Abstract
Cytolysin A (ClyA) is a pore-forming cytotoxic protein encoded by the clyA gene that has been characterized so far only in Escherichia coli. Using DNA sequence analysis and PCR, we established that clyA is conserved in the human-specific typhoid Salmonella enterica serovars Typhi and Paratyphi A and that the entire clyA gene locus is absent in many other S. enterica serovars, including Typhimurium. The gene products, designated ClyASTy and ClyASPa, show ≥90% amino acid identity to E. coli cytolysin A, ClyAEC, and they are immunogenically related. The Salmonella proteins showed a pore-forming activity and are hence functional homologues to ClyAEC. The chromosomal clyASTy gene locus was expressed at detectable levels in the serovar Typhi strains S2369/96 and S1112/97. Furthermore, in the serovar Typhi vaccine strain Ty21a, expression of clyASTy reached phenotypic levels, as detected on blood agar plates. The hemolytic phenotype was abolished by the introduction of an in-frame deletion in the clyASTy chromosomal locus of Ty21a. Transcomplementation of the mutant with a cloned clyASTy gene restored the hemolytic phenotype. To our knowledge, Ty21a is the first reported phenotypically hemolytic Salmonella strain in which the genetic determinant has been identified.
Pathogenic bacteria produce many substances that are directly or indirectly toxic to host cells, and such toxins often play a central role in the pathogenesis of microbial disease (9). Salmonella enterica colonizes and infects a wide range of different organisms and causes a broad spectrum of diseases, such as gastroenteritis and bacteremia, as well as life-threatening conditions, such as typhoid fever (32). Although some S. enterica serovars, including Typhimurium, can infect a broad range of hosts, other serovars, such as Typhi and Paratyphi A (humans) and Dublin (cattle), have a narrow host range. The molecular bases for the host restriction, however, are not well understood. Virulent serovars possess an array of genes that encode proteins required for invasion and that are secreted by a specialized type III secretion apparatus (10). Cytotoxic activity towards cultured macrophages of Salmonella, independent of host restriction, is evident (6), and it has also been established that serovar Typhimurium and serovar Typhi induce macrophage apoptosis via a mechanism involving the invasin SipB (17, 29). Up to now, however, there has been no experimental evidence for expression of an RTX-like toxin or cytolysin in Salmonella, although cytotoxic activity associated with a cholera toxin (CT)-like toxin and an Escherichia coli heat-labile enterotoxin (LT-I)-like toxin has been reported (7, 31). Members of the RTX toxin family, and especially the HlyA hemolysin of uropathogenic E. coli, are among the most thoroughly studied cytolytic toxins of gram-negative bacteria (3, 48, 49). Using a model for experimental infections in mice, it has been established that HlyA is directly involved in the pathogenesis of E. coli infection (14). ClyA (also called HlyE or SheA), a 34-kDa protein, is the prototype of a growing family of non-RTX cytolysins that was first identified in E. coli (8, 13, 35; B. E. Uhlin and Y. Mizunoe, abstract from the Keystone Symposia on Molecular and Cellular Biology: Molecular Events in Microbial Pathogenesis, J. Cell. Biochem. 18A(Suppl.):71, 1994). X-ray crystallography has shown that ClyA has unusual structural features and does not resemble any previously studied cytotoxin (47). In contrast to RTX toxins, ClyA does not require calcium for activity, nor does it contain the conserved fatty acylation sites typical of RTX toxins or a C-terminal secretion signal (8, 34). Purified E. coli cytolysin A (ClyAEC) protein has been shown to have a cytotoxic and cell-detaching activity towards murine-derived macrophage-like cells (34). This effect caused apoptosis in the macrophage-like cells (22), and in addition, ClyAEC was recently shown to act by forming pores in target membranes (25, 34, 47). Expression of the clyA gene is silenced in nonpathogenic E. coli K-12 laboratory strains by the abundant nucleoid protein H-NS (50). Thus, in H-NS-deficient E. coli K-12 derivatives, expression of ClyA is derepressed, resulting in cytotoxicity towards cultured mammalian cells, e.g., HeLa cells (Uhlin and Mizunoe, J. Cell. Biochem. 18A(Suppl.):71, 1994; J. M. Gómez-Gómez, J. Blazquez, F. Baquero, and J. L. Martinez, Letter, Mol. Microbiol. 19:909-910, 1996). Furthermore, phenotypic expression of ClyA is observed in strains carrying any of the genes slyA, mprA, and hlyX on plasmids (8, 13, 25, 26, 35). Recent genome sequence analysis of serovar Typhi CT18 and serovar Typhimurium LT2 revealed the presence of an open reading frame, highly homologous to clyAEC, in serovar Typhi CT18, which was apparently absent in the LT2 strain analyzed (27, 39). We have screened different S. enterica serovars for the presence and expression of clyA, and here we report that the gene locus encoding ClyA cytotoxin is conserved in typhoid serovars while it is absent in many other serovars, including Typhimurium. As judged by our experimental findings, the Salmonella proteins showed a pore-forming activity, and hence they are the first examples of such a factor expressed by Salmonella.
(A preliminary account of part of this work has been presented [37]).
MATERIALS AND METHODS
Bacterial strains and culture media.
Serovar Typhimurium LT2 strain GT3258 was obtained from Dan Andersson, Department of Bacteriology, Swedish Institute for Infectious Disease Control, Solna, Sweden. Serovar Typhimurium SVA16 was isolated from a domesticated French quail (Coturnix coturnix). Serovar Typhimurium 4576 is a nonsubtypeable, highly antibiotic-resistant clinical isolate acquired in Ethiopia by a Swedish tourist. Serovar Havana St 63/1992 was isolated from animal feed ingredients in Sweden. Serovar Havana SR 15 was isolated from an Antarctic fur seal (Arctocephalus gazella) (38). Serovar Enteritidis PT4 was isolated from a Gentoo penguin (Pygoscelis papua) (33). Serovar Enteritidis 2894 is a human isolate of subtype PT4 acquired in Greece by a Swedish tourist. Serovar Newport Arg6, serovar Anatum Arg10, and serovar St. Paul Arg11 are all Argentinean human isolates and a kind gift from the Department of Microbiology, Hospital Rawson, Córdoba, Argentina. Serovar Dublin 2229 has been described previously (53). Serovar Typhi S2369/96, S1112/97, S2060/97, S1735/97, S1087/98, and S0200/99 and serovar Paratyphi A S3068/99 were isolated from clinical patients in Sweden who had all acquired typhoid fever abroad at different times and places and hence are epidemiologically unrelated. Ty2 is a wild-type strain of serovar Typhi isolated in 1916. Ty21a is a virulence-attenuated derivative of Ty2 that is used in a human live vaccine against typhoid fever (Vivotif, Berna, Italy) (12). JON42 is Ty21a ΔclyA. The E. coli strains YMZ16 and YMZ19 are derivatives of MC1061 (5) and BEU616 (MC1061 hns::cat) (50), respectively, that have a kanamycin cassette inserted into the clyA locus. DH5α (recA1) (15) was used in cloning experiments. The bacterial strains were grown aerobically at 37°C in Luria-Bertani (LB) broth or on LB broth solidified with 1.5% (wt/vol) agar. Unless stated otherwise, blood agar plates consisted of 5% horse erythrocytes solidified with 1% (wt/vol) Columbia agar (base) (Merck), which, according to the manufacturer, contains 2.3% (wt/vol) special nutrient substrate. When needed, antibiotic selection was carried out using 50 μg of carbenicillin ml−1, 50 μg of kanamycin ml−1, 15 μg of tetracycline ml−1, or 10 μg of chloramphenicol ml−1.
Genetic techniques and sequence analysis.
Standard procedures were used in all general molecular applications (42). Generalized bacteriophage P1 transduction was performed as described previously (52). Oligonucleotides were made on an Applied Biosystems 394 synthesizer or obtained from DNA Technology, Aarhus, Denmark, or from TAG Copenhagen, Copenhagen, Denmark. General purpose DNA sequencing was performed using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit With AmpliTaq DNA polymerase and an ABI PRISM 377 DNA sequencer. Salmonella genome sequence data were obtained from the following sources: sequence data for serovar Typhi strain CT18 were produced by the Microbial Genomes Sequencing Group at the Sanger Center. Serovar Paratyphi A ATCC 9510 and serovar Typhimurium LT2 SGSC1412 sequence data were produced by the Genome Sequencing Center, Washington University, St. Louis, Mo. During revision of this article, sequence data of serovar Typhi CT18 and serovar Typhimurium SGSC1412 were also published (27, 39). The following oligonucleotide primers based on serovar Typhi DNA sequences were used for screening by PCR and cloning of clyA-like sequences in Salmonella: stm1 (5′-CGCAGGTTCTGAATGCGGAA-3′), stm3 (5′-TAATACCTGCTGTAGCAAGG-3′), sal1 (5′-CTCGTCAGCCCGGTAACGAC-3′), sal2 (5′-GAGGTAATAGGTAAGAATAC-3′), and sal6 (5′-CGGTACCGATATCACCGATG-3′). The approximate annealing positions of these primers in the clyA locus are indicated below (see Fig. 2A). The primers sal1 and sal2 amplify a 1,101-bp DNA fragment that starts 73 bp upstream of the clyA start codon and ends 116 bp downstream of the clyA stop codon. The primers sal1 and sal6 amplify a 1,381-bp DNA fragment that starts 353 bp upstream of the clyA start codon and ends 116 bp downstream of the clyA stop codon. The primer stm1 anneals 170 bp downstream of the serovar Typhi clyA (clyASTy) stop codon, and the primer stm3 anneals 1,568 bp upstream of the clyASTy start codon.
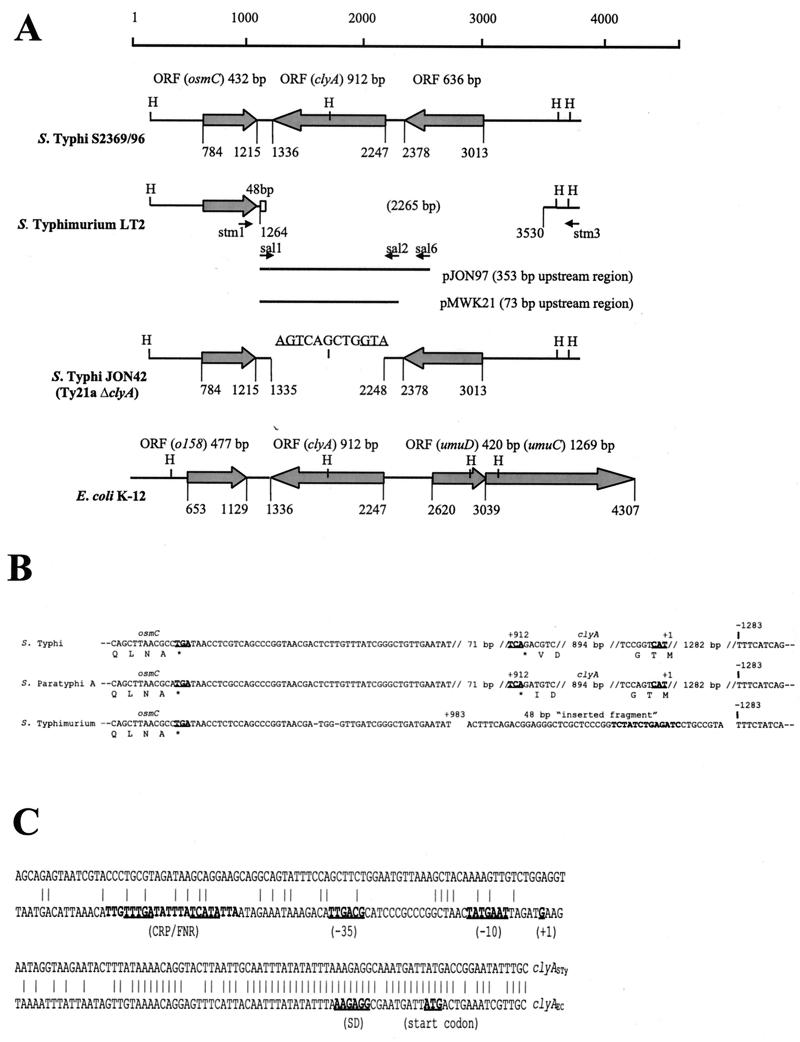
FIG. 2.
(A) Schematic map of the gene arrangement in a 4.5-kb region containing the clyA locus in serovar Typhi S2369/96, serovar Typhimurium LT2, serovar Typhi JON42, and E. coli K-12. The regions were aligned using the clyASTy gene sequence as a reference. Base pair coordinates are indicated relative to the arbitrarily chosen end point. Open reading frames are shown as shaded arrows, and the 48-bp unique sequence in serovar Typhimurium is shown as an open box. The 12 bp of the ΔclyASTy allele in JON42 are shown with start and stop codons underlined. The approximate positions of recognition sequences for the restriction endonuclease HincII (H) and of sequences corresponding to the primers stm1, stm3, sal1, sal2, and sal6 (small arrows) are indicated. The extents of the clyASTy DNA regions cloned in the constructs pMWK21 and pJON97 are shown as solid bars. ORF, open reading frame. (B) Alignment of nucleotide sequences flanking the osmC clyA DNA region of serovar Typhi S2369/96, serovar Paratyphi A ATCC9510, and serovar Typhimurium LT2. Start and stop codons are shown in boldface and are underlined. The 13 bp of the 48-bp “inserted fragment” that were absent in the serovar Havana strains SR15 and St 63/1992 (see Results) are shown in boldface. Positions are indicated relative to the clyA start codon (+1). (C) Alignment of clyA upstream sequences of serovar Typhi S2369/96 and the clyA promoter region of E. coli K-12. Promoter elements [(−35) and (−10)], the transcriptional start point (+1), the Shine-Dalgarno sequence (SD), the translational start codon (ATG), and binding sites for the CRP and FNR regulatory proteins (CRP/FNR), described for the clyAEC locus (13, 25, 50), are indicated.
Plasmid and strain construction.
The relevant genotypes of plasmids used in this work are listed in Table 1. To construct a 900-bp in-frame deletion mutant of the clyA gene locus in serovar Typhi, we used a PCR-based strategy with DH5α as the host strain. Serovar Typhi S2369/96 was used as the template. The primers p100 (5′-AGCAAATGACGAGCATGCGCTACGTGGATCCGATGAAAGAGAAC-3′) and p101 (5′-TTGTTCTGCAAATATGTCGACCATAATCATTTGCCT-3′) amplify a 1,344-bp PCR fragment comprising the DNA from 1,320 bp upstream to 23 bp downstream of the clyASTy start codon. This fragment contains a new restriction site for each of the enzymes BamHI (sequence in boldface) and SphI and SalI (underlined sequences). The SalI restriction site in this PCR fragment overlaps bp 4 to 9 in clyASTy. The primers p102 (5′-CTTTTCGAGGTTCCTGTCGACTGATACATTTTCATT-3′) and p103 (5′-GATGTCACTGAGGAGCTCATTGTCGGGATCCTCAACCCATTTG-3′) amplify a 1,252-bp PCR fragment comprising the DNA from 21 bp upstream to 1,230 bp downstream of the clyASTy stop codon. This fragment contains a new restriction site for each of the enzymes BamHI (sequence in boldface) and SacI and SalI (underlined sequences). The SalI restriction site in this PCR fragment overlaps bp 904 to 909 in clyASTy. The PCR fragments were digested with SphI-SalI and SalI-SacI, respectively, and were simultaneously ligated into an SphI-SacI-digested pUC18 cloning vector. The resulting construct, designated pJON101, was used as the source for a 2.6-kb BamHI fragment containing the clyASTy in-frame deletion, which was subsequently ligated into a BamHI-digested pKO3 suicide donor plasmid, resulting in the construct pJON103. Using pJON103, the clyASTy in-frame deletion was introduced onto the chromosome of Ty21a, as previously described (24), to generate the strain JON42. That JON42 carried the ΔclyA allele in the chromosome was confirmed by PCR analysis and DNA sequence determination (data not shown). For overexpression of Salmonella ClyA proteins, PCR-amplified DNA sequences were cloned directly into pGEM-T Easy using DH5α as the host strain. To avoid enhanced expression from the lac promoter carried on the pGEM-T Easy cloning vector, we used only plasmid constructs in which the amplified PCR fragments had been ligated in the orientation opposite to that of the lac promoter. The constructs pMWK21 and pJON94 (clyASTy) and pJON80 (serovar Paratyphi A clyA [clyASPa]) contain the PCR fragments obtained by using the oligonucleotide primers sal1 and sal2 with the strains S2369/96, Ty21a, and S3068/99, respectively, as templates. The construct pJON97 (clyASTy) contains the PCR fragment obtained by using the oligonucleotides sal1 and sal6 with S2369/96 as the template. The strain YMZ16 (clyAEC::kan) was created by the use of an allelic-exchange system involving the plasmid pPM103, with temperature-sensitive replication properties. The plasmid clone pYMZ71 contains the clyAEC locus within a 1.3-kb KpnI-SspI restriction fragment from the E. coli K-12 chromosome. A 1.2-kb BamHI restriction fragment containing the kanamycin resistance gene from the plasmid pUC4K was ligated into BglII-digested pYMZ71, resulting in the construct pYMZ103, with the kanamycin cassette inserted 71 bp into the clyAEC coding sequence. A 3.4-kb EcoRI-ScaI restriction fragment from pYMZ103, containing the insertionally inactivated clyAEC locus, was subsequently ligated into EcoRI-SmaI-digested pPM103. The resulting temperature-sensitive replicon carrying the inactivated clyA locus (clyA::kan) was then introduced by transformation into BEU616, which has a ClyA-positive phenotype and is hemolytic on blood agar plates (50). Tetracycline-resistant transformants were selected at 30°C on LB agar plates containing tetracycline. ClyA-negative colonies were found after the temperature was raised to 42°C, and the colonies were plated on blood agar plates. An isolate designated YMZ16 was selected for further studies from among colonies that were sensitive to tetracycline, indicating loss of the plasmid. That YMZ16 carried the clyA::kan allele in the chromosome was confirmed by PCR analysis and by phage P1-mediated transduction experiments. The strain MC1061 was transduced with P1 grown on YMZ16, and transductants were isolated by selection for kanamycin resistance, resulting in the strain YMZ19.
TABLE 1.
Plasmids used in this work
| Plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| pGEM-T Easy | T vector for cloning of PCR fragments; Cbr | Promega |
| pJON80 | 1.1-kb subclone of clyASPa from serovar Paratyphi A S3068/99 in pGEM-T Easy | This work |
| pJON94 | 1.1-kb subclone of clyASTy from serovar Typhi Ty21a in pGEM-T Easy | This work |
| pJON97 | 1.4-kb subclone of clyASTy from serovar Typhi S2369/96 in pGEM-T Easy | This work |
| pJON103 | clyASTy in-frame deletion in pKO3 | This work |
| pKO3 | Gene replacement vector; Cmr | 24 |
| pMWK21 | 1.1-kb subclone of clyASTy from serovar Typhi S2369/96 in pGEM-T Easy | This work |
| pMMkatF2 | 4.2-kb subclone of rpoSEC in pAT153 | 30 |
| pPM103 | Gene replacement vector; Tcr | 28 |
| pUC4K | Kanamycin resistance gene cartridge plasmid; Kanr | 46 |
| pUC18 | Cloning vector; Cbr | 55 |
| pYMZ71 | 1.3-kb subclone of clyAEC in pUC18 | This laboratory |
| pYMZ80 | 1.6-kb subclone of clyAEC in pUC18 | This laboratory |
| pYMZ81 | 1.6-kb subclone of clyAEC in pUC18 | This laboratory |
| pYMZ103 | clyAEC::kan in pUC18 | This work |
Determination of cytolytic activity.
Bacterium-mediated lysis of erythrocytes was scored by a clearance zone on blood agar plates incubated for 16 to 17 h at 37°C. As a convenient assay for cytolysin activity, we quantified the release of hemoglobin from erythrocytes essentially as described previously (43). The bacteria were grown on LB agar plates at 37°C for 16 to 17 h, and the cells were then harvested and suspended in 1× phosphate-buffered saline (PBS). Fifty microliters of a bacterial suspension in 1× PBS (final densities, 1 × 109, 4 × 108, and 2 × 108 bacterial cells ml−1 as specified in the figure and table legends) was mixed with 50 μl of an erythrocyte suspension (final density, ∼2 × 109 erythrocyte cells ml−1 in 1× PBS) in a 96-well microtiter plate. The microtiter plate was incubated for 45 min at 37°C prior to determination of the release of hemoglobin at 545 nm as described previously (43). For osmotic protection, saccharides were added at a final concentration of 30 mM in the assay wells as described previously (34). Saccharides were assumed to have the following molecular diameters according to published data (44, 45): maltose, 1.0 nm; dextrin 20, 1.6 nm; and dextrin 15, 2.2 nm. In accordance with a previous study (2), the mean molecular diameters of raffinose and dextran 4 were assumed to be 1.2 to 1.4 nm and 3.0 to 3.5 nm, respectively, on the basis of published data (44).
Subcellular fractionation of serovar Typhi cells and protein analysis.
For subcellular localization of ClyASTy, serovar Typhi cells were grown on LB agar plates at 37°C for 16 to 17 h and subsequently harvested in saline (final density, 3 × 1010 bacterial cells ml−1). The cells were pelleted, and the culture supernatants were filtered through a 0.45-μm-pore-size filter (Millipore) and used as the extracellular fraction. Periplasmic proteins were isolated from the sediment by osmotic shock, essentially as described earlier (25). The pelleted bacteria were washed with 20 mM Tris-HCl (pH 8.0) and resuspended in 0.25 volume (compared to the starting volume of harvested bacteria) of a solution containing 20% (wt/vol) sucrose, 20 mM Tris-HCl (pH 8.0), and 1 mM Na-EDTA. The mixture was incubated for 10 min at room temperature. Subsequently, the bacteria were pelleted and resuspended in 1 volume of ice-cold H2O. After incubation on ice for 10 min, the cells were removed by centrifugation at 12,000 × g. The supernatant was used as the periplasmic protein extract. The cell pellet was then disrupted by sonication in 1 volume of 20 mM Tris-HCl (pH 8.0). The cell debris and unbroken cells were removed by centrifugation at 5,000 × g for 10 min at 4°C, and the supernatant was subsequently fractionated into the membrane and cytosolic fractions by centrifugation at 10,000 × g for 30 min at 4°C. The supernatant was used as the cytosolic fraction, and the sediment was used as the membrane fraction. Extracellular, periplasmic, and cytosolic proteins were precipitated by addition of ice-cold trichloroacetic acid (final concentration, 10%), pelleted by centrifugation at 12,000 × g, washed with acetone, and dried under vacuum. The total protein concentration in the different subcellular fractions was subsequently adjusted to approximately 0.6 mg ml−1 by dissolving the fractions in 1× Laemmli lysis buffer. The protein concentrations in the fractions were determined using the bicinchoninic acid protein assay kit (Pierce) following the instructions of the manufacturer. For analysis of protein contents in bacterial strains, the strains were grown on LB agar plates at 37°C and harvested after 16 to 17 h of incubation. The bacteria were resuspended in saline, and the concentration was estimated by measuring the optical density at 600 nm using a spectrophotometer. The samples were then adjusted to 1010 cells ml−1 in 1× Laemmli lysis buffer. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting were performed as previously described (20, 21). For immunoblot anlaysis, we used a monoclonal antibody raised in mice against purified ClyAEC (S. N. Wai and B. E. Uhlin, unpublished data) or polyclonal antisera raised in rabbits against the purified proteins ClyAEC (34), SlyAEC (50), H-NS (19), β-lactamase (our unpublished data), and CRP (our unpublished data). The polyclonal ClyAEC and SlyAEC antisera were subtraction purified using a method that was described earlier (54). The monoclonal anti-ClyAEC antibody was used at a final dilution of 1:5,000 or 1:1,000 (as specified in the figure legends) with anti-mouse horseradish peroxidase-conjugated secondary antibody at a final dilution of 1:20,000. Polyclonal anti-ClyAEC and anti-SlyAEC antisera were used at final dilutions of 1:1,000 with horseradish peroxidase-conjugated anti-rabbit secondary antibody at a final dilution of 1:5,000. The polyclonal anti-H-NS, anti-CRP, and anti-β-lactamase antisera were used at a final dilution of 1:3,000 with horseradish peroxidase-conjugated anti-rabbit secondary antibody at a final dilution of 1:20,000. Immunoreactive bands were visualized using the ECL+ chemiluminiscence Western blotting detection system from Amersham Pharmacia Biotech, following the instructions of the manufacturer. Expression of catalase activity was evidenced by formation of bubbles on the addition of a few drops of 3% hydrogen peroxide to bacterial colonies on agar plates as described previously (51).
Nucleotide sequence accession numbers.
Our nucleotide sequence data for the clyASTy locus in serovar Typhi S2369/96 and Ty21a and the clyASPa locus in serovar Paratyphi A S3068/99 have been deposited in the EMBL database as entries AJ313032, AJ313033, and AJ313034.
RESULTS
Presence of clyA-homologues in human-specific Salmonella serovars.
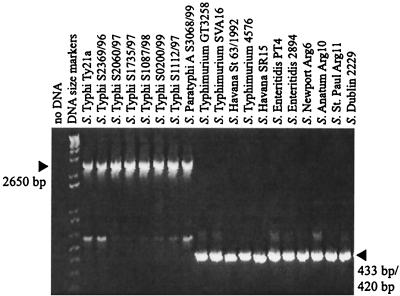
In a previous study, no clyA gene could be detected in a clinical isolate of serovar Typhimurium by Southern hybridization (8). Similarly, by using PCR and oligonucleotide primers based on the E. coli K-12 sequence, umu1 (5′-AATATTTGTCGCTGC-3′) and p79 (5′-TGTCAACAGGTAACTCTC-3′), or by performing Southern hybridization, we found that no clyA-like sequence could be amplified from or detected in the serovar Typhimurium LT2 strain GT3258 (our unpublished data). However, recent genome sequence analysis showed that there is a clyA-like sequence in the serovar Typhi strain CT18 but not in the serovar Typhimurium LT2 strain SGSC1412 (27, 39). To extend this analysis, we tested for the presence of a homologue to the clyAEC gene in various Salmonella genomes by means of PCR using the oligonucleotide primers stm1 and stm3, which are based on the serovar Typhi genome sequence (Fig. 1). Our results clearly showed that clyA-like sequences are present in the genomes of all seven tested serovar Typhi isolates and in the tested serovar Paratyphi A isolate. On the other hand, all tested isolates of other serovars—three serovar Typhimurium isolates, two serovar Enteritidis isolates, two serovar Havana isolates, one serovar Anatum isolate, serovar Dublin, serovar Newport, and serovar St. Paul—were all assumed to lack the clyA locus, since a much shorter DNA fragment was obtained by PCR. A weaker band corresponding to ∼500-bp fragments appeared in all cases and presumably represents partial unspecific base pairing of the oligonucleotide primers used.
FIG. 1.
PCR analysis of the osmC clyA DNA regions of various Salmonella isolates. PCR products were resolved by agarose gel electrophoresis following amplification using the primers stm1 and stm3, which amplify a 433- or a 420-bp fragment in strains lacking both clyA and osmC and a 2,650-bp fragment in clyA+ osmC+ strains (arrowheads on right and left, respectively).
DNA sequence analysis showed that there was 100% sequence identity between the serovar Typhi strains S2369/96 and CT18 over the 2,650-bp region between the primers stm1 and stm3, which encompasses the clyA locus. Furthermore, we found that the clyA-like open reading frame of serovar Typhi S2369/96 and CT18 shows 100% nucleotide sequence identity with the clyA coding sequence of the serovar Typhi vaccine strain Ty21a and 87% nucleotide sequence identity with clyAEC. Sequence analysis of the clyA homologue (clyASPa) of serovar Paratyphi A S3068/99 revealed that this gene shows 98 and 86% nucleotide sequence identity with clyASTy and clyAEC, respectively, and 100% nucleotide sequence identity with the clyA locus of the serovar Paratyphi A strain ATCC 9510. In E. coli K-12, the clyA locus is located in close proximity and transcribed in the opposite direction to the umuCD operon, which is involved in resistance to UV-mediated stress (8). However, in typhoid Salmonella, other sequences were found in close proximity to the clyA locus (Fig. 2). In serovar Typhi, a 432-bp open reading frame shows 79% nucleotide sequence identity with the E. coli gene osmC, encoding an osmotically inducible protein. The osmC homologue starts 552 and ends 121 bp downstream of the clyASTy stop codon, i.e., it is transcribed in the direction opposite to that of clyASTy. A 636-bp open reading frame in the same direction as clyASTy, showing no significant similarity to that encoding any known protein according to computer searches, starts 766 and ends 131 bp upstream of the clyASTy sequence. Sequence comparisons showed the same arrangement of open reading frames around the clyA locus in serovar Paratyphi A as in serovar Typhi (Fig. 2). When we analyzed the shorter PCR fragments (433 bp) amplified from the serovar Typhimurium strains SVA16, 4576, and GT3258 obtained using the oligonucleotides stm1 and stm2, we found that both the clyA locus and the 636-bp open reading frame (see above) were absent in all the isolates. Although these strains carry an osmC-like sequence, a 2,265-bp DNA region spanning from 1,282 bp upstream of what would correspond to the clyASTy start codon to 71 bp downstream of the clyASTy stop codon was found to be missing. We also noted the presence of a novel 48-bp DNA sequence positioned at the junction of this apparent 2,265-bp deletion (Fig. 2B). Computer searches, however, did not reveal any significant similarity that could suggest the origin of this 48-bp DNA sequence. DNA sequence analysis confirmed that there was 100% sequence identity over the region encompassed by the oligonucleotides stm1 and stm3 in SVA16, 4576, and GT3258. According to sequence analysis, the same region was absent in the other nontyphoid S. enterica serovars. The two serovar Havana isolates SR15 and St 63/1992 additionally lacked 13 bp in the 48-bp unique DNA fragment (Fig. 2B). Hence, the PCR product obtained from the serovar Havana strains using stm1 and stm3 was slightly smaller (420 bp), which was also supported by gel electrophoresis (Fig. 1). In conclusion, these findings expand the host range of ClyA to typhoid Salmonella and show that the clyA locus and a novel 636-bp open reading frame found directly upstream of clyASTy and clyASPa are absent in many Salmonella serovars, including Typhimurium.
Activity and secretion of ClyA cytotoxin in typhoid Salmonella.
Analogous to clyAEC, the clyASTy and clyASPa genes were predicted to encode polypeptides of 303 amino acids (mass, ∼34,000 Da) that show 91 and 90% amino acid identity to ClyAEC, respectively. To assess whether ClyASTy and ClyASPa, similarly to ClyAEC, exhibit lytic activity, we amplified the clyASTy and clyASPa gene loci by PCR using the oligonucleotides sal1 and sal2 with the strains S2369/96 and S3068/99, respectively, as templates. The resulting 1,101-bp PCR products were cloned into the expression vector pGEM-T Easy, resulting in the constructs pMWK21 (clyASTy) and pJON80 (clyASPa). The clyASTy locus was also amplified as a 1,381-bp fragment by PCR using the primers sal1 and sal6 and cloned into pGEM-T Easy, resulting in the construct pJON97. When introduced by transformation into the E. coli strain YMZ19 (clyA::kan), pMWK21, pJON80, and pJON97 all induced a strong lytic phenotype when cultured on blood agar plates, indicating that clyASTy and clyASPa have cytolytic activity per se (Table 2). We subsequently tested whether the ClyASTy and ClyASPa proteins could also be recognized by a monoclonal antibody raised against ClyAEC, since they are very similar to the E. coli protein. As evidenced by Western immunoblot analysis, the anti-ClyAEC antibody could be used to monitor ClyASTy and ClyASPa (Fig. 3A). The strains YMZ19/pMWK21 and YMZ19/pJON97 were found to express high levels of ClyASTy protein, and YMZ19/pJON80 was found to express large amounts of ClyASPa protein, in contrast to the vector control strain YMZ19/pGEM-T Easy, which did not.
TABLE 2.
Expression of phenotypical levels of ClyA as evidenced by lysisa of horse erythrocytes in agar
| Strain | Relevant characteristics | Lysis activity |
|---|---|---|
| E. coli | ||
| YMZ19/pGEM-T Easy | clyA::kan; vector | − |
| YMZ19/pMWK21 | clyA::kan clyASTy+ | ++ |
| YMZ19/pJON97 | clyA::kan clyASTy+ | ++ |
| YMZ19/pJON80 | clyA::kan clyASPa+ | ++ |
| MC1061/pYMZ80 | clyA+clyAEC+ | ++ |
| MC1061/pUC18 | clyA+; vector | − |
| M182/pGEM-T Easy | crp+; vector | − |
| M182crp/pGEM-T Easy | crp; vector | − |
| M182/pJON97 | crp+clyASTy+ | ++ |
| M182crp/pJON97 | crp clyASTy+ | ++ |
| Serovar Typhi | ||
| Ty21a | clyASTy+ | + |
| JON42 | ΔclyA | − |
| JON42/pJON94 | ΔclyA clyASTy+ | ++ |
| JON42/pGEM-T Easy | ΔclyA; vector | − |
| Ty21a/pGEM-T Easy | clyA+; vector | + |
| Ty21a/pMWK21 | clyA+clyASTy+ | ++ |
| Ty21a/pJON97 | clyA+clyASTy+ | ++ |
| Ty21a/pUC18 | clyA+; vector | + |
| Ty21a/pMMkatF2 | clyA+rpoSEC+ | + |
| S2369/96 | clyASTy+ | − |
| S1112/97 | clyASTy+ | − |
| Serovar Paratyphi A S3068/99 | clyASPa+ | − |
Lysis was scored on blood agar plates containing horse erythrocytes as follows: ++, large lysis zones around individual colonies; +, small lysis zones around individual colonies; −, no lysis.
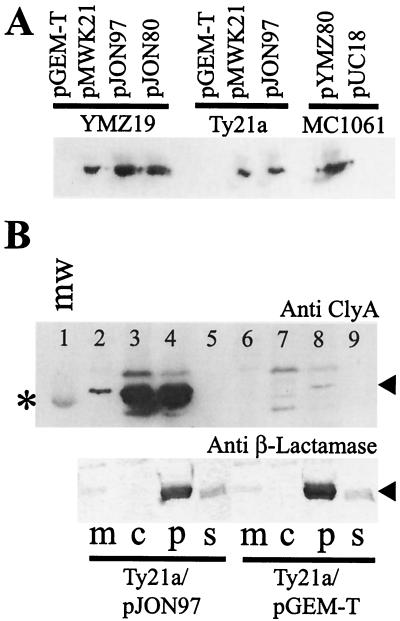
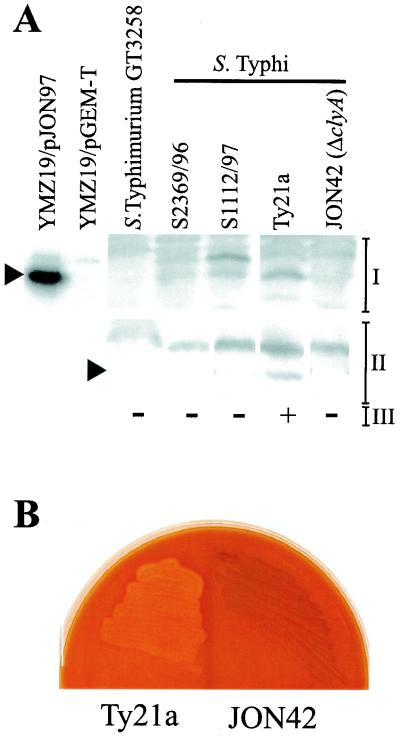
FIG. 3.
Expression of cloned serovar Typhi, serovar Paratyphi A, and E. coli ClyA determinants and subcellular localization of ClyASTy in serovar Typhi. Immunoreactive bands were visualized using the ECL+ blotting detection system (see Materials and Methods). (A) Immunoblot analysis, using a monoclonal ClyAEC antibody (final dilution, 1:5,000), of expression of cloned clyASTy (constructs pJON97 and pMWK21), clyASPa (construct pJON80), and clyAEC (construct pYMZ80) gene loci in E. coli YMZ19 (clyA::kan) and MC1061 (clyA+) and serovar Typhi Ty21a. Approximately 5 × 107 cells of each strain were used for the extracts applied on the gel. (B) Subcellular fractionation of serovar Typhi Ty21a/pJON97 (lanes 2 to 5) and Ty21a/pGEM-T Easy (lanes 6 to 9) in membrane (m), cytosolic (c), periplasmic (p), and supernatant (s) fractions. Above, immunoblot analysis using a monoclonal antibody raised against ClyAEC (final dilution 1:5,000) is shown. The position of the band corresponding to the 30-kDa protein of the molecular-mass marker (mw; lane 1) is indicated by an asterisk, and the position of the immunoreactive band representing the ClyA protein is indicated by an arrowhead. Below, immunoblot analysis of the same subcellular fractions (corresponding to lanes 2 to 9 in the upper blot) using a polyclonal antiserum against β-lactamase is shown. The position of the immunoreactive band representing β-lactamase is indicated by an arrowhead. An amount of the subcellular fraction equal to approximately 3 μg of protein was applied in each lane of the gel.
To test whether ClyASTy could be expressed and secreted in serovar Typhi, we introduced the constructs pMWK21 and pJON97 into the serovar Typhi strain Ty21a by transformation. Similar to E. coli K-12, overexpression of clyASTy in serovar Typhi resulted in a strong hemolytic phenotype, as detected on blood agar plates (Table 2). As shown by Western immunoblotting, a significant amount of ClyASTy protein was produced by Ty21a/pJON97 and Ty21a/pMWK21 (Fig. 3A), and the lytic-plate phenotype suggested that ClyASTy reached the serovar Typhi cell surface. How ClyA is translocated out of the bacterial cell is currently unclear in E. coli as well as Salmonella. Like ClyAEC, the Salmonella homologues did not show a canonical signal sequence according to computer analysis.
We performed subcellular fractionation (see Materials and Methods) to localize ClyASTy in the strains Ty21a/pGEM-T Easy, Ty21a/pMWK21, and Ty21a/pJON97 (Fig. 3B). Large amounts of the ClyASTy protein were found in both the cytosolic and periplasmic fractions of Ty21a/pJON97 and Ty21a/pMWK21 (Fig. 3B, lanes 3 and 4, and data not shown). This is similar to what has been found previously with ClyAEC in E. coli strains (25). Also, in the case of the vector control strain Ty21a/pGEM-T Easy there was an immunoreactive band presumably representing a small amount of ClyASTy in the periplasmic fraction (Fig. 3B, lane 8). As ClyA was present in both cytosolic and periplasmic fractions, the possibility could not be excluded that a fraction of periplasm was mixed in with the cytosol during the preparation of the extracts. Hence, we used the periplasmic enzyme β-lactamase as a marker protein and analyzed the same extracts using a polyclonal antiserum raised against β-lactamase. As shown in Fig. 3B, bottom, we found that the β-lactamase indeed was present in periplasmic fractions but not in cytosolic fractions, indicating that there was no or very minor contamination of the cytosol from the periplasm. We also analyzed the same extracts using polyclonal antisera against the global regulatory proteins CRP and H-NS. As expected, these cytosolic proteins were mainly present in the cytosolic fractions, although there were also minor but detectable amounts of the proteins in the periplasmic fractions (data not shown). Taken together, these findings indicated that ClyASTy was efficiently translocated through the inner membrane in serovar Typhi.
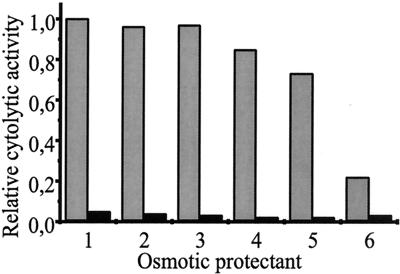
To assess if, similarly to ClyAEC, the Salmonella cytotoxins act on erythrocytes by pore formation, we used an osmotic protection assay (see Materials and Methods). For this assay, it has been reported that the saccharide raffinose affords no protection against HlyA hemolysin- or ClyAEC-induced lysis whereas the larger molecule dextran 4 prevents lysis at 30 mM (2, 34). In hemolysis assays using a suspension of the serovar Typhimurium LT2 strain GT3258/pJON97 (clyASTy), lysis could be significantly reduced (approximately five times) in the presence of the osmoprotectant dextran 4 but not by the other, smaller saccharides (Fig. 4). Similar results were obtained with ClyASPa as overexpressed in GT3258/pJON80 (data not shown). Based on these findings, we concluded that ClyASTy and ClyASPa, like ClyAEC, form pores in erythrocyte membranes that are smaller than 3.0 to 3.5 nm (the molecular diameter of dextran 4) but larger than 2.2 nm (the molecular diameter of dextrin 15). To summarize, we found that the clyA loci of the human-specific typhoid Salmonella encode pore-forming proteins that can be released or secreted from the bacteria.
FIG. 4.
Pore-forming activity of ClyASTy evidenced by osmotic protection using different sugar solutions in combination with suspensions of the serovar Typhimurium LT2 strains GT3258/pJON97 (clyASTy+) (shaded bars) and GT3258/pGEM-T Easy (vector control) (solid bars). Cytolytic activity was determined as described in Materials and Methods. The bacteria were grown on LB agar plates at 37°C for 16 to 17 h before being harvested. Approximately 108 bacterial cells were used for the samples applied in each well of the microtiter plate (final density, 109 bacterial cells ml−1). The activity of GT3258/pJON97 in 1× PBS was arbitrarily set to 1.0. Osmotic protectants (lane 1, 1× PBS; lane 2, maltose; lane 3, raffinose; lane 4, PEG1000; lane 5, PEG1500; and lane 6, dextran 4) were added at a final concentration of 30 mM.
ClyASTy is phenotypically expressed by serovar Typhi strain Ty21a.
The presence of a pore-forming determinant in the genomes of typhoid Salmonella prompted us to investigate whether these gene loci are phenotypically expressed or, as in clyAEC, are repressed and phenotypically silent. Interestingly, upon cultivation on agar plates containing horse erythrocytes, the serovar Typhi strain Ty21a exhibited zones of hemolysis around the colonies while serovar Typhi S2369/96 and S1112/97 and serovar Paratyphi A S3068/99 did not (Table 2). To monitor the protein expression of clyASTy from the chromosome, the strains were subsequently harvested from the blood agar plates and analyzed by Western immunoblotting, using an enhanced detection system (Fig. 5A). Consistent with the phenotypes on blood agar, there was a detectable amount of ClyASTy protein accumulated in Ty21a, at a slightly higher level than in serovar Typhi S2369/96 and S1112/97, which could explain the difference in phenotype among the strains (Fig. 5A).
FIG. 5.
Phenotypic expression of ClyA cytotoxin by serovar Typhi Ty21a and loss of lytic phenotype upon inactivation of the clyASTy gene locus. (A) Western immunoblotting detection of ClyASTy protein content in serovar Typhi strains using a polyclonal ClyAEC antiserum (I) or a monoclonal antibody raised against ClyAEC (final dilution, 1:1,000) (II) and the ECL+ Western blotting detection system for visualization of immunoreactive bands (see Materials and Methods). The following approximate numbers of bacterial cells were used for the extracts loaded in the lanes. For the Salmonella strains, 3 × 108 cells were used for the polyclonal antiserum and 1 × 108 cells were used for the monoclonal antibody. For YMZ19/pJON97 (clyA::kan clyASTy+) and YMZ19/pGEM-T Easy (clyA::kan; vector), approximately 8 × 106 bacterial cells were used. The positions of reactive bands corresponding to ClyA are indicated with arrowheads on the left. The phenotypes of the strains on blood agar plates containing horse erythrocytes are indicated at the bottom (III) and were scored after 16 h of growth at 37°C (+, small lysis zones around individual colonies; −, no lysis). (B) Effect of inactivation of the clyASTy locus in serovar Typhi Ty21a. Shown are the phenotypes of Ty21a and the clyASTy in-frame deletion mutant JON42 on a blood agar plate containing horse erythrocytes after incubation at 37°C for 17 h.
We noted that Ty21a, although able to lyse horse erythrocytes, did not exhibit lysis zones when grown on agar plates containing human or rabbit erythrocytes, and only a weak cytolytic activity was observed on blood agar containing sheep erythrocytes. Using cloned clyA genes from serovar Typhi and E. coli, we extended the analysis of the lysis of different erythrocytes by quantifying the release of hemoglobin by means of the assay described in Materials and Methods. E. coli DH5α cells containing various plasmid constructs were harvested and resuspended in 1× PBS and subsequently mixed with horse, sheep, human, goat, hen, and rabbit erythrocytes (Table 3). Evidently, both the ClyASTy- and ClyAEC-expressing cells showed the highest lytic activity towards horse erythrocytes, compared to, e.g., sheep and human erythrocytes, while the activity was very low against rabbit erythrocytes.
TABLE 3.
Lysis of various erythrocytes by ClyASTy and ClyAEC
| Strain | Plasmid charac- teristics | Lysis (% of maximum)a
|
|||||
|---|---|---|---|---|---|---|---|
| Horse | Sheep | Human | Goat | Hen | Rabbit | ||
| DH5α/pUC18 | p(vector) | 1.6 | 1.6 | 1.1 | 0.7 | 1.0 | 0.1 |
| DH5α/pYMZ81 | p(ClyAEC) | 96 | 82 | 69 | 46 | 43 | 10 |
| DH5α/pGEM-T Easy | p(vector) | 2.5 | 0.9 | 0.6 | 0.9 | 2.7 | 0.1 |
| DH5α/pJON97 | p(ClyASTy) | 57 | 37 | 35 | 30 | 21 | 0.5 |
Lysis of erythrocytes was quantified using the hemolysis assay described in Materials and Methods. Shown are the percentage as of the maximal release of hemoglobin which was obtained by the addition of 1% Triton X-100 in the reaction wells. The bacteria were grown on LB agar plates at 37°C for 16 to 17 h before being harvested. Approximately 2 × 107 bacterial cells were used for the samples applied in each assay well (final density, 2 × 108 bacterial cells ml−1).
To confirm that the hemolytic phenotype of Ty21a was caused by expression of ClyASTy, we introduced by allelic replacement an in-frame deletion in the clyASTy gene locus on the chromosome of Ty21a (Fig. 2A) (see Materials and Methods). The resulting derivative, JON42 (ΔclyASTy), was tested on blood agar plates containing horse erythrocytes and compared to the parent strain, Ty21a. As shown in Fig. 5B, the hemolytic phenotype was completely lost after the mutagenesis. Furthermore, our Western immunoblot analyses of these strains demonstrated that ClyASTy protein expression was abolished in JON42 (Fig. 5A). The hemolytic phenotype was restored by introduction of the plasmid pJON94, containing the clyA gene locus cloned from Ty21a, into JON42 (Table 2). Hence, we could conclude that the hemolytic phenotype of Ty21a was indeed caused by expression of the ClyASTy protein from the clyASTy locus.
H-NS-mediated repression of ClyASTy production in E. coli.
Recently, the clyA promoter in E. coli K-12 has been characterized as an H-NS-silenced class I promoter that can be transcriptionally activated by CRP and/or FNR (50). An alignment of sequences 400 bp upstream of the clyA start codon revealed high (>90%) sequence identity between serovar Typhi and serovar Paratyphi A, but the clyA upstream regions were very different from that of E. coli K-12 (Fig. 2C). Sequence identity was observed only very close to the clyA start codon in E. coli K-12, overlapping with the Shine-Dalgarno sequence. We noted that a CRP/FNR DNA site seemingly is absent in the clyASTy and clyASPa upstream sequences. Hence, a direct involvement of CRP and/or FNR as a modulator(s) of the expression of clyA in typhoid Salmonella appears less likely. To test whether there was a requirement for CRP for expression of clyASTy in E. coli, similar to what has been observed with clyAEC (50), we introduced the construct pJON97 into the E. coli K-12 strains M182 and M182 crp. As indicated in Table 2, we found that there was no apparent difference in ClyASTy expression in these strains, indicating that there was no requirement for CRP.
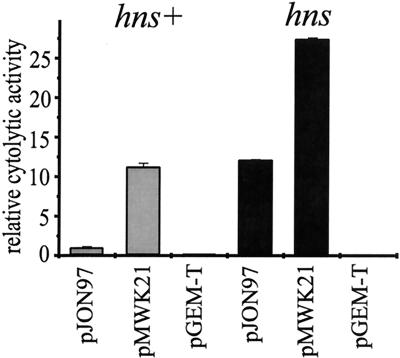
The DNA of the upstream region (the 276 bp immediately upstream of the clyASTy start codon), like clyAEC, is rich in AT basepairs (65%), a hallmark of H-NS-regulated genes (1), and according to our data it may also be subject to silencing by H-NS. We used the constructs pJON97, containing the clyASTy structural gene and the DNA up to 353 bp upstream of the start codon, and pMWK21, containing the clyASTy locus, including a shorter part of the upstream region (73 bp). To measure the expression of cytolytic activity, we used bacterial suspensions and erythrocytes in a quantifying assay (see Materials and Methods). As shown in Fig. 6, when introduced into the E. coli K-12 strains YMZ19 (clyA) and YMZ16 (hns clyA), the expression of cytolytic activity from pJON97 was substantially higher (more than ninefold) in the hns strain YMZ16 than in the hns+ strain YMZ19. Derepression of clyASTy expression in the hns strain YMZ16 was also observed using the construct pMWK21, but the increase in cytolytic activity was less pronounced (approximately twofold). Hence, it appears that the clyASTy locus, similar to the clyA locus in E. coli K-12, may be subject to silencing by H-NS and that this feature may keep the gene in a repressed state. To investigate whether the enhanced expression of ClyA cytotoxin in Ty21a could be due to a reduced expression of H-NS in this strain, we performed Western immunoblotting using a polyclonal anti-H-NS antiserum. Our findings showed that H-NS was expressed at similar levels in all of the serovar Typhi strains Ty21a, S2369/96, and S1112/97 (data not shown).
FIG. 6.
H-NS-dependent repression of clyASTy in the E. coli K-12 strains YMZ19 (clyA hns+) (shaded bars) and YMZ16 (clyA hns) (solid bars) carrying different constructs. The bacteria were grown on LB agar plates at 37°C for 16 to 17 h before being harvested. Shown is the relative cytolytic activity as determined by quantification of the release of hemoglobin from erythrocytes (see Materials and Methods). Approximately 4.0 × 107 bacterial cells were used for the samples applied in each well of the microtiter plate, giving a final density of 4 × 108 cells per ml. The activity of YMZ19/pJON97 was arbitrarily set to 1.0. The error bars indicate standard errors of the mean from two separate experiments.
In E. coli K-12 strains, the H-NS-mediated silencing of clyA expression has been shown to be relieved upon overexpression of the SlyA protein, possibly by competing with H-NS for binding at the clyA locus, although clyA expression does not require SlyA per se (25, 35, 50). Examination of available genomic data for serovar Typhi CT18 (39) revealed the presence of a 435-bp open reading frame showing 99 and 81% nucleotide sequence identity with slyA of serovar Typhimurium and E. coli K-12, respectively. Using an anti-SlyAEC antiserum, we performed immunoblot analyses of SlyA protein in serovar Typhi strains. The results showed that SlyA was expressed at a low level in all tested strains, i.e., Ty21a, S2369/96, and S1112/97, and the levels were similar to that observed in E. coli K-12 (data not shown). Since there was little or no difference among the serovar Typhi strains, we find it less likely that expression of SlyA was responsible for the increase in ClyA cytotoxin seen in Ty21a.
Serovar Typhi Ty21a has been shown to be defective for the alternative sigma factor σS (RpoS) (41). Since expression of many RpoS-dependent genes is modulated by global regulators, such as H-NS and CRP in E. coli (reference 16 and references therein), we tested whether the RpoS deficiency could be a possible reason for the hemolytic ClyA expression in Ty21a. To restore RpoS in Ty21a, we introduced the rpoSEC+ allele carried on the plasmid pMMkatF2 into Ty21a by transformation. As evidenced by the expression of catalase activity (see Materials and Methods), Ty21a/pMMkatF2 was considered to be rpoS+ in contrast to the vector control strain Ty21a/pUC18. As indicated in Table 2, we found that the hemolytic phenotype of Ty21a was retained in this rpoS+ background, suggesting that the rpoS mutation per se is not responsible for the enhanced expression of ClyA in Ty21a.
DISCUSSION
Previous research with purified ClyA protein from E. coli K-12 demonstrated that ClyA is a pore-forming protein, with cytotoxic and apoptotic activities towards cultured mammalian cells (22, 25, 34, 47). As no experimental evidence of pore-forming factors in Salmonella had previously been presented, we aimed our experiments at elucidating features of the serovar Typhi and serovar Paratyphi A clyA homologues. Based on the strong similarity to clyAEC, both of the gene products (ClyASTy and ClyASPa) were assumed to share features with ClyAEC. To initially characterize these proteins, the clyASTy and clyASPa determinants were cloned and overproduced from an expression vector. The proteins could be immunogenically recognized by a monoclonal antibody raised against ClyAEC and, analogous to ClyAEC, their molecular masses were determined to be ∼34 kDa. The results from hemolysis assays using various saccharides as osmoprotectants demonstrated that ClyASTy and ClyASPa, similar to ClyAEC, have a pore-forming activity towards erythrocytes. Hence, ClyASTy and ClyASPa are the first examples of a pore-forming cytolysin encoded by Salmonella. It was earlier reported by Libby and coworkers (23) that they had occasionally observed weak zones of hemolysis around some colonies, especially in low-passage clinical isolates of Salmonella. However, it was not mentioned which Salmonella serovars were studied. In a cloning experiment using serovar Typhimurium 14028s, these authors obtained a clone containing a gene designated slyA (23). We and others subsequently showed that slyA, which also occurs in E. coli, encodes a regulatory protein that can activate clyA expression (26, 35). It remains to be elucidated whether the clinical isolates mentioned by Libby et al. (23) represent serovar Typhimurium, serovar Typhi, or other serovars.
The lytic phenotype of serovar Typhi Ty21a on horse erythrocytes, caused by natural expression of ClyA cytotoxin from the chromosomal gene locus, could not be detected on blood agar plates containing human or rabbit erythrocytes, and we observed only a weak cytolytic activity towards sheep erythrocytes. These findings were supported by our hemolysis assays, in which both ClyASTy and ClyAEC exhibited the strongest lysis activity against horse erythrocytes, while there was a lower activity against, e.g., sheep, human, and especially rabbit erythrocytes (Table 3). The reason for this difference in lysis of different erythrocytes is not known. In neither Salmonella nor E. coli is it established how ClyA is released from the bacterial cell. When overexpressed in E. coli strains, the ClyAEC protein was previously shown by subcellular localization to accumulate in periplasmic fractions (25). We obtained similar results upon subcellular fractionation of serovar Typhi Ty21a expressing ClyASTy naturally from the chromosomal gene locus or from an expression vector, indicating that the cytolysin was efficiently translocated across the inner membrane (Fig. 3B).
To our knowledge, Ty21a is the first reported phenotypically hemolytic Salmonella strain in which the genetic determinant has been identified. We observed that the serovar Typhi vaccine strain Ty21a lysed horse erythrocytes and had a hemolytic phenotype on blood agar plates (Fig. 5B). The lytic activity was abolished upon inactivation of the clyASTy gene locus in Ty21a and could be restored by the introduction of the clyASTy gene carried on a plasmid. We could therefore conclude that the hemolytic phenotype of Ty21a was caused by expression of ClyA cytotoxin. None of the other tested serovar Typhi isolates were detectably hemolytic on blood agar plates. It should be noted that the absence of hemolysis zones around colonies on blood agar does not necessarily mean that the clyA locus is silent. Recent findings using a sensitive system for Western immunoblot detection have demonstrated that, although nonhemolytic, some E. coli strains may contain a small amount of ClyA protein (36). Our present experiments, using the same system, demonstrated that there was a somewhat enhanced expression of ClyASTy from the chromosomal gene locus of Ty21a compared to the nonhemolytic serovar Typhi isolates S2369/96 and S1112/97. Previous studies have shown that the threshold for appearance of hemolysis on blood agar plates is attained by relatively small increases in clyAEC expression (40), and thus the smaller quantities of ClyASTy produced by S2369/96 and S1112/97 could explain their nonhemolytic phenotypes.
At present, we do not know why Ty21a expresses a higher level of ClyASTy than other tested serovar Typhi isolates. Strain Ty21a has multiple mutations due to nitrosoguanidine-induced mutagenesis. The genetic alterations include mutations in genes affecting (i) Vi antigen biosynthesis, (ii) isoleucine-valine metabolism, (iii) H2S production, and (iv) growth rate (11, 12, 18). Although the clyASTy and clyAEC genes are highly homologous and both genes appear not to be fully derepressed, our sequence alignments showed that their promoter regions are very different (Fig. 2C). It appeared unlikely that CRP and/or FNR, as in E. coli K-12, is directly involved in the transcriptional activation of clyA expression in serovar Typhi or in serovar Paratyphi A, and no DNA site for these global regulators was identified upstream of the clyASTy and clyASPa coding sequences. Experiments with the cloned clyASTy locus in clyA and clyA hns mutant E. coli K-12 strains demonstrated that clyASTy, similarly to clyAEC, may be subject to silencing by H-NS (Fig. 6). Our constructs used in these experiments contained the clyASTy locus with the 73 (pMWK21) or the 353 (pJON97) bp directly upstream of the clyASTy start codon, and our findings suggested that at least the 73 bp upstream of the clyA start codon are required for the regulation of clyA expression. This implies that clyA, analogous to the case in E. coli, may constitute a monocistronic operon in typhoid Salmonella. Even if clyASTy is a putative regulatory target for H-NS, the enhanced expression of the ClyASTy protein in Ty21a appeared not to be due to a reduced expression of H-NS in this strain, as evidenced by Western immunoblot analysis. Although our study showed that there was H-NS repression of clyASTy in E. coli, the results must be viewed with caution with respect to the situation in serovar Typhi. Further studies using, e.g., hns mutant serovar Typhi will hopefully reveal if H-NS plays a similar role in Salmonella.
Overproduction of the SlyA protein is known to activate ClyA expression in E. coli (25, 26, 35). As judged by our experimental determination of SlyA protein content, the phenotypic expression of ClyA cytotoxin in Ty21a is not likely to be caused by a higher level of SlyASTy in that strain. Although it has been suggested to contribute to the virulence of serovar Typhimurium (4), which lacks the clyA locus, the role of SlyA is not yet fully understood. Further research should reveal if SlyA plays a role in the activation of clyA expression in typhoid Salmonella.
In the present work, we establish that clyA is conserved in serovar Typhi and serovar Paratyphi A, while the gene locus is apparently absent in many other Salmonella serovars, including Typhimurium. Our PCR results and DNA sequencing showed that the coding regions are highly homologous among serovar Typhi (clyASTy), serovar Paratyphi A (clyASPa), and E. coli (clyAEC). However, when the corresponding DNA regions in isolates of other Salmonella serovars were analyzed, a large apparent deletion (2,265 bp) was observed, removing the entire clyA locus and, interestingly, also a 636-bp open reading frame found directly upstream of clyASTy and clyASPa (Fig. 2). The clyA locus was replaced by a 48-bp DNA fragment that did not show any significant similarity to the sequences available in databases. Upon completion of the genome sequences of serovar Typhi CT18 (39) and serovar Typhimurium LT2 (27), many differences were found between the two serovars that may explain their different host specificities, e.g., several genes present in serovar Typhi are missing in serovar Typhimurium and vice versa. The absence of the clyA locus obviously reflects the fact that the clyA gene product is not correlated with virulence in some serovars. However, in the case of certain Salmonella serovars, we may consider that expression of ClyA cytotoxin could play a role at some point in particular types of infections. Hence, there could be selection for clyA in certain narrow-host-range serovars like Typhi and Paratyphi A that are pathogenic in humans only. Several lines of evidence support the idea that cytolytic factors are important for the pathogenesis of virulent bacteria (9), and the fact that typhoid Salmonella can express a protein with a pore-forming activity towards mammalian cells is intriguing. Our findings suggest that we must consider that ClyA may be a component of the arsenal of virulence factors possessed by typhoid Salmonella. The expression of ClyA by such strains may cause sublytic effects analogous to the host cell responses recently shown to be triggered by the α-hemolysin from uropathogenic E. coli (45a). Whether this is the case with typhoid Salmonella and the vaccine strain Ty21a is an intriguing question, and the construction of a clyASTy mutant derivative (JON42) should allow the assessment of such aspects.
Acknowledgments
We thank the Genome Sequencing Center, Washington University, for communication of DNA sequence data prior to publication. We thank Anna Aspán, Sven Bergström, Tina Broman, the Swedish Polar Research Secretariat, and the Center for Environmental Research for valuable assistance with sampling of Salmonella strains. We thank Hans Wolf-Watz for critical reading of the manuscript, Margareta Ramberg at SMI for valuable technical assistance, Dan Andersson for providing serovar Typhimurium LT2 GT3258, and Roland Rosqvist for providing serovar Dublin 2229.
This work was supported by grants from the Swedish Science Research Council (projects K2001-06X-10383-09C and B5101-1435) and the Swedish Foundation for International Cooperation in Research and Higher Education (STINT reference no. 99/966). S.N.W. was supported in part by a Visiting Scientist Fellowship from the Wenner-Gren Foundations. J.O. was supported by a Research Associate Fellowship and research funds from the Faculty of Medicine and Odontology, Umeå University.
Editor: V. J. DiRita
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Bhakdi, S., N. Mackman, J. M. Nicaud, and I. B. Holland. 1986. Escherichia coli hemolysin may damage target cell membranes by generating transmembrane pores. Infect. Immun. 52:63-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, V., and T. Focareta. 1991. Pore-forming bacterial protein hemolysins (cytolysins). Crit. Rev. Microbiol. 18:115-158. [DOI] [PubMed] [Google Scholar]
- 4.Buchmeier, N., S. Bossie, C. Y. Chen, F. C. Fang, D. G. Guiney, and S. J. Libby. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. M., K. Kaniga, and J. E. Galan. 1996. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 21:1101-1115. [DOI] [PubMed] [Google Scholar]
- 7.Chopra, A. K., J. H. Huang, X. Xu, K. Burden, D. W. Niesel, M. W. Rosenbaum, V. L. Popov, and J. W. Peterson. 1999. Role of Salmonella enterotoxin in overall virulence of the organism. Microb. Pathog. 27:155-171. [DOI] [PubMed] [Google Scholar]
- 8.del Castillo, F. J., S. C. Leal, F. Moreno, and I. del Castillo. 1997. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted hemolysin. Mol. Microbiol. 25:107-115. [DOI] [PubMed] [Google Scholar]
- 9.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galan, J. E. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263-271. [DOI] [PubMed] [Google Scholar]
- 11.Germanier, R., and E. Furer. 1983. Characteristics of the attenuated oral vaccine strain S. typhi Ty 21a. Dev. Biol. Stand. 53:3-7. [PubMed] [Google Scholar]
- 12.Germanier, R., and E. Furer. 1975. Isolation and characterization of Gal E mutant Ty 21a of Salmonella typhi: a candidate strain for a live, oral typhoid vaccine. J. Infect. Dis. 131:553-558. [DOI] [PubMed] [Google Scholar]
- 13.Green, J., and M. L. Baldwin. 1997. The molecular basis for the differential regulation of the hlyE-encoded haemolysin of Escherichia coli by FNR and HlyX lies in the improved activating region 1 contact of HlyX. Microbiology 143:3785-3793. [DOI] [PubMed] [Google Scholar]
- 14.Hacker, J., C. Hughes, H. Hof, and W. Goebel. 1983. Cloned hemolysin genes from Escherichia coli that cause urinary tract infection determine different levels of toxicity in mice. Infect. Immun. 42:57-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 16.Hengge-Aronis, R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 17.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hone, D. M., S. R. Attridge, B. Forrest, R. Morona, D. Daniels, J. T. LaBrooy, R. C. Bartholomeusz, D. J. Shearman, and J. Hackett. 1988. A galE via (Vi antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect. Immun. 56:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson, J., and B. E. Uhlin. 1999. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary-phase survival of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:10776-10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston, K. H., and J. B. Zabriskie. 1986. Purification and partial characterization of the nephritis strain-associated protein from Streptococcus pyogenes, group A. J. Exp. Med. 163:697-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lai, X. H., I. Arencibia, A. Johansson, S. N. Wai, J. Oscarsson, S. Kalfas, K. G. Sundqvist, Y. Mizunoe, A. Sjöstedt, and B. E. Uhlin. 2000. Cytocidal and apoptotic effects of the ClyA protein from Escherichia coli on primary and cultured monocytes and macrophages. Infect. Immun. 68:4363-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Libby, S. J., W. Goebel, A. Ludwig, N. Buchmeier, F. Bowe, F. C. Fang, D. G. Guiney, J. G. Songer, and F. Heffron. 1994. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. USA 91:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig, A., S. Bauer, R. Benz, B. Bergmann, and W. Goebel. 1999. Analysis of the SlyA-controlled expression, subcellular localization and pore-forming activity of a 34 kDa haemolysin (ClyA) from Escherichia coli K-12. Mol. Microbiol. 31:557-567. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig, A., C. Tengel, S. Bauer, A. Bubert, R. Benz, H. J. Mollenkopf, and W. Goebel. 1995. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol. Gen. Genet. 249:474-486. [DOI] [PubMed] [Google Scholar]
- 27.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 28.Meacock, P. A., and S. N. Cohen. 1979. Genetic analysis of the interrelationship between plasmid replication and incompatibility. Mol. Gen. Genet. 174:135-147. [DOI] [PubMed] [Google Scholar]
- 29.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 93:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulvey, M. R., P. A. Sorby, B. L. Triggs-Raine, and P. C. Loewen. 1988. Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene 73:337-345. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien, A. D., and R. K. Holmes. 1996. Protein toxins of Escherichia coli and Salmonella, p. 2788-2802. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 32.Ohl, M. E., and S. I. Miller. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259-274. [DOI] [PubMed] [Google Scholar]
- 33.Olsen, B., S. Bergström, D. J. McCafferty, M. Sellin, and G. Wiström. 1996. Salmonella enteritidis in Antarctica: zoonosis in man or humanosis in penguins? Lancet 348:1319-1320. [DOI] [PubMed] [Google Scholar]
- 34.Oscarsson, J., Y. Mizunoe, L. Li, X. H. Lai, Å. Wieslander, and B. E. Uhlin. 1999. Molecular analysis of the cytolytic protein ClyA (SheA) from Escherichia coli. Mol. Microbiol. 32:1226-1238. [DOI] [PubMed] [Google Scholar]
- 35.Oscarsson, J., Y. Mizunoe, B. E. Uhlin, and D. J. Haydon. 1996. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol. Microbiol. 20:191-199. [DOI] [PubMed] [Google Scholar]
- 36.Oscarsson, J., M. Westermark, L. Beutin, and B. E. Uhlin. 2002. The bacteriophage-associated Ehly1 and Ehly2 determinants from Escherichia coli O26:H− strains do not encode enterohemolysins per se but cause release of the ClyA cytolysin. Int. J. Med. Microbiol. 291:625-631. [DOI] [PubMed] [Google Scholar]
- 37.Oscarsson, J., M. Westermark, S. Löfdahl, and B. E. Uhlin. 2000. Expression of a pore-forming cytotoxin (ClyA, HlyE, SheA) by typhoid Salmonella. Med. Microbiol. Immunol. 189:44. [Google Scholar]
- 38.Palmgren, H., D. McCafferty, A. Aspán, T. Broman, M. Sellin, R. Wollin, S. Bergström, and B. Olsen. 2000. Salmonella in sub-Antarctica: low heterogeneity in Salmonella serotypes in South Georgian seals and birds. Epidemiol. Infect. 125:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 40.Ralph, E. T., J. R. Guest, and J. Green. 1998. Altering the anaerobic transcription factor FNR confers a hemolytic phenotype on Escherichia coli K12. Proc. Natl. Acad. Sci. USA 95:10449-10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robbe-Saule, V., C. Coynault, and F. Norel. 1995. The live oral typhoid vaccine Ty21a is a rpoS mutant and is susceptible to various environmental stresses. FEMS Microbiol. Lett. 126:171-176. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sansonetti, P. J., A. Ryter, P. Clerc, A. T. Maurelli, and J. Mounier. 1986. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect. Immun. 51:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scherrer, R., and P. Gerhardt. 1971. Molecular sieving by the Bacillus megaterium cell wall and protoplast. J. Bacteriol. 107:718-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schönherr, R., M. Hilger, S. Broer, R. Benz, and V. Braun. 1994. Interaction of Serratia marcescens hemolysin (ShlA) with artificial and erythrocyte membranes. Demonstration of the formation of aqueous multistate channels. Eur. J. Biochem. 223:655-663. [DOI] [PubMed] [Google Scholar]
- 45a.Uhlén, P., A. Laestadius, T. Jahnukainen, T. Söderblom, F. Backhed, G. Celsi, H. Brismar, S. Normark, A. Aperia, and A. Richter-Dahlfors. 2000. Alpha-hemolysin of uropathogenic E. coli induces Ca2+ oscillations in renal epithelial cells. Nature 405:694-697. [DOI] [PubMed] [Google Scholar]
- 46.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 47.Wallace, A. J., T. J. Stillman, A. Atkins, S. J. Jamieson, P. A. Bullough, J. Green, and P. J. Artymiuk. 2000. E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell 100:265-276. [DOI] [PubMed] [Google Scholar]
- 48.Welch, R. A. 1991. Pore-forming cytolysins of gram-negative bacteria. Mol. Microbiol. 5:521-528. [DOI] [PubMed] [Google Scholar]
- 49.Welch, R. A., C. Forestier, A. Lobo, S. Pellett, W. Thomas, Jr., and G. Rowe. 1992. The synthesis and function of the Escherichia coli hemolysin and related RTX exotoxins. FEMS Microbiol. Immunol. 5:29-36. [DOI] [PubMed] [Google Scholar]
- 50.Westermark, M., J. Oscarsson, Y. Mizunoe, J. Urbonaviciene, and B. E. Uhlin. 2000. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 182:6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whittenbury, R. 1964. Hydrogen peroxide formation and catalase activity in the lactic acid bacteria. J. Gen. Microbiol. 35:13-26. [DOI] [PubMed] [Google Scholar]
- 52.Willets, N. S., A. J. Clark, and B. Low. 1969. Genetic locations of certain mutations conferring recombination deficiency in Escherichia coli. J. Bacteriol. 97:244-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williamson, C. M., G. D. Pullinger, and A. J. Lax. 1988. Identification of an essential virulence region on Salmonella plasmids. Microb. Pathog. 5:469-473. [DOI] [PubMed] [Google Scholar]
- 54.Woods, J. P., J. F. Dempsey, T. H. Kawula, D. S. Barritt, and J. G. Cannon. 1989. Characterization of the neisserial lipid-modified azurin bearing the H.8 epitope. Mol. Microbiol. 3:583-591. [DOI] [PubMed] [Google Scholar]
- 55.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]