Abstract
Post-translational histone modifications regulate epigenetic switching between different chromatin states. Distinct histone modifications, such as acetylation, methylation and phosphorylation, define different functional chromatin domains, and often do so in a combinatorial fashion. The centromere is a unique chromosomal locus that mediates multiple segregation functions, including kinetochore formation, spindle-mediated movements, sister cohesion and a mitotic checkpoint. Centromeric (CEN) chromatin is embedded in heterochromatin and contains blocks of histone H3 nucleosomes interspersed with blocks of CENP-A nucleosomes, the histone H3 variant that provides a structural and functional foundation for the kinetochore. Here, we demonstrate that the spectrum of histone modifications present in human and Drosophila melanogaster CEN chromatin is distinct from that of both euchromatin and flanking heterochromatin. We speculate that this distinct modification pattern contributes to the unique domain organization and three-dimensional structure of centromeric regions, and/or to the epigenetic information that determines centromere identity.
The flexible N-terminal tails of the four core histones (H2A, H2B, H3 and H4) undergo a range of post-translational modifications, including acetylation, methylation, phosphorylation and ubiquitination1,2. Histone modifications are indicators of active or repressed chromatin, and the proposed ‘histone code’ hypothesis suggests that combinations of specific histone modifications create a complex, functional hierarchy for chromatin regulation2. For example, acetylation of histones H3 and H4, and H3 methylation at Lys4, have been predominantly correlated with euchromatin and gene activity. Methylation of H3 at Lys4 (H3 Lys4-Me) is typically associated with transcriptionally active chromatin, whereas methylation at Lys9 (H3 Lys9-Me) correlates with transcriptionally silent chromatin1,2. Notably, different methylated states of the same amino acid residue provide additional tiers of regulation to epigenetic inheritance of chromatin domains. For example, H3 Lys4 dimethylation (H3 Lys4-diMe) is associated with ‘permissive’ chromatin that is either active or potentially active, and H3 Lys4 trimethylation (H3 Lys4-triMe) is linked with transcriptional activity3–5. Conversely, H3 Lys9 di- and trimethylation (H3 Lys9-diMe and H3 Lys9-triMe) mark facultative and constitutive heterochromatin, respectively, in mammals6,7. These heterochromatic modifications are also associated with stochastic silencing (position effect variegation) of euchromatic genes experimentally positioned within or near heterochromatin in, for example, D. melanogaster and Schizosaccharomyces pombe8–11.
The specification and propagation of the site of centromere and kinetochore formation (centromere identity) seem to be determined by epigenetic mechanisms12,13. The centromere-specific histone H3-like protein CENP-A (CID in D. melanogaster) is a conserved marker for centromeric chromatin14,15. Previous studies have provided evidence that CENP-A proteins are structural and functional foundations for the kinetochore, making them excellent candidates for an epigenetic mark that specifies centromere identity. In vertebrates and invertebrates, CENP-A is required for recruitment of kinetochore proteins, mitotic progression and chromosome segregation, and may initiate the kinetochore assembly pathway12,16.
CENP-A/CID chromatin also seems to be the structural foundation for the specialized three-dimensional structure of the kinetochore. We have previously demonstrated that centromeric chromatin in humans and the fruit fly, D. melanogaster, contains interspersed blocks of CENP-A/CID and H3 nucleosomes, which we will refer to here collectively as CEN chromatin15. CENP-A- and H3-containing subdomains are located in close proximity in two dimensions on a ‘linear’ chromatin fiber and on polynucleosomes. However, in mitotic chromosomes, CENP-A/CID subdomains merge to form a three-dimensional cylindrical structure that largely excludes H3 nucleosomes. Blocks of CENP-A nucleosomes are oriented on the poleward face of the chromosome, and blocks of H3 nucleosomes are located toward the inner chromatid region. To reconcile two-dimensional interspersion of CENP-A/CID and H3 blocks with separation in three-dimensional mitotic chromosomes, we proposed that CEN DNA may spiral or loop through the cylindrical structure, leading to alignment or stacking of nucleosomes with the same composition15. Recent studies in rice indicate that the presence of H3 nucleosomes in CEN chromatin is conserved in plants17.
Centromeres are usually embedded in heterochromatin, and thus might be expected to contain heterochromatic histone modifications. However, epistasis studies and cytological localizations suggest that CEN chromatin and flanking (pericentromeric) heterochromatin are structurally and functionally distinct18,19. Are H3 subdomains within CEN chromatin of multicellular eukaryotes modified like heterochromatin or euchromatin, or are they uniquely marked? In S. pombe, CENP-A (Cnp1) is uniformly distributed across the 5–7 kilobase ‘central core’ regions, and large heterochromatic domains that contain H3 Lys9-methylation and heterochromatin proteins flank the cores12,16. Are larger eukaryotic centromeres composed of multimers of units equivalent to S. pombe centromeres, such that the interspersed H3 domains are modified like heterochromatin? To address these questions, we studied histone modifications within centromere regions as markers for the chromatin states of these domains, using extended chromatin fibers and mitotic chromosomes from cultured human and D. melanogaster cells, and from D. melanogaster larval neuroblasts.
RESULTS
CEN chromatin fibers lack heterochromatic modifications
We used immunofluorescence with mono-specific antibodies to localize modified histones within CEN chromatin and the flanking heterochromatin. First, we asked whether CEN chromatin contains modifications that typically mark pericentromeric heterochromatin, specifically H3 Lys9 di- and trimethylation6,7 (Fig. 1). On extended chromatin fibers from interphase human cells, CEN chromatin was identified by the full extent of CENP-A antibody staining. H3 Lys9-diMe was not present in the spaces between CENP-A spots, which we know from previous studies contains blocks of H3 nucleosomes15. Instead, H3 Lys9-diMe antibody staining was present in the pericentromeric regions that flanked CEN chromatin (Fig. 1a). Similarly, on every interphase chromatin fiber from either D. melanogaster S2 cells or third-instar larval neuroblasts, H3 Lys9-diMe staining was not observed within CEN chromatin, but was present in the flanking regions (Fig. 1b). We carried out semiquantitative analysis of fluorescent signals, and determined that H3 Lys9-diMe did not overlap, or overlapped minimally, with the edges of the CENP-A-containing domain on chromatin fibers (Fig. 2; see Methods).
Figure 1.
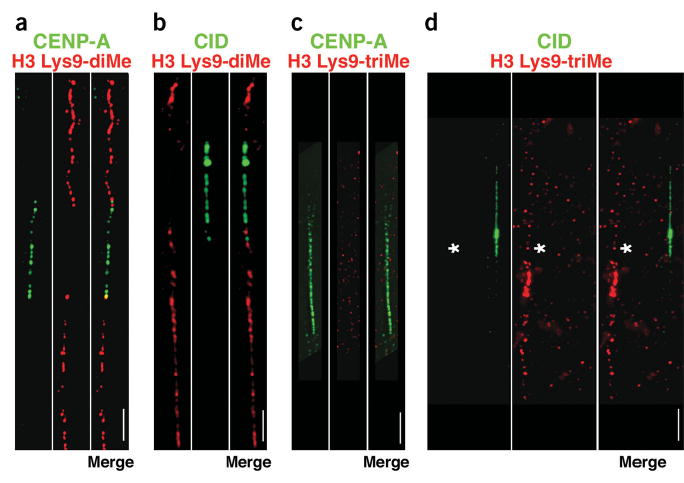
H3 is not di- or trimethylated at Lys9 in CEN chromatin. (a–d) Extended chromatin fibers from human (a,c) or D. melanogaster (b,d) interphase cells were stained with antibodies to CENP-A or CID (green), and H3 Lys9-diMe or H3 Lys9-triMe (red), detected with FITC- and Cy3-conjugated secondary antibodies. Merged images are to the right of the single-color images. Spaces between CENP-A/CID blocks denote H3-containing nucleosome blocks15, and did not stain for H3 Lys9-diMe or H3 Lys9-triMe. H3 Lys9-diMe, typically found in heterochromatin, was present on one side (67%, n = 30) or both sides (33%; n = 30) of the human CENP-A region (a). Lys9 was not dimethylated on H3 within the CID domain on chromatin fibers from D. melanogaster third-instar larval neuroblasts and S2 cells, and always flanked the CID region on both sides (b). In contrast, H3 Lys9-triMe was not present in the regions flanking CENP-A/CID (c,d). To demonstrate that staining for H3 Lys9-triMe was present in noncentromeric regions in these preparations, two different fibers are shown in d, one that is CID-negative but H3 Lys9-triMe-positive (denoted by asterisk), and one that is CID-positive and H3 Lys9-triMe-negative. Quantitative evaluations of the overlaps are presented in Figure 2. Scale bars, 15 μm in a,c; 5 μm in b,d.
Figure 2.
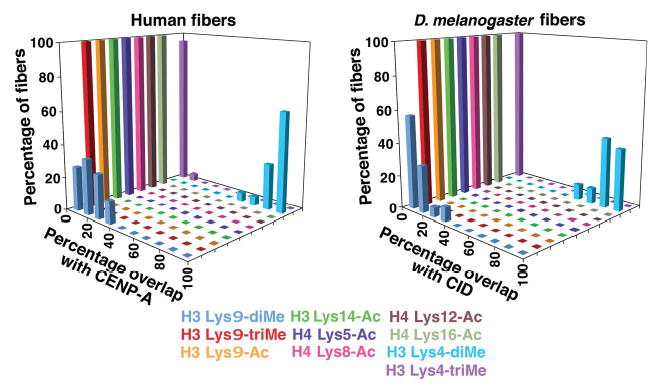
Quantification of overlap between histone modifications and CENP-A/CID on chromatin fibers from flies and humans. Extended chromatin fibers from human cells and flies were analyzed by fluorescence line plots to determine the amount of overlap between CENP-A/CID and modified histone antibody signals. Chromatin fibers from human and flies showed <40% overlap of regions containing H3 Lys9-diMe with areas containing CENP-A or CID. Conversely, >60% overlap was seen between H3 Lys4-diMe and CENP-A or CID regions. Acetylated residues in H3 and H4, and H3 Lys4-triMe, did not overlap with CENP-A or CID regions.
We conclude that the H3 Lys9-diMe modification is present in the pericentric heterochromatin immediately flanking both D. melanogaster and human CEN chromatin. The slight overlap observed at the edges of human and fly CEN chromatin probably reflects the resolution limitations of the fiber technique, rather than real intermixing of the CENP-A/CID and H3 Lys9-diMe domains. Specifically, individual elements in the fibers contain blocks of nucleosomes, and are not stretched to reveal individual nucleosomes. Most importantly, in both species H3 Lys9-diMe was not present between CENP-A/CID blocks, indicating that the absence of this heterochromatic modification in CEN chromatin is conserved.
H3 Lys9-triMe is enriched in pericentric regions of mouse and human chromosomes6,7. In our studies, H3 Lys9-triMe antibody staining did not overlap at all with CEN chromatin on human and D. melanogaster fibers, indicating that the absence of this heterochromatic modification in CEN chromatin is also conserved. Notably, H3 Lys9-triMe staining was not present in the regions immediately flanking CENP-A/CID (Figs. 1c,d and 2), as was observed for H3 Lys9-diMe. In fact, H3 Lys9-triMe was present on only one side of the CEN domain on many fibers in both flies and humans, and was observed 5–30 μm away from CENP-A or CID antibody staining. These results suggest that H3 Lys9-triMe is present in pericentric sequences located farther away from CEN chromatin in humans and flies, in comparison to H3 Lys9-diMe (see Discussion).
Heterochromatin is excluded from CEN chromatin at metaphase
To determine the distribution of H3 Lys9-diMe within the three-dimensional centromere and kinetochore, we examined unfixed mitotic chromosomes by indirect immunofluorescence and deconvolution microscopy15 (Fig. 3). H3 Lys9-diMe was present between sister kinetochores and in chromatin immediately flanking CENP-A/CID in both human and D. melanogaster chromosomes (Fig. 3a–f). Three-dimensional modeling and semiquantitative analysis confirmed that CENP-A/CID and H3 Lys9-diMe regions did not overlap substantially in mitosis (Figs. 3c,f and 4).
Figure 3.
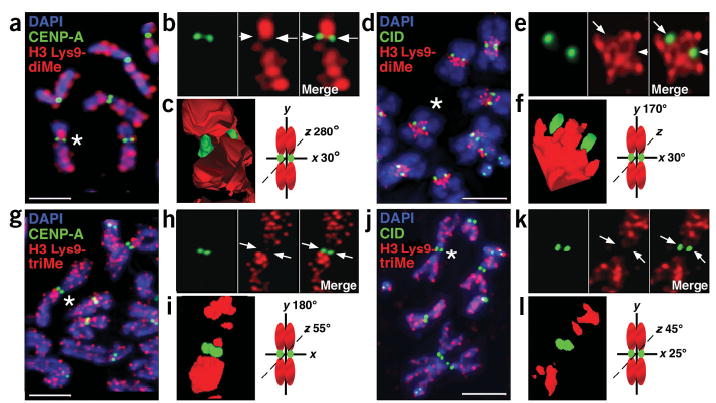
H3 is not di- or trimethylated at Lys9 in CEN chromatin at metaphase. (a–j) Immunofluorescence patterns on human (a,g) and D. melanogaster (d,j) metaphase chromosomes show localization of H3 Lys9-diMe or H3 Lys9-triMe (red) relative to CENP-A and CID staining (green). Chromosomal DNA was stained with DAPI (blue). (b,e,h,k) Enlarged views of chromosomes marked with asterisks in a, d, g (human chromosome 1) and j (D. melanogaster chromosome 3), with merged images to the right of the single-color images. Arrows point to the areas on individual chromosomes where CENP-A/CID is located. (c,f,i,l) Three-dimensional modeling of antibody staining on the same human and D. melanogaster metaphase chromosomes from b, e, h, and k. Chromosome cartoons depict the degree to which each three-dimensional model was rotated around the x-, y- and/or z-axes. In all cases, CENP-A/CID shows minimal overlap with H3 Lys9-diMe or tri-Me staining. H3 Lys9-diMe was present in the spaces between the CENP-A/CID cylinders15 on the two sister chromatids, as well as on either side of the centromere region along the chromosome arms. In contrast, H3 Lys9-triMe was offset significantly from CENP-A/CID staining along the chromosome arms, and was not present in the spaces between the sister centromeres. Quantifications of overlaps are shown in Figure 4. Scale bars, 15 μm in a,g; 5 μm in b,j.
Figure 4.
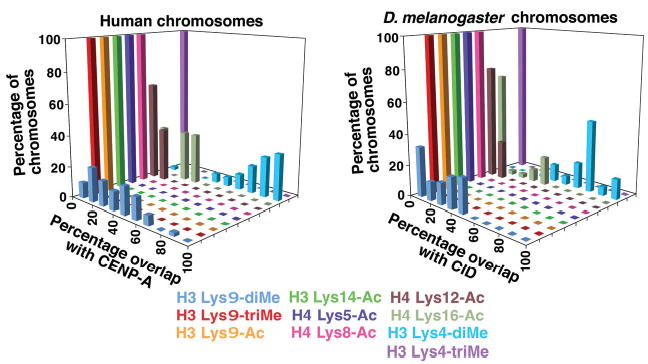
Quantification of overlap between histone modifications and CENP-A/CID on metaphase chromosomes from humans and flies. Metaphase chromosomes from human cells and flies were analyzed by fluorescence line plots to determine the amount of overlap between CENP-A/CID and modified histone antibody signals. Overlap between CENP-A or CID staining, and H3 Lys9-diMe, was less than that seen for H3 Lys4-diMe. More overlap was observed in metaphase chromosomes in comparison with fibers for Lys9 methylation and some of the acetylated H3 and H4 residues, reflecting the lower resolution obtained in condensed chromosomes versus extended fibers.
H3 Lys9-triMe also did not overlap substantially with CENP-A/CID in human or D. melanogaster metaphase chromosomes (Figs. 3g–l and 4). However, the pattern of H3 Lys9-triMe associations with the flanking heterochromatin was less consistent than observed for H3 Lys9-diMe. On human chromosomes, H3 Lys9-triMe was concentrated in the pericentromeric regions of chromosomes 1, 9 and Y, where the large blocks of classical satellite DNA are located (Fig. 3g–i). Unlike H3 Lys9-diMe, H3 Lys9-triMe was present between sister CENP-A spots on only some human chromosomes. The higher concentration of H3 Lys9-triMe in pericentromeric regions of many human chromosomes is in agreement with the distribution observed in mouse chromosomes6,7. However, in the mouse studies low levels of H3 Lys9-triMe were observed in the chromosome arms, whereas we observed more H3 Lys9-triMe in many human chromosome arms. It is possible that the relative concentrations of this modification in heterochromatin and euchromatin differ between human and mouse chromosomes (see Discussion). However, we think it is more likely that these differences can be attributed to chromosome preparation methods. In a recent study6, cells were treated with colchicine and hypotonic buffer before fixation, whereas we have analyzed untreated, unfixed chromosomes (see Methods). Determination of which preparation method more accurately reflects the distribution of H3 Lys9-triMe in native chromosomes awaits more detailed studies.
The distribution of H3 Lys9-triMe in D. melanogaster mitotic chromosomes has not previously been reported. We observed that H3 Lys9-triMe was present in pericentromeric regions of chromosomes 2 and 3, but was not enriched in the proximal heterochromatic regions of the X chromosome (Fig. 3j–l and data not shown)
From the fiber and three-dimensional studies, we conclude that centromeric chromatin in flies and humans, as defined by CENP-A/CID, is not enriched for H3 Lys9 di- or trimethylation during interphase or mitosis. Notably, the absence of these modifications distinguishes the H3 nucleosome blocks present in centromeric chromatin from the flanking heterochromatin.
CEN chromatin contains hypoacetylated histones
Because heterochromatic modifications were absent from CEN chromatin, we tested whether modifications associated with euchromatin and/or active genes were present in CEN chromatin, specifically acetylation of H3 and H4. On extended fibers and mitotic chromosomes from flies and humans, CEN chromatin did not contain detectable amounts of H3 Lys9 acetylation, or H4 Lys5, Lys8, Lys12 or Lys16 acetylation (Fig. 5, quantification in Figs. 2 and 4). These modifications, in general, were absent from regions immediately flanking CEN chromatin, in agreement with previous studies of pericentromeric heterochromatin20,21. Therefore, the interspersed H3 in CEN chromatin exhibits typical histone hypoacetylation associated with heterochromatin, but lacks heterochromatic H3 Lys9 di-and trimethylation. It has been shown that CENP-A/CID nucleosomes do contain H4 dimers14,15, so we conclude that the CENP-A/CID nucleosomes are also not acetylated at these H4 lysine residues.
Figure 5.
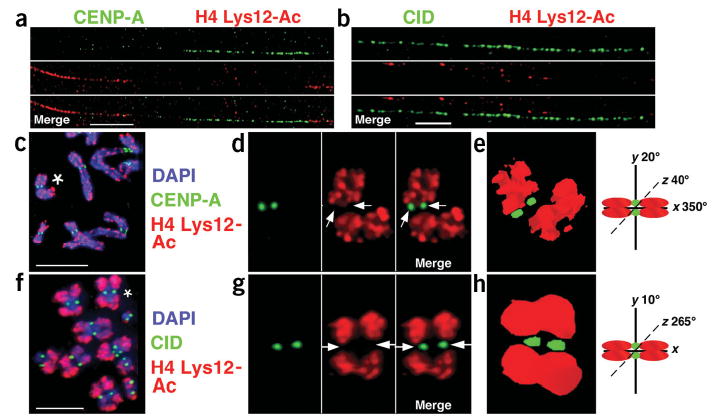
H3 and H4 are hypoacetylated in CEN chromatin. (a,b) Immunofluorescence patterns on human (a) and D. melanogaster (b) chromatin fibers stained with H4 Lys12-Ac antibodies. Chromatin fibers and metaphases from human and D. melanogaster cells did not show enrichment for H3 or H4 lysine acetylation in CEN chromatin. For example, CENP-A is located close to, but does not overlap with, H4 Lys12-Ac. Single-color images are shown separated from the merged image. (c–h) Antibody localization on metaphase chromosomes (c,f) shows that CENP-A/CID and H4 Lys12-Ac staining do not colocalize, and that pericentromeric regions are also hypoacetylated at H4 Lys12. (d,g) Enlarged view of chromosomes marked with asterisks in c and f, with single-color images shown to the right of the merged images. Arrows point to the areas on individual chromosomes where CENP-A/CID, but not H4 Lys12-Ac, were located. (e,h) Three-dimensional modeling of individual metaphase chromosomes from d and g demonstrate the separation of CENP-A/CID and acetylated histones. The separation is particularly pronounced on D. melanogaster chromosomes (f), which contain large amounts of pericentromeric heterochromatin. Chromosome cartoons depict the degree to which each three-dimensional model was rotated around the x-, y- and/or z-axes in e and h. Similar results were obtained after staining fibers and metaphases with antibodies to five other H3 and H4 acetylated residues, which are quantified in Figures 2 and 4.
CEN chromatin contains a euchromatic modification
The methylation status (mono-, di- and trimethylation) of histone H3 at Lys4 has been shown to be biologically important for transitioning to active chromatin states. The H3 Lys4-diMe modification has been associated with ‘poised’ but not necessarily ‘active’ euchromatin4,5, and has been identified at promoter and transcribed genomic regions. Thus, it was surprising that H3 Lys4-diMe staining was observed between CENP-A/CID subdomains in both human and fly centromeres on extended fibers (Figs. 2 and 6a,b). Mitotic chromosomes from human cell lines contained H3 Lys4-diMe on chromosome arms, as previously reported22, and was largely excluded from pericentromeric regions, particularly in chromosomes 1, 9, 16 and most of the Y chromosome (Fig. 6c–h). However, H3 Lys4-diMe and CENP-A staining frequently colocalized (Figs. 4 and 6d). Three-dimensional modeling showed that H3 Lys4-diMe staining was positioned near the inner chromatid portion of the CENP-A cylinder (Fig. 6e). In D. melanogaster S2 tissue culture cells, H3 Lys4-diMe also colocalized with CID on mitotic chromosomes (Figs. 4 and 6f–h).
Figure 6.
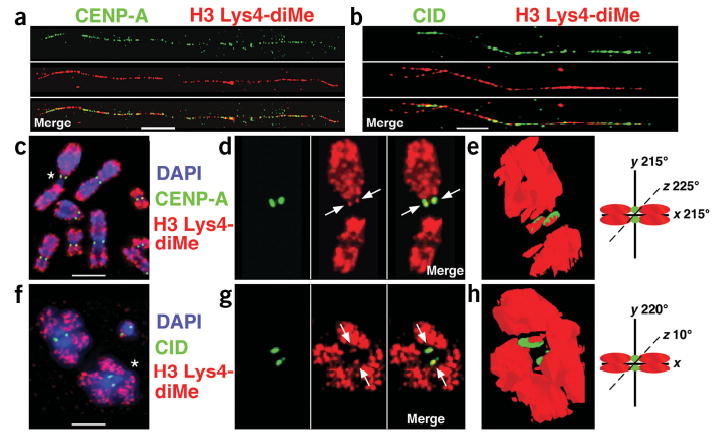
H3 is dimethylated at Lys4 in CEN chromatin. (a,b) Extended chromatin fibers from human cells (a) and D. melanogaster S2 cells (b) were stained with antibodies to CENP-A/CID and H3 Lys4-diMe, detected with secondary antibodies conjugated with FITC (green) and Cy3 (red), respectively. Single-color images are shown separated from the merged image. H3 nucleosomes interspersed with CENP-A/CID nucleosomes are dimethylated at Lys4 in both humans and flies. (c–h) Immunofluorescence patterns on human (c) and D. melanogaster (f) metaphases stained with H3 Lys4-diMe antibodies (red). DNA was stained with DAPI (blue). Antibody localization on metaphase chromosomes shows that H3 Lys4-diMe is located to close to, and partially overlapping with, CENP-A/CID (green). (d,g) Enlarged view of chromosomes marked with asterisks in c and f, with single-color images to the right of the merged images. (e,h) Three-dimensional modeling of antibody staining on individual metaphase chromosomes from d and g shows that H3 Lys4-diMe is located in the region between sister kinetochores, partially overlapping with CENP-A/CID staining (arrows), consistent with the coiling or looping model proposed for CEN chromatin higher ordering packaging at metaphase15. The asterisk in d denotes human chromosome 1, which is particularly under-represented for H3 Lys4-diMe in the 1q region. Chromosome cartoons depict the degree to which each three-dimensional model was rotated around the x-, y- and/or z-axes in e and h. Fiber and metaphase results are quantified in Figures 2 and 4. Bars, 15 μm in a,c; 5 μm in b,f.
The amount of H3 Lys4-diMe at the centromere seemed to be less than the staining on euchromatic chromosome arms. This is probably due to the fact that regions of H3 Lys4-diMe in euchromatin are continuous, whereas in CEN chromatin, H3 blocks are less concentrated owing to interspersion with CENP-A. Clearly, there are resolution limitations to the three-dimensional immunofluorescence analysis; nevertheless, our results suggest that CENP-A and H3 Lys4-diMe are closely associated, with some spatial distinction. In addition, this finding is consistent with our previous results showing that H3 is located internally to the CENP-A/CID cylinder at mitosis, with CENP-A/CID oriented toward the outside of the chromosome15.
Another ‘euchromatic’ modification, H3 Lys4 trimethylation (H3 Lys4-triMe), has been associated with active gene expression4, and was not observed within the CENP-A/CID regions on human and D. melanogaster chromatin fibers (Figs. 2 and 7a,b). In human mitotic chromosomes, H3 Lys4-triMe was present in most flanking heterochromatin regions (Fig. 7c–e), but was under-represented in the large satellite regions of chromosomes 1, 9, 16, and Y (Fig. 7c-e; data not shown). Likewise, H3 Lys4-triMe was also excluded from the CID staining region in D. melanogaster mitotic chromosomes (Figs. 4 and 7f–h). In pericentromeric regions that did not contain substantial stretches of satellite DNA, such as human chromosome 1p, and chromosome 2 of D. melanogaster, H3 Lys4-triMe staining was present within 1–10 μm of CENP-A/CID. In fact, the under-representation of H3 Lys4-triMe on mitotic chromosomes often occurred on one side of the centromeric region (Fig. 7c,f), particularly on human chromosome 1q and chromosome 3 of D. melanogaster (Fig. 7c,f). Overall, our studies of H3 Lys4 methylation indicate that H3 within CEN chromatin is dimethylated at Lys4, a modification previously associated with euchromatin, but is not trimethylated at this residue.
Figure 7.
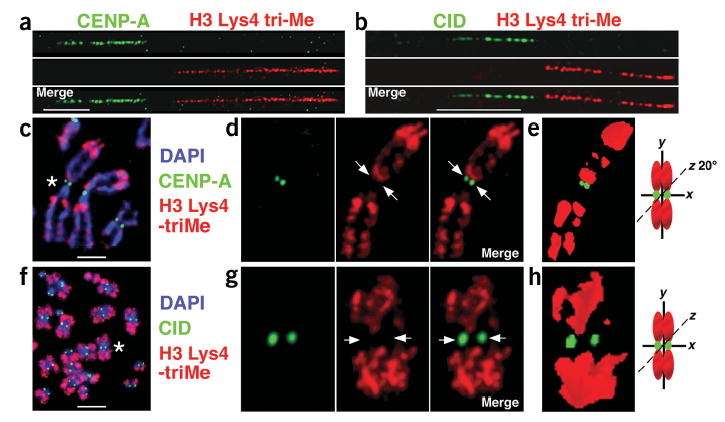
H3 is not trimethylated at Lys4 in CEN chromatin. (a–f) Metaphases and extended chromatin fibers were stained with antibodies to CENP-A or CID (green) and H3 Lys4-triMe (red), detected with FITC and Cy3-conjugated antibodies, respectively. Single-color images are shown separated from the merged image. On chromatin fibers from human and D. melanogaster interphase cells (a,b), H3 Lys4-triMe did not overlap with CENP-A/CID. A gap of 1–20 μM between CENP-A and H3 Lys4-triMe staining was observed on most human chromatin fibers (80%, n = 18). In addition, H3 Lys4-triMe antibody staining was present on only one side of CENP-A antibody staining in 70% of fibers (n = 16), and was observed on both sides in 30% of fibers (n = 7). On D. melanogaster fibers, H3 Lys4-triMe antibody staining was observed on both sides of the CID antibody staining, but the gap between H3 Lys4-triMe and CID on every fiber was at least 5 μm (average = 15 μm; n = 15). The pericentromeric regions that were negative for H3 Lys4-triMe staining may correspond to H3 Lys9-diMe modified nucleosomes (Fig. 1). (c,f) Immunofluorescence patterns on human (c) and D. melanogaster (f) metaphases stained with H3 Lys4-diMe antibodies (red). DNA was stained with DAPI (blue). Antibody localization on metaphase chromosomes shows that H3 Lys4-triMe did not coincide with CENP-A/CID (green). (d,g) Enlarged view of chromosomes marked with asterisks in c (human chromosome 1) and f (D. melanogaster chromosome 2), with single-color images to the right of the merged images. Arrows point to the areas on individual chromosomes where CENP-A/CID, but not H3 Lys4-triMe, were located. (e,h) Three-dimensional modeling of individual metaphase chromosomes from d and g shows the separation of CENP-A/CID and H3 Lys4-triMe into distinct domains. Chromosome cartoons depict the degree to which each three-dimensional model was rotated around the x-, y- and/or z-axes in e and h. Fiber and metaphase results are quantified in Figures 2 and 4. Bars, 15 μm in a,c; 5 μm in b,f.
DISCUSSION
CEN chromatin contains a distinct combination of modified histones
Post-translational modifications of histones are known to be biologically important in defining chromatin states, such as silent or active gene expression. Centromeric chromatin in flies and humans is defined as the full extent of staining for the centromere-specific histones CENP-A and CID, which contain interspersed subdomains of the CENP-A/CID and H3 nucleosomes. Immunofluorescence analysis of two-dimensional extended chromatin fibers and three-dimensional mitotic chromosomes demonstrated that H3 subdomains present within CEN chromatin were enriched for H3 Lys4-diMe, a modification associated with open but not active euchromatin. H3 subdomains within CEN chromatin did not contain the H3 Lys9 di- or trimethylation associated with heterochromatin, and lacked acetylations at H3 Lys9 and H4 Lys5, Lys8, Lys12 and Lys16 that are generally found in euchromatin. Finally, the H3 Lys4-trimethylation associated with actively transcribed regions was also not present in CEN chromatin. We conclude that the interspersed H3 present in fly and human CEN chromatin contains individual H3 and H4 modifications previously associated with both euchromatin and heterochromatin, but in a combined pattern that is distinct from each chromatin state individually. These results are unexpected; the fact that eukaryotic centromeres are embedded in heterochromatin has suggested that CEN chromatin should contain heterochromatic epigenetic imprints. This distinct pattern of histone modifications, which we term ‘centrochromatin,’ may contribute to the unique structure and function of the centromere, in combination with the presence of CENP-A/CID (see below).
The link between CEN and flanking chromatin
The regions that flank CEN chromatin in fly and human samples contained H3 Lys9-diMe and triMe, and hypoacetylation of H3 and H4, consistent with previous studies of pericentric heterochromatin6,7,21,22. In fission yeast, tRNA genes seem to be associated with boundaries between CEN and flanking chromatin18. It is unclear at this time whether the separation of CENP-A/CID and flanking heterochromatin domains in humans and D. melanogaster reflects the presence of a sequence-specific boundary, or a sequence-independent balance between the two epigenetic states12,23.
The pericentromeric regions of human metaphase chromosomes contained H3 Lys9-triMe, although this modification was under-represented, but was not completely deficient, in D. melanogaster pericentromeric heterochromatin. Fly pericentromeric regions showed a much more substantial enrichment for H3 Lys9-diMe in metaphase chromosome and chromatin fiber analysis. These results are consistent with previous reports showing that H3 Lys9-diMe is concentrated at heterochromatic chromocenters in D. melanogaster salivary glands9. Pericentric regions in flies contain essential genes24, whereas few genes have been reported in the pericentric regions of human chromosomes. Perhaps higher-order heterochromatin is regulated differently between humans and flies by other histone-modifying enzymes and heterochromatin proteins, to allow the expression of heterochromatic genes. Further investigations of the distributions of different histone modifications in pericentric heterochromatin are necessary to validate these observations, and to determine whether they have functional consequences.
Two recent studies have discovered correlations between distinct heterochromatic domains and different degrees of H3 Lys9 methylation in mouse embryonic stem cells and embryonic fibroblasts6,7. It was demonstrated by indirect immunofluorescence that H3 Lys9-triMe is enriched in pericentric regions of mouse chromosomes and at DAPI-bright regions in interphase nuclei. These results agree with our findings that H3 Lys9 methylation was present in pericentric regions of human and fly chromosomes. These authors also used chromatin immunoprecipitation (ChIP) analysis to identify patterns of H3 methylation within mouse chromosomes. In mice, the centromere and pericentromeric regions contain distinct, expansive arrays of satellite DNA. Major satellite comprises the largest region and is immediately adjacent to the functional kinetochore, and minor satellite is the region where mouse kinetochore proteins are located25,26. It was recently reported that major and minor satellite DNAs were enriched primarily for H3 Lys9-triMe, and that both satellites were associated with H3 Lys9-diMe to a lesser extent6.
The presence of H3 Lys9 methylation in mouse minor satellite suggests that CEN chromatin may be modified differently in mice, in comparison with the results reported here for humans and D. melanogaster. However, human centromeres contain expansive, megabase-sized arrays of α-satellite DNA, and CENP-A localizes to only a portion of these arrays27 (data not shown). Our demonstration that CEN chromatin in humans and flies lacks H3 Lys9 methylation is consistent with a similar model for centromere organization in mice, in which minor satellite DNA contributes partly to CEN chromatin and partly to heterochromatin formation. Additional studies on mouse centromeres are necessary to specifically map H3 Lys9 methylation with respect to CENP-A and CEN chromatin.
CEN chromatin in other eukaryotes
Our studies have focused on centromeric chromatin structure in human cells and D. melanogaster cultured cells and larval brains. Is CEN chromatin in other organisms marked by the same histone modifications? Centromeres in S. pombe consist of a basic unit of central core chromatin that contains CENP-A (Cnp1), flanked on both sides by heterochromatin that is marked by H3 Lys9 methylation18. Our initial finding that subdomains of CENP-A/CID and H3 are interspersed in fly and human centromeres produced the hypothesis that centromeres in larger eukaryotes might represent amplification of the basic CEN domain unit (heterochromatin–CENP-A–heterochromatin) found in S. pombe. However, in the present study, we did not observe H3 Lys9 methylation in the regions between CENP-A/CID subdomains with the CEN regions. Thus, our results argue that fly and human centromeres are not composed of multimers of units equivalent to S. pombe centromeres. However, the overall organization of the centromere region is conserved, such that the entire CENP-A/CID chromatin domain is flanked by heterochromatin that contains H3 Lys9 methylation.
Alternatively, it is possible that CEN chromatin does differ among organisms. Recently, ChIP analysis of rice centromeric regions suggested that H3 Lys9 di-Me is present within the CEN chromatin (defined by the presence of the CENP-A homolog CenH3)17. This result may reflect differences in CEN chromatin composition and organization between plants and flies, humans and S. pombe. However, a more extensive analysis of the spectrum of modifications, including cytological studies of the distributions of modifications in extended fibers and mitotic chromosomes, needs to be carried out in different plant species to test this hypothesis.
Specification of CEN chromatin by distinct modifications
What are the functional roles of histone modifications in CEN and flanking chromatin? First, distinct chromatin states in the CEN region may contribute to the diverse properties of centromeric domains, such as differential replication timing of the CEN and flanking heterochromatin15,28–30. Heterochromatic modifications may also maintain centromere size by creating a barrier against expansion of CEN chromatin. In D. melanogaster, CEN chromatin readily spreads into neighboring sequences when flanking heterochromatin is removed, allowing neocentromere activation23.
Second, the stacking and self-association of CENP-A nucleosomes, distinctly modified interspersed H3 nucleosomes and flanking heterochromatin may be responsible for the three-dimensional structure of CEN chromatin in mitosis15. This organization could facilitate kinetochore assembly by orienting CENP-A/CID chromatin toward the outside of the chromosome, where it can interact with kinetochore proteins (Fig. 8). CEN-specific combinations of histone modifications and the three-dimensional organization could also be important for recruitment of cohesion complexes to heterochromatin near sister kinetochores, while ensuring spatial separation of cohesion and kinetochore domains31,32.
Figure 8.
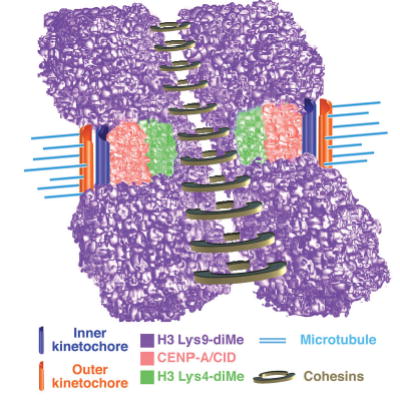
Model for three-dimensional organization of centromeric (CEN) chromatin in D. melanogaster and humans. Incorporation of the two-dimensional and three-dimensional histone modification patterns extends our understanding of the chromatin composition and organization of the CEN region, and suggests that interspersed CENP-A/CID and H3 Lys4-dimethyl nucleosome blocks comprise a unique chromatin state that is distinct from the flanking heterochromatin. Associations between similarly modified nucleosome blocks are proposed to contribute to the formation of distinct three-dimensional structures in CEN and flanking chromatin. Interspersed CENP-A/CID and distinctly modified H3 and H4 may mediate formation of the ‘cylindrical’ three-dimensional structures observed in metaphase chromosomes15. H3 Lys9-diMe chromatin, which recruits heterochromatin proteins such as HP1 and cohesion proteins such as RAD21/SCC1, is present in the inner kinetochore space between mitotic sister chromatids and in regions that flank CEN chromatin. This arrangement may position CENP-A toward the poleward face of the mitotic chromosome and facilitate recruitment of outer kinetochore proteins, and promote HP1 self-interaction and proper chromosome condensation and cohesion. Cohesins are presented as ringed structures, in accord with recent models36.
Finally, distinctly modified, interspersed H3 nucleosomes could participate in epigenetic propagation of centromere identity. As observed for other histone ‘variants’33, CENP-A/CID assembly can be replication-independent, unlike that of canonical H3 nucleosomes30,34. Specifically modified interspersed H3 subdomains could create a ‘permissive’ chromatin structure necessary for assembly of new CENP-A16.
A new model for deposition of CENP-A specifically in centromeric chromatin is suggested by these observations. Perhaps the modification pattern of interspersed H3 nucleosomes and histone modification proteins (such as acetyltransferases, methyltransferases and kinases) helps propagate centromere identity, in lieu of (or in addition to) CENP-A-associated proteins. Future studies are necessary to address mechanisms responsible for formation, maintenance and separation of these distinct chromatin states, and well as their roles in centromere structure and function. It is also important to determine whether other functional domains embedded within heterochromatin, such as the nucleolus organizer–ribosomal DNA, show distinct patterns of histone modifications.
METHODS
Cell culture and antibodies
S2 cells were maintained as described19. HT1080 cells containing FLAG-tagged CENP-A15 and primary human cell lines were used for these studies. Chicken antibodies against CID have been described15,19. CENP-A was detected in HT1080 lines using antibodies against FLAG M2 (Sigma; 1:200 for metaphases; 1:50 for fibers) or mouse monoclonal antibodies against CENP-A (1:1,000 for metaphases; 1:200 for fibers), which were provided by K. Yoda (Nagoya, Japan)35. Additional antibodies were: anti-dimethyl H3 Lys9 (which does not cross-react with other H3 modifications, (Upstate), anti-trimethyl H3 Lys9 (Abcam), anti-dimethyl H3 Lys4 (Upstate), anti-trimethyl H3 Lys4 (GeneTex), anti-acetyl H3 Lys9 and anti-acetyl H4 Lys5, Lys8, Lys12 and Lys16-specific antibodies (all from Upstate). A second set of anti-trimethyl H3 Lys9 antibodies (anti-2X-trimethyl H3 Lys9 (no. 4861)) were provided by T. Jenuwein and A. Peters6.
Cytological preparations
Unfixed S2 and human metaphase chromosomes and chromatin fibers used for immunofluorescence were prepared as described15. Metaphase chromosomes from D. melanogaster third-instar larval neuroblasts were used for immunofluorescence as described and to confirm antibody staining patterns observed in S2 cells29. Chromatin fibers were prepared from neuroblasts as follows: six to eight individual brains from third-instar wandering larvae were dissociated in PBS or 0.5% (w/v) sodium citrate containing collagenase (Invitrogen) and dispase (Sigma), then passed five times through a 27-gauge needle. This mixture (500 μl) was loaded into a single chamber cytofunnel (Shandon), and centrifuged in a Cytospin 3 (Shandon) at 800 r.p.m. (g value is: 90g) for 4 min onto clean glass microscope slides. Immediately after centrifugation, slides were placed in lysis buffer15 for 12–15 min. Slides were then processed for immunofluorescence as described15. After immunofluorescence, cells were counterstained with SloFade Light containing DAPI (Molecular Probes).
Microscopy and image analysis
All images were captured using the Deltavision Spectris Restoration Microscope System (Applied Precision) using an Olympus IX70 inverted microscope and a SONY ICX285 ER Progressive Scan CCD. Images were acquired using Deltavision SoftWoRx Resolve 3D capture program and collected as a stack of 0.1–0.2 μm increments in the z-axis. Each metaphase image consisted of 1.5–3.6 μm (15–36 sections). Images were deconvolved using the conservative algorithm with 10 iterations, and stacked images were viewed using quick projection or volume view options. Chromatin fibers often extended through multiple fields of view and were captured as a series of time points and z sections using the ‘collect panels’ option in Experiment Designer. Stacked, deconvolved images were superimposed using the ‘stitch image’ option and viewed as quick projections. Volume renderings were verified using 1- and 4-μm fluorescent beads (Molecular Probes) as described15. Volume models were made using the 2D Polygon and 3D Model option of softWoRx (Applied Precision), and all models were constructed directly from the data presented. Images were imported into Adobe Photoshop for presentation.
Fluorescence quantification of CENP-A/CID and histones on metaphases from single plane-of-focus images was done as described15. For quantification of overlap between CENP-A/CID and modified histones on chromatin fibers, the amount of histone antibody staining (in micrometers) that overlapped with CENP-A/CID was measured against the entire CENP-A/CID staining region (in micrometers) using the ‘measure distances’ tool in softWoRx or using a custom histogram-line plot macro generated using IP Lab (Scanalytics). Results were plotted as bar graphs using Kaleidograph (Synergy Software).
Acknowledgments
We thank A. Skora for technical assistance, and K. Scott, C. Vaziri, A. Dernburg, D. Allis, S. Erhardt and C. Yan for discussions and advice. We acknowledge K. Maggert for originally coining the term “centro-chromatin.” We are indebted to T. Jenuwein and A. Peters for use of their unique H3 Lys9-triMe antibodies. This work was supported by the following grants: US Department of Energy LDRD 366987 and US National Institutes of Health (NIH) R01 GM66272 (G.H.K.), and American Cancer Society IRG-72-001-29 and NIH R01 GM069514 (B.A.S.).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests
References
- 1.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein BE, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos-Rosa H, et al. Active genes are trimethylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 5.Schneider R, et al. Histone H3 Lys 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 6.Peters AHFM, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 7.Rice JC, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1599. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 8.Nakayama JI, Rice JC, Strahl BD, Allis CD, Grewal SIS. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 9.Schotta G, et al. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller HJ. Types of visible variations induced by X-rays in Drosophila. J Genet. 1930;22:299–334. [Google Scholar]
- 11.Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 13.Cleveland DW, Mao Y, Sullivan KF. Centromere and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 14.Yoda K, et al. Human centromere protein A (CENP-A) can replace histone H3 in nucleosomes reconstitution in vitro. Proc Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellone BG, Allshire RC. Stretching it: putting the CEN(P-A) in centromere. Curr Opin Genet Dev. 2003;13:191–198. doi: 10.1016/s0959-437x(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 17.Nagaki K, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 18.Partridge JF, Borgstrøm B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 19.Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 21.O’Neill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boggs BA, et al. Differentially methylated forms of histone H3 show unique associations patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 23.Maggert KA, Karpen GH. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 2001;158:1615–1628. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoskins RA, et al. Heterochromatin sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002;3:0085.1–0085.16. doi: 10.1186/gb-2002-3-12-research0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell AR, Jeppesen P, Nicol L, Morrision H, Kipling D. Epigenetic control of mammalian centromere proteins binding: does DNA methylation have a role? J Cell Sci. 1996;109:2199–2206. doi: 10.1242/jcs.109.9.2199. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell AR, Nicol L, Malloy P, Kipling D. Novel structural organization of a Mus musculus DBA/2 chromosome shows a fixed position for the centromere. J Cell Sci. 1993;106:79–85. doi: 10.1242/jcs.106.1.79. [DOI] [PubMed] [Google Scholar]
- 27.Warburton PE, et al. Immunolocalization of CENP-A suggests a distinct nucleosomes structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 28.Ahmad K, Henikoff S. Centromeres are specialized replication domains in heterochromatin. J Cell Biol. 2001;153:101–110. doi: 10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan B, Karpen G. Centromere identity in Drosophila is not determined in vivo by replication timing. J Cell Biol. 2001;154:683–690. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SM, Dubey DD, Huberman JA. Early-replicating heterochromatin. Genes Dev. 2003;17:330–335. doi: 10.1101/gad.1046203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 32.Bernard P, et al. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 33.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 34.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhlmann F. The mechanism of sister chromatid cohesion. Exp Cell Res. 2004;296:80–85. doi: 10.1016/j.yexcr.2004.03.005. [DOI] [PubMed] [Google Scholar]


