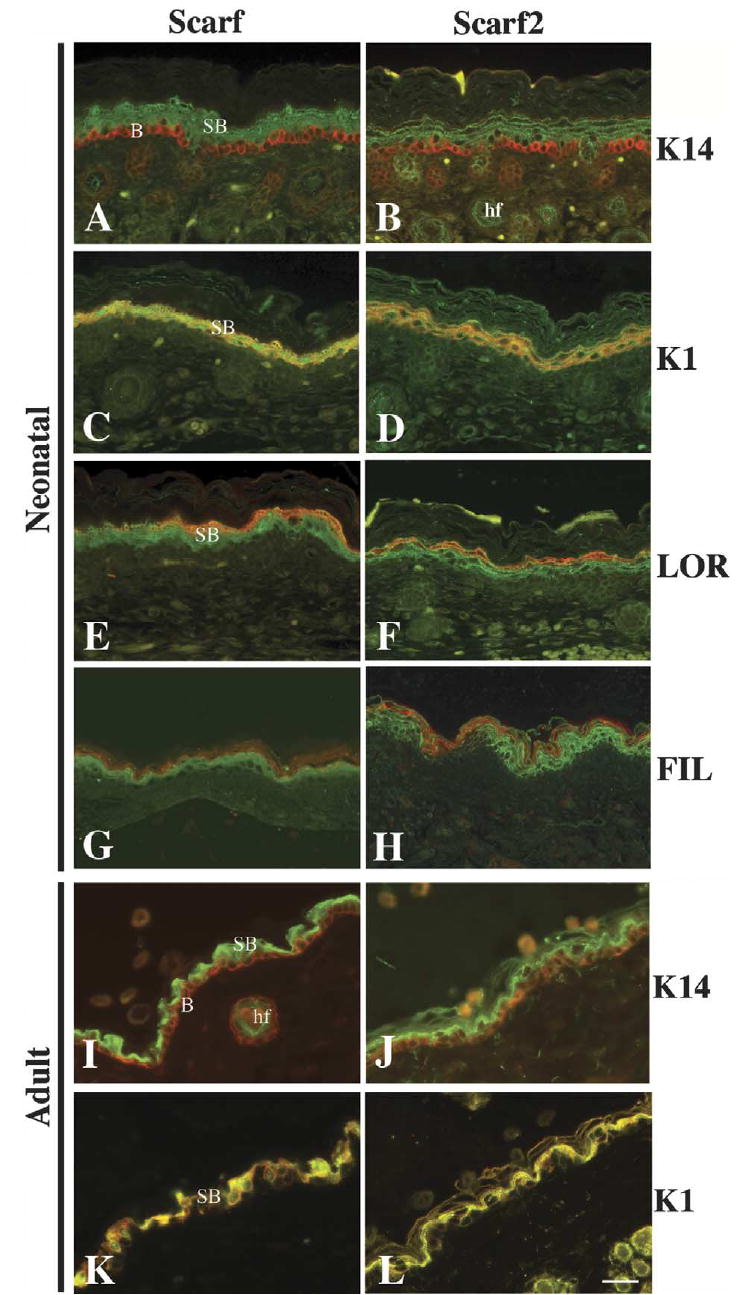
Immunohistochemical analysis of Scarf and Scarf2 expression in the mouse neonatal and adult skin. Neonatal skin sections were stained with anti-Scarf (A, C, E and G) and anti-Scarf2 (B, D, F and H) (green). Co-localization with epidermal differentiation markers was studied using anti-K14 (A and B), anti-K1 (C and D), anti-LOR (E and F) and FIL (G and H) (red). Adult skin sections were stained with anti-Scarf (I and K) and anti-Scarf2 (J and L) (green). Co-localization was performed using anti-K14 (I and J) and anti-K1 (K and L) (red). Merged images of anti-K1 and anti-Scarf or anti-Scarf2 are shown in yellow (C, D, K, L). Scale bar 50 μm. B, basal; SB, suprabasal; hf, hair follicle.
Taken together, the presence of Scarf2 transcript and protein in the stratifying epidermis, and its close resemblance in spatial and temporal expression patterns with Scarf raises the interesting possibility that these novel Ca++ -binding proteins are involved in the Ca++ -dependent epidermal differentiation process. Further investigations as to the specific role and target proteins for each of these CaM-like proteins will provide new insights into the overall mechanisms necessary for the continuously renewing skin in the adult.
2. Experimental procedures
2.1. In situ hybridization
RNA probes corresponding to the sense and antisense strand of mouse Scarf2 cDNA (from 17 to 423 bp) were prepared using T7 and Sp6 RNA polymerases. In situ hybridization was performed at high stringency on consecutive sagittal sections of 13-, 15- and 16-day mouse embryos and neonatal skin as described by Mackem and Mahon (1991).
2.2. Antibodies and immunofluorescence
Immunohistochemical analysis was performed on paraffin sections of 16-day mouse embryos, neonatal and adult mouse skin, and thymus. Anti-Scarf2 antibody against a C-terminal peptide (LSHYIETVYKVHISD) was generated in hens (Aves Laboratories). The deparaffinized sections were treated with chicken anti-Scarf antibody (1:500), chicken anti-Scarf2 (1:200), rabbit anti-K14 (1:1000, Covance), rabbit anti-K1 (1:1000, Covance) rabbit anti-LOR (1:500, Covance), rabbit anti-FIL (1:2000, Covance) and goat anti-OC (1:40) overnight at 4 °C. For secondary antibody, an anti-chicken fluorescein isothiocyanate (FITC)-conjugated antibody (1:500, Cappel) and anti-rabbit rhodamine-conjugated antibody (1:500, Jackson Immunoresearch) were used for 1 h at room temperature. Sections were visualized by fluorescence microscopy.
2.3. RNA sources and Northern blot analysis
Basal (B) and suprabasal (SB) keratinocytes from neonatal skins (epidermis) were separated on Percoll gradients (Lichti and Yuspa, 1988) and hair follicles were also isolated from neonatal skins (dermis) by centrifugation on 9% Ficoll solution (Rogers et al., 1987). We used TRIZOL (Invitrogen) to isolate total RNA. Messenger RNA was isolated utilizing an mRNA isolation kit (Ambion). The mRNA samples (0.5 μg) were run by electrophoresis in 1.2% agarose methylmercury hydroxide gels, electroblotted to nylon membranes, and hybridized with 32P-labeled probes according to Church and Gilbert (1984). Commercial Northern blots for adult mouse tissues were purchased from Clontech and used according to the instructions of the manufacturer. The mouse-specific probes for Scarf and Scarf2 corresponded to the full-length coding regions. A 2 kb β-actin cDNA probe was used as control for RNA integrity. K1 probe corresponded to the 3′ untranslated region and was kindly provided by Dr Compton. Expression was quantified by densitometry (NIH IMAGE 1.61) and normalized with β-actin.
2.4. Western blot analysis
Recombinant CaM, Clp, Clsp, Scarf and Scarf2 proteins (1 μg) and keratinocyte whole cell extract were resolved by SDS-PAGE, transferred onto a nitrocellulose membrane, and probed using anti-Scarf2 primary antibody (1:200). The membranes were incubated with anti-chicken horseradish peroxidase-conjugated secondary antibody (1:5000; Aves Laboratories). Secondary antibody binding was visualized using chemiluminescence detection methods (Amersham). The detection of the endogenous protein in whole cell extracts required incubation of the sample in SDS loading buffer plus β-mercaptoethanol (12.5 mM Tris base pH 6.8, 0.4% SDS, 2% glycerol, 0.004% bromophenol blue, 1% β-mercaptoethanol) at 98 °C for at least 10 min. Recombinant Clp was kindly provided by Dr Strehler. All other recombinant proteins were generated in the laboratory from plasmids in which the full length open reading frame was subcloned into pET-28a(+) vector (Novagen) (Hwang and Morasso, 2003).
Acknowledgments
The authors would like to thank members of the Morasso laboratory for useful comments and assistance during the preparation of this manuscript; to members of the Tuan Laboratory and Ms S. Everett for photographic assistance.
References
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signaling. Nature Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Bidder M, Latifi T, Towler DA. Reciprocal temporospatial patterns of Msx2 and Osteocalcin gene expression during murine odontogenesis. J Bone Miner Res. 1998;13:609. doi: 10.1359/jbmr.1998.13.4.609. [DOI] [PubMed] [Google Scholar]
- Byrne C, Tainsky M, Fuchs E. Programming gene expression in developing. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;8:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- Dizhoor AM. Regulation of cGMP synthesis in photoreceptors: role in signal transduction and congenital diseases of the retina. Cell Signal. 2000;12:711–719. doi: 10.1016/s0898-6568(00)00134-0. [DOI] [PubMed] [Google Scholar]
- Fisher SM, Nelson KDG, Reiners JJ, Jr, Viaie A, Pelling JC, Slaga TJ. Separation of epidermal cells by density centrifugation: a new technique for studies on normal and pathological differentiation. J Cutan Pathol. 1982;91:43–49. doi: 10.1111/j.1600-0560.1982.tb01040.x. [DOI] [PubMed] [Google Scholar]
- Haeseleer F, Palczewski K. Calmodulin and Ca2+-binding proteins (CaBPs): variations on a theme. Adv Exp Med Biol. 2002;514:303–317. doi: 10.1007/978-1-4615-0121-3_18. [DOI] [PubMed] [Google Scholar]
- Haeseleer F, Sokal I, Verlinde CLMJ, Erdjumnet-Bromage H, Tempst P, Pronin AN, et al. Ca (2+)-binding proteins in the retina: structure, function, and the etiology of human visual diseases. J Biol Chem. 2000;275:1247–1260. doi: 10.1074/jbc.275.2.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennings H, Michael D, Cheng C, Steinert P, Holbrook K, Yuspa SH. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 1980;19:245–254. doi: 10.1016/0092-8674(80)90406-7. [DOI] [PubMed] [Google Scholar]
- Hwang M, Morasso MI. The novel murine Ca2+-binding protein, Scarf, is differentially expressed during epidermal differentiation. J Biol Chem. 2003;278:47827–47833. doi: 10.1074/jbc.M306561200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti U, Yuspa SH. Modulation of tissue and epidermal transglutaminases in mouse epidermal cells after treatment with 12-O-tetradecanoylphorbol-13-acetate and/or retinoic acid in vivo and in culture. Cancer Res. 1988;48:74–81. [PubMed] [Google Scholar]
- Mackem S, Mahon KA. Ghox 4.7: a chick homeobox gene expressed primarily in limb buds with limb-type differences in expression. Development. 1991;112:791–806. doi: 10.1242/dev.112.3.791. [DOI] [PubMed] [Google Scholar]
- Marks Jr., S.C., Herney, D.C., 1996. Principles of Bone Biology. Academic Press, NY, USA, pp. 3–14.
- McGowan KM, Coulombe PA. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J Cell Biol. 1998;143:486–496. doi: 10.1083/jcb.143.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehul B, Bernard D, Simonetti L, Bernard MA, Schmidt R. Identification and cloning of a new calmodulin-like protein from human epidermis. J Biol Chem. 2000;275:12841–12847. doi: 10.1074/jbc.275.17.12841. [DOI] [PubMed] [Google Scholar]
- Mehul B, Bernard D, Schmidt R. Calmodulin-like skin protein: a new marker of keratinocyte differentiation. J Invest Dermatol. 2001;116:905–909. doi: 10.1046/j.0022-202x.2001.01376.x. [DOI] [PubMed] [Google Scholar]
- Nelson, M.R., Chazin, W.J., 1998. Calmodulin and Signal Transduction. Academic Press, London.
- Rogers G, Martinet N, Steinert P, Wynn P, Roop D, Kilenny A, et al. Cultivation of murine hair follicles as organoids in a collagen matrix. J Invest Dermatol. 1987;89:369–379. doi: 10.1111/1523-1747.ep12471760. [DOI] [PubMed] [Google Scholar]
- Rogers MS, Kobayashi T, Pittelkow MR, Strehler EE. Human calmodulin-like protein is an epithelial-specific protein regulated during keratinocyte differentiation. Exp Cell Res. 2001;267:216–224. doi: 10.1006/excr.2001.5254. [DOI] [PubMed] [Google Scholar]
- Shezen E, Okon E, Ben-Hur H, Abramsky O. Cytokeratin expression in human thymus: immunohistochemical mapping. Cell Tissue Res. 1995;279:221–231. doi: 10.1007/BF00300707. [DOI] [PubMed] [Google Scholar]
- Steven AC, Bisher ME, Roop DR, Steinert PM. Biosynthetic pathways of filaggrin and loricrin-two major proteins expressed by terminally differentiated epidermal keratinocytes. J Struct Biol. 1990;104:150–162. doi: 10.1016/1047-8477(90)90071-j. [DOI] [PubMed] [Google Scholar]
- Yuspa SH, Kilkenny AE, Steinert PM, Roop DR. Expression of murine epidermal differentiation markers is tightly regulated by restricted extracellular calcium concentration in vitro. J Cell Biol. 1989;109:1207–1217. doi: 10.1083/jcb.109.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]


