Abstract
Lysyl oxidase (LO) catalyzes crosslinking of collagen and elastin essential for maintaining the structural integrity of the lung extracellular matrix (ECM). To understand mechanisms of cigarette smoke (CS)-induced emphysema, we investigated effects of cigarette smoke condensate (CSC), the particulate matter of CS, on LO mRNA expression in cultured rat fetal lung fibroblasts (RFL6). Exposure of RFL6 cells to 0–120 μg CSC/ml for 24 h induced a dose-dependent inhibition of LO steady-state mRNAs, for example, reducing transcript levels to below 10% of the control in cells incubated with 80–120 μg CSC/ml. Nuclear run-on assays indicated a marked reduction in LO relative transcriptional rates amounting to 27.7% of the control in cells treated with 120 μg CSC/ml. The actinomycin D-chase assay showed that CSC enhanced the instability of LO transcripts. The t1/2 for LO mRNA decay was decreased from 24 h in the control to 4.5 h in cells treated with 120 μg CSC/ml. Moreover, 80–120 μg CSC/ml also inhibited LO promoter activity as revealed by suppression of reporter gene expression in cells transfected with LO promoter-luciferase vectors. Thus, inhibition of LO transcription initiation and enhancement of LO mRNA instability both contributed to downregulation of LO steady-state mRNA in CSC-treated cells. Note that inhibition of LO mRNA expression by CSC was closely accompanied by markedly decreased levels of transcripts of collagen type I and tropoelastin, two substrates of LO. Thus, transcriptional perturbation of LO and its substrates may be a critical mechanism for ECM damage in CS-induced emphysema.
Keywords: cigarette smoke condensate, lysyl oxidase relative transcriptional rate, lysyl oxidase mRNA stability, lysyl oxidase promoter activity, collagen type I, elastin
INTRODUCTION
The structural and functional integrity of the lung extracellular matrix (ECM) is largely dependent on the conversion of soluble collagen and elastin to insoluble, fibrous aggregates catalyzed by lysyl oxidase (LO), a copper dependent enzyme. This catalyst oxidizes specific lysine residues within these proteins to generate covalent cross-linkages stabilizing the ECM. Thus, LO is a key extracellular enzyme functioning in lung morphogenesis and tissue repair (Kagan and Li, 2003).
Cigarette smoke (CS) contains more than 4,000 different constituents which could act individually or collectively as pathogenic agents for pulmonary diseases (Hoffmann and Wynder, 1999). The particulate phase or cigarette smoke condensate (CSC) is composed of major toxicants such as nicotine, phenol, anthracyclic hydrocarbons, nitrosamines, heavy metals and chemical carcinogens such as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). CS accounted for 81.8% of deaths resulting from chronic obstructive pulmonary disease, mainly manifest as emphysema (Hoffmann et al., 2001; American Lung Association of Washington, 2004). Although clinical correlation studies have established a causal relationship between smoking and emphysema (Kimbel, 1985), the precise mechanisms and cellular events leading to the abnormal enlargement of the airspace in the lung by CS remain unclear (Wright and Churg, 1995; D’Armiento et al., 1990).
Our studies have shown that exposure of rat fetal lung fibroblasts (RFL6) to CSC induced down-regulation of LO at protein and catalytic levels (Chen et al., 2005). LO deficiency is intrinsically relevant to the pathogenesis of emphysema as evidenced by reduced levels of LO activities in rats, hamsters or chicks fed with food deficient in copper, a co-factor of LO, associated with the reduction of elastin deposition and evolution of emphysematous lesions in the lung (Harris, 1986; O’Dell et al., 1978; Soskel et al., 1984).
To further define cellular and molecular events in downregulation of LO by CSC in an effort to understand mechanisms for CS-induced emphysema, we have examined modulation of the mRNA expression of LO as well as its substrates, i.e., collagen and elastin by CSC in cultured rat fetal lung fibroblasts. In this paper, we report that CSC strongly inhibited mRNA expression of LO and its substrates in exposed cells and that CSC-induced reduction of LO mRNA stability combined with suppression of LO transcriptional initiation collectively leading to downregulation of LO at the mRNA level.
MATERIALS AND METHODS
Materials
CSC was purchased from Murty Pharmaceuticals (Lexington, KY, USA). It was prepared using a Phipps-Bird 20-channel smoking machine. The particulate matter from Kentucky standard cigarette 1R3F (University of Kentucky, KY, USA) was collected on Cambridge glass fiber filters and the amount obtained determined by the weight increase of the filter. CSC so prepared was dissolved in dimethyl sulfoxide (DMSO) to give a final concentration of 4% (w/v). The average yield of CSC was 26.1 mg/cigarette. Aliquots of CSC were kept at −80° C before experimental use. [32P]dCTP and [32P]UTP were from NEN (Boston, MA). All tissue culture products were from GIBCO (Grand Island, NY).
Cell culture
Rat fetal lung fibroblasts (RFL6) obtained from ATCC were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) as previously described (Chen et al., 2005). Stock cultures were derived from the freezing cell line and passaged every 4 days. A total of 6 passages were used for experiments. To obtain quiescent cultures, cells were plated at 3–4 × 103 cells/cm2 in 10% FBS/DMEM, incubated for 24 h and then growth-arrested by incubation of cells in DMEM containing 0.3% FBS for 3 days (Li, et al., 1995). Cells were then changed to fresh 0.3% FBS/DMEM before experimental use.
The cell viability assay
Growth-arrested cells were exposed to CSC for 24 h at indicated doses. Control cells were treated only with vehicle at a final concentration of 0.003 (v/v) and the same control was included in all assays in this study. Cell viability was determined by the trypan blue exclusion assay. Briefly, control and treated cells were detached from dishes by trypsinization and resuspended in PBS at the concentration of 106 cells/ml. Aliquots of cell suspension mixed with the same volume of 0.4% trypan blue/PBS (Sigma, St. Louis, MO) were transferred into the counting chamber of the hemocytometer. The number of membrane-injured cells stained by trypan blue and the total number of cells were counted under the microscope as described (Elia et al., 1993).
Northern blot analysis
Total RNA was isolated from control and CSC-treated cells by TRIzol reagent (Invitrogen, Carlsbad, CA). The steady-state concentrations of mRNA for LO, collagen type I, tropoelastin, β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were determined by Northern blot, using the corresponding 32P-labeled cDNA probe as described (Li et al., 1995). Briefly, equal amounts of RNA (15 μg), quantified by spectrophotometry at 260 nm, were size-fractionated by electrophoresis on a 1% (wt/vol) agarose gel. RNA was transferred to a GeneScreen nylon membrane (NEN, Boston, MA) and immobilized by ultraviolet crosslinking. Approximately 25 ng of the cDNA probe for LO, collagen type I, tropoelastin, β-actin or GAPDH were labeled with 32P using Ready-to-Go DNA labeling beads (dCTP) from Amersham Biosciences (Piscataway, NJ). The RNA blots on the membrane were hybridized with 32P-labeled cDNA at 68° C in an ExpressHyb hybridization buffer (Clontech, Palo Alto, CA). The membrane was washed with 2 × SSC at room temperature, then with 0.2 × SSC at 50° C and exposed to Kodak XAR-5 film at −80° C with an intensifying screen. Autoradiograms were analyzed with the 1D Scan EX software for densities of mRNA bands (Scananalytics, Fairfax, VA) (Chen et al., 2005).
The nuclear run-on assay
The nuclear run-on assay was employed to determine relative rates of LO transcription in control and CSC-treated cells as described (Schubeler and Bode, 1997). For isolation of nuclei, cell pellets were gently resuspended in a lysis buffer [0.32 M sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 0.1 mM EDTA, 10 mM Tris-HCl, pH 8.0, 1 mM DTT, 0.5 mM PMSF, 0.5% (v/v) NP-40] and incubated on ice with intermittent microscopic examination for nuclear integrity. The nuclei were centrifuged at 500 × g and resuspended in a nuclear freezing buffer [50 mM Tris-HCl, pH 8.0, 40% (v/v) glycerol, 5 mM MgCl2, 0.1mM EDTA, 1 mM DTT, 0.5 mM PMSF] either for direct use or for storage in liquid nitrogen (Dyer and Herzog, 1995). For the nuclear run-on reaction, 100 μl of thawed nuclei were mixed with 30 μl of a 5 × run-on buffer (25 mM Tris-HCl, pH 8.0, 12.5 mM MgCl2, 0.75 M KCl, 1.25 mM each of ATP, GTP, and CTP) containing 100 μCi [α-32P]UTP and 5 μl of the Sarkosyl stock to give a final concentration of 0.06%. The mixture was incubated for 30 min at 30°C, then 15 μl of DNase I (1U/μl) were added and the incubation continued for another 15 min. RNA was isolated by a single step TRIzol extraction and the incorporation of 32P determined by γ–counting. Plasmids containing LO cDNA, GAPDH cDNA or histone 3B cDNA, or without insert, were slot-blotted onto the nitrocellulose membrane using a BioRad BioDot SF apparatus. The blots were prehybridized in 1% SDS/10% dextran sulfate, 1.4 M NaCl and 325 μg/ml each of herring sperm DNA and yeast tRNA for 2 h at 60°C followed by treatment with RNasin plus DTT. Radiolabeled RNAs were hybridized onto filters for 2 days. The filters were then washed, dried and autoradiographed on preflashed film.
The assay for LO mRNA stability
The half-life of LO mRNAs was assessed by an inhibitor-chase assay (Dani et al., 1984). Cells were incubated with 5 μg/ml of actinomycin D (Sigma, Saint Louis, MO) in the absence or presence of CSC at indicated doses. Cultures were harvested at various times during the actinomycin D-chase. Total RNA was isolated and analyzed by Northern blots using the 32P-labeled rat LO cDNA probe. After hybridization and autoradiography, band densities were measured by analysis with the 1D Scan EX software. For time course studies, using mRNA levels at time zero of actinomycin D treatment as 100%, the t1/2 values for mRNA decay were determined from semilog plots of integrated densities vs time. For dose-response assays, using mRNA levels after a 24 h actinomycin D-chase in cells incubated in the absence of CSC as a control, mRNA levels in cells treated with various doses of CSC were measured and expressed as % of the control.
LO promoter-luciferase construct
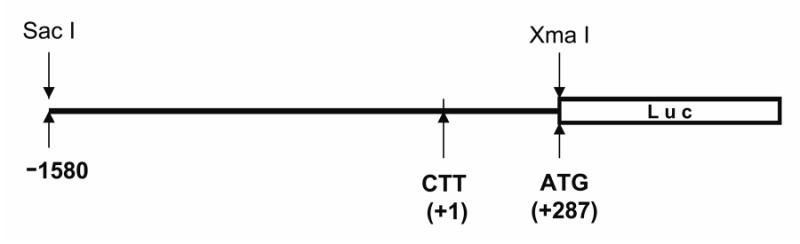
To make LO promoter-reporter gene construct, using the rat genomic DNA extracted from RFL6 cells as a template, PCR amplifications of the rat LO promoter were carried out with following primers (5′-GACTGAGCT CGAATGCACTAGGAAAGTCTGGAGGA-3′) and (5′-GTCACCCGGGATGCTCCCGGCTCGTCCCTTCT-3′) in the presence of high fidelity Taq DNA polymerase (Invitrogen, Carlsbad, CA), yielding an approximately 1.9 kb fragment of the LO promoter. The LO-specific sequences for the primer design as described here are based on the rattus norvegicus chromosome 18 WGS supercontig NW_047513 (Rat Genome Sequencing Consortium, 2003). The amplified LO promoter fragment was restricted with Sac I and Xma I and ligated into similarly restricted plasmid pGL3-basic (Promaga, Madison, WI) upstream of the luciferase gene. The predicted initiation site (CTT) for transcription was designated as +1 whereas the start codon (ATG) for translation was located at +287. This fragment of the LO promoter covers 1,579 bp before the predicted initiation site of transcription (Figure 4A).
Figure 4A. A schematic linear map of the cloned LO promoter.
Cell transfection and assays for reporter gene products
RFL6 cells were plated at 5 × 105 cells per 60 mm dish containing 5 ml of 10% FBS/DMEM. After 24 h incubation, cells were co-transfected with LO promoter-luciferase constructs as well as pSV-β-galactosidase (β-gal) expression vectors (0.5 μg, Promega), the latter used as an internal control to monitor the transfection efficiency, by using lipofectamine reagent (Invitrogen, Carlsbad, CA) as described (Ausubel et al., 1995) . Note that cells co-transfected with pGL3-basic vectors containing the luciferase gene without the LO promoter and β-gal expression vectors were always included in any experiments to evaluate the background. Following 6 h posttransfection incubation, cells were incubated in 10% FBS/DMEM for an additional 18 h-period, washed, and then exposed to CSC for 24 h at indicated doses (0, 20 40, 80 and 120 μg/ml). Luciferase and β-gal activities in cell lysates extracted from control and CSC-treated cells were measured by luminometry and spectrophotometry, respectively, as recommended by supplier (Promega). LO promoter-luciferase activities in transfected cells were corrected by subtracting the background value, and normalized to transfection efficiency as revealed by the β-gal assay. Results are expressed as cpm/OD β-gal.
Statistical analysis
In cell viability and LO promoter activity assays, data are presented as mean ± standard deviation (SD) of three separate experiments in which each control or variable was assessed in triplicate dishes. A one-way analysis of variance, followed by Dunnett test was performed. Differences were considered significant at p < 0.05.
RESULTS
Cell viabilities in control and CSC-treated cells
Prior to assessing perturbation of LO transcription by CSC, cell viabilities in control and treated cultures were examined to evaluate the cytotoxicity of CSC. Sub-confluent growth arrested RFL6 cells were incubated for 24 h in the absence or the presence of CSC at indicated doses. Cell viabilities were determined by the trypan blue exclusion assay. After exposure to CSC at doses of 0, 20, 40, 80, and 120 μg/ml, 95.6 ± 6.2, 94.0 ± 8.0, 93.5 ± 12.3, 90.5 ± 9.2 and 85.0 ± 14.2% (mean ± SD), respectively, of cells excluded trypan blue, indicating that CSC at indicated doses did not significantly affect cell viabilities. Thus, this dose range of CSC was used to determine its effects on the expression of LO and its substrates at transcriptional levels in the following studies.
Steady-state mRNA levels in RFL6 cells treated with CSC
To assess LO transcriptional expression in RFL6 cells exposed to CSC, steady-state mRNA levels of LO were determined by Northern blot. As shown (Figure 1), consistent with prior observations, two LO transcripts were seen in the blot at approximately 5.8 and 4.5 kb, presumably due to alternative polyadenylation of the transcript (Li et al., 1995). Treatment of cells with CSC induced a dose-dependent inhibition of LO steady-state mRNAs. As revealed by density analysis, after 24 h incubation with CSC at 20, 40, 80 and 120 μg/ml, LO transcript levels normalized to internal GAPDH controls were reduced to 65.3, 37.4, 3.6 and 0.8% of the control, respectively, with IC50 = 30 μg/ml of CSC. Thus, CSC specifically inhibited LO expression at the transcriptional level.
Figure 1. Northern blot analysis for steady-state LO mRNA levels in control and CSC-treated cells.
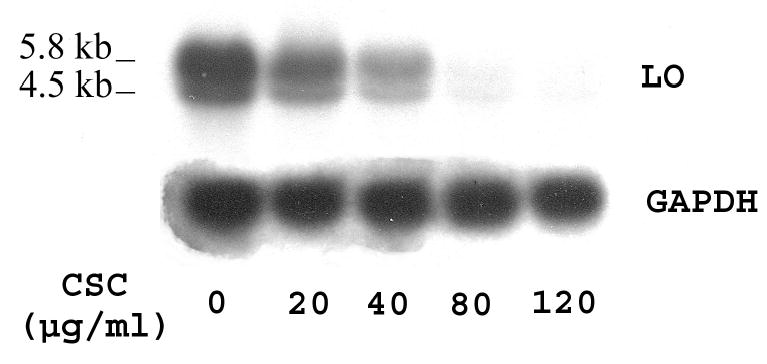
Total RNA (15 μg) was derived from growth-arrested cells treated with or without CSC for 24 h at indicated doses. Transcripts for LO and GAPDH, an internal control, migrated and labeled as shown. Densities of LO bands were normalized to internal controls following analysis by the 1D Scan EX software. Experiments shown here and below were repeated at least three times with reproducible results, and a representative one is presented, unless otherwise indicated.
Relative transcription rates of LO in RFL6 cells treated with CSC
The steady-state concentrations of mRNA determined by Northern blot actually reflect a composite of both the transcriptional rate and the mRNA stability (Schubeler and Bode, 1997). To assess mechanisms of CSC downregulation of LO expression at the mRNA level, we carried out a nuclear run-on assay to evaluate changes in the LO transcription initiation. As shown (Figure 2), following density normalization to the internal control of GAPDH, we found that levels of transcripts hybridized to the LO cDNA were markedly diminished in nuclei of CSC treated cells amounting to 90.5, 79.7, 39.8 and 27.7% of the control, respectively, in cells treated with CSC at 20, 40, 80 and 120 μg/ml (IC50 = 65 μg/ml of CSC). Therefore, reduction of transcriptional rates by CSC may be a critical mechanism for downregulation of the steady-state LO mRNA expression in treated cells.
Figure 2. Nuclear run-on assay for relative transcription rates of LO in control and CSC-treated cells.
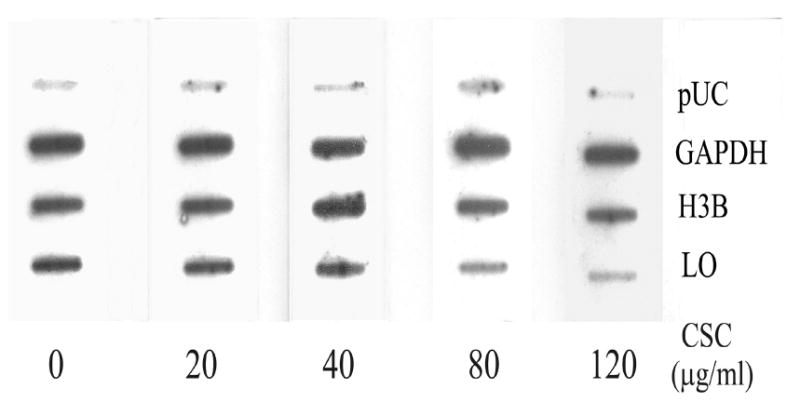
Nuclei were freshly isolated from control and CSC-treated cells under the same conditions as described in Figure 1. Nascent transcripts were labeled with 32P-UTP and hybridized to a previously prepared filter containing cDNAs for LO, histone 3B (H3B) and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) and pUC plasmids without insert (pUC).
Stabilities of LO mRNAs in RFL6 cells treated with CSC
The half-life of LO mRNAs was determined by an inhibitor-chase assay as described (Dani et al., 1984). Total RNA was extracted from RFL6 cells incubated in the absence or presence of 120 μg/ml of CSC before and at various times after treatment with actinomycin D to block essentially all transcription (Dani et al., 1984). Samples were then analyzed by Northern blot for the content of LO and GAPDH mRNAs, an internal control. As shown (Figure 3A), control RFL6 cells incubated in the absence of CSC displayed a relatively stable mRNA level after actinomycin D chase. The t1/2 for LO mRNA decay is about 24 h. In contrast, in cells treated with 120 μg/ml CSC plus actinomycin D, the level of LO mRNAs was significantly reduced by 80% at 6-h chase, and completely disappeared after 12-h chase while the level of GAPDH mRNA was nearly unchanged under the same conditions (Figure 3B). The t1/2 for LO mRNA decay in the presence of 120 μg/ml CSC was decreased to 4.5-h. Furthermore, the dose-response assays showed that following the 24 h actinomycin D-chase, CSC at 20, 40, 80 and 120 μg/ml reduced mRNA levels to 80.0, 57.5, 26.3 and 0.1% of the control, respectively, in treated cells (Figure 3C). The 50% LO mRNA decay occurred at 45 μg/ml CSC. These data indicate that CSC facilitated the LO mRNA degradation, thus enhancing its instability.
Figure 3. Actinomycin D-chase assay for LO mRNA stabilities in control and CSC-treated cells.
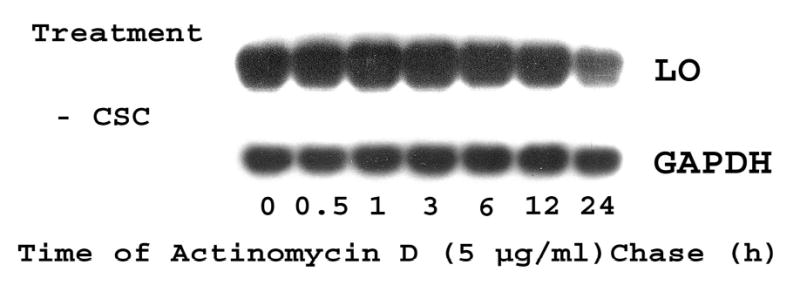

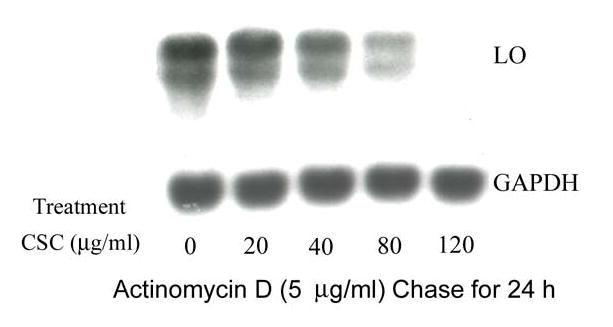
For time course studies, growth-arrested cells were incubated in the presence of 5 μg/ml of actinomycin D without (Panel A) or with CSC at 120 μg/ml (Panel B). At indicated time points, total RNA (15 μg) was extracted from each sample and analyzed by Northern blot. LO transcript levels were quantitated by the 1D Scan EX software and normalized to the internal control (GAPDH) using the level at zero time of actinomycin D treatment as 100%. For dose-response assays (Panel C), cells were incubated for 24 h with 5 μg/ml of actinomycin D in the absence (control) or presence of CSC at various doses. mRNA levels in control and treated cells were measured as described above and expressed as % of the control.
LO promoter activities in RFL6 cells treated with CSC
To assess the effects of CSC on LO transcriptional control, we have prepared rat LO promoter-luciferase constructs (Figure 4A) and examined the transient expression of the reporter gene in transfected RFL6 cells. β-Gal expression vectors as an internal control were co-transfected into cells. As shown (Figure 4B), although CSC at 20 and 40 μg/ml did not induce marked alterations in the expression of the reporter gene, high doses of CSC (> 80 μg/ml) significantly inhibited luciferase activities in treated cells amounting to 42% and 33% of the control at 80 and 120 μg/ml of CSC, respectively. This is consistent with CSC effects on the LO mRNA expression in these cells under the same conditions. Thus, downregulation of LO by CSC was observed at the promoter level.
Figure 4B. LO promoter activities in control and CSC-treated cells.
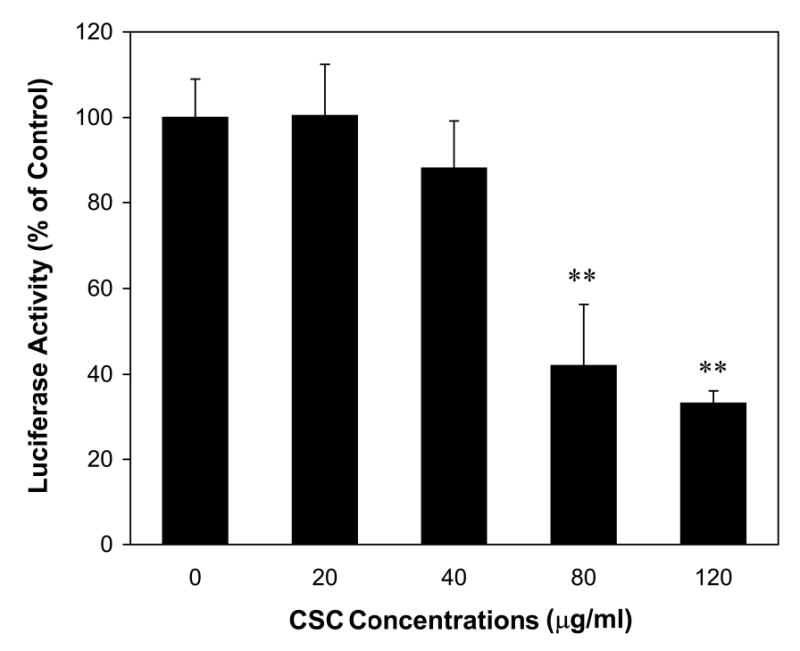
RFL6 cells were co-transfected with LO promoter-reporter plasmids and pSV-β-galactosidase (β-gal) vectors. After 6 h posttransfection, cells were washed, incubated in 10% FBS/DMEM for an additional 18 h period, washed again, and exposed to CSC at indicated doses for 24 h. Luciferase and β-gal activities in control and treated cells were measured according to manufacturer’s instructions. LO-promoter activities in transfected cells were normalized to β-gal activities and expressed as cpm/OD β-gal (100% luciferase activity in the control = 3,528 ± 312 cpm/OD β-gal). ** p < 0.01 vs. control for cells treated with 80 and 120 μg/ml of CSC .
The expression of collagen type I and elastin, substrates of LO, in RFL6 cells treated with CSC
Since collagen and elastin are substrates of LO (Kagan and Li, 2003), downregulation of LO mRNAs by CSC may be accompanied by altered metabolism of collagen and elastin substrates. To test this possibility, we examined the expression of collagen type I, selected as a representative of collagen protein family, and elastin at mRNA levels in control and CSC-treated cells. Northern blot analysis demonstrated that steady-state mRNA levels of collagen type I and tropoelastin were both decreased dramatically in CSC treated cells (Figures 5A and 5B). The density analysis showed that CSC at 20, 40, 80 and 120 μg/ml reduced transcript levels of collagen type I to 90, 75, 50 and 10% of the control (Figure 5A) and of tropoelastin to 90, 78, 61 and 33% of the control (Figure 5B), respectively. These results indicate that downregulation of LO by CSC was concomitant with inhibition of the expression of collagen and elastin substrates, major components of the lung ECM.
Figure 5. Northern blot analysis for steady-state collagen type I and tropoelastin mRNA levels in control and CSC-treated cells.
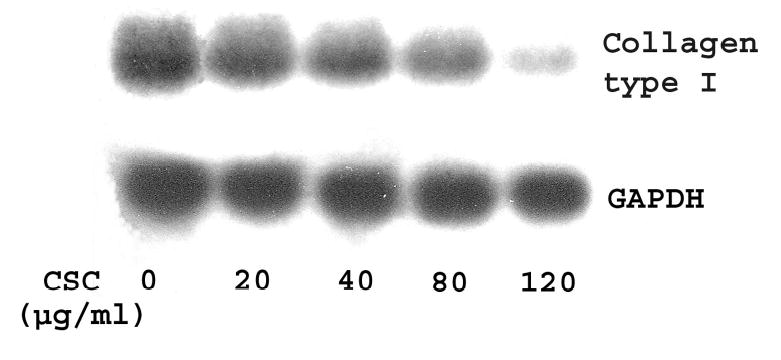

Total RNA (15 μg) was derived from growth-arrested cells treated with or without CSC for 24 h at indicated doses. Transcripts for collagen type I and GAPDH (Panel A) and for tropoelastin and β-actin (Panel B) were size-fractionated and labeled as shown, respectively. β-Actin and GAPDH were used as internal controls.
DISCUSSION
In this study, we have demonstrated transcriptional and posttranscriptional perturbation of LO expression induced by CSC in cultured rat fetal lung fibroblasts (RFL6). Exposure of cells to CSC induced a dose-dependent decrease in LO steady-state mRNA levels (Figure 1) which occurred as a collective result of suppression of LO promoter activity (Figure 4B), inhibition of transcription initiation (Figure 2) and enhancement of mRNA instability (Figures 3A-3C), but not due to a decrease in cell viability. Furthermore, reduction of LO steady-state mRNA levels by CSC was accompanied by downregulation of transcripts of collagen type I (Figure 5A) and tropoelastin (Figure 5B), substrates of LO and also essential components of the lung ECM, pertinent to lung ECM diseases such as emphysema elicited by CS in humans.
Apparently, CSC-induced downregulation of LO mRNA as shown (Figure 1) did not result from overt CSC toxicity since doses of CSC used in this study ranging from 20–120 μg/ml did not induce significant damage to the cell membrane as revealed by trypan blue staining. Notably, is the maximum exposure dose of CSC relevant to the human exposure situation? Yes, according to the following calculation. There are 5 × 109 particles/ml with diameters of 0.1–1 μm in the mainstream CS aerosol. Thus, all of the particles in cigarette smoke are within the size range reaching the alveolar space, known to be 0.1– 3 μm (Hoffmann and Wynder, 1999). Furthermore, each cigarette yields 26.1 mg CSC of which at least 50% presumably deposit in the respiratory tract (IARC, 1985). At a half pack to one pack per day (10–20 cigarettes), the inhaled CSC amounts to 130.5–261 mg. Since the wet weight of a human lung approximates 1,200 g, upon homogenization, it could produce 1.2 L lung homogenate. Thus, the 24 h-adjusted lung burden of CSC of such a smoker is 130.5–261 mg/1.2 L (= 108.8–217.5 μg/ml) in comparison to the maximum dose of 120 μg/ml CSC exposure for 24 h to RFL6 cells. This is the rationale for using the range from 20 to 120 μg/ml of CSC in this cell culture study.
LO is a copper-dependent enzyme secreted by fibrogenic cells such as fibroblasts. This enzyme oxidizes peptidyl lysine residues to form peptidyl α-aminoadipic-δ-semialdehyde resulting in spontaneous, covalent crosslinking of collagen and elastin in the ECM. Therefore, LO plays a critical role in ECM morphogenesis, stabilization and repair (Kagan and Li, 2003). LO is synthesized by fibroblasts as a 46 kD preproenzyme. Following signal peptide cleavage and N-glycosylation, the resulting 50 kD proenzyme is secreted and then proteolytically cleaved to the 32 kD functional species in the extracellular space (Trackman et al., 1992; Li et al, 1995). CS, a complex chemical mixture, contains more than 4,000 different compounds including oxidants, heavy metals, and carcinogens that individually or collectively initiate or promote lung injuries. Our recent studies have indicated that LO is an important cellular target for CS insult. Exposure of rat fetal lung fibroblasts to CSC, the particulate phase of CS, induced decreased levels of all LO protein species including the 46 kDa preproenzyme, the 50 kDa proenzyme and the 32 kDa mature enzyme, as well as the catalytic activity in a dose-dependent manner (Chen et al., 2005). Consistent with these findings, the present data indicate that transcriptional and posttranscriptional regulation of LO are important targets for CSC.
The steady-state concentrations of mRNA as determined by Northern blot are dependent on both the transcriptional rate and the mRNA stability (Schubeler and Bode, 1997). The nuclear run-on assay (Figure 2) showed that CSC treatment induced decreased transcriptional rates of LO such that the level of nascent LO transcripts was decreased to 27.7% of the control in cells exposed to 120 μg/ml CSC. Such inhibition of LO transcriptional initiation may result from perturbing by CSC the processes of transcription complex assembly and/or binding of RNA polymerase II as well as elongation of RNAs (Parent et al., 2004; Howe, 2002 ). Furthermore, the actinomycin D chase assay indicated that CSC also inhibited LO mRNA stability in a dose-dependent manner with an IC50 = 45 μg/ml CSC (Figure 3C). The half-life of LO mRNA decay decreased from 24 h in the control to 4.5 h in cells incubated in the presence of 120 μg/ml of CSC (Figures 3A and 3B). It is established that mammalian mRNAs contain stability elements at either termini including the 5′m7 G cap and the poly (A) tail. Specific protein factors such as the cap binding proteins and the poly (A) binding proteins bind to these corresponding cis elements preventing an mRNA from exonucleolytic degradation (Guhaniyogi and Brewer, 2001). CSC reduced LO mRNA stability as shown in this study possibly by blocking the binding and the interaction of these stability-related cis elements with their protein factors. By further data analysis, we found that under our cell culture conditions, for example, 40 μg/ml CSC decreased the LO mRNA stability to 57.5% of the control (Figure 3C) while the same dose of CSC reduced the LO relative transcription rate only to 79.7% of the control (Figure 2). Furthermore, 80 μg/ml CSC induced the mRNA decay to 26.3% of the control (Figure 3C) whereas the same dose of CSC resulted in the transcription initiation only to 38.9% of the control (Figure 2). Based on these parameters, the calculated overall decrements induced by 40 and 80 μg/ml CSC in LO mRNA levels reached 45.8% (79.7% × 57.7%) and 10.2% (38.9% × 26.3%) of the controls, respectively, that are close to the actual steady-state LO mRNA levels, i.e., 37.4% and 3.6% of the controls, respectively, measured by Northern blots under the same conditions (Figure 1). These results suggest that lower doses of CSC were sufficient to perturb the LO mRNA stability whereas higher doses of CSC were required to interfere with its transcriptional initiation. Thus, the decreased transcriptional rate and the enhanced mRNA instability both collectively led to decreased levels of steady-state concentrations of LO transcripts in cells exposed to CSC.
To further investigate CSC modulation of LO transcription, we cloned the rat LO promoter into the reporter gene construct, pGL3-basic (Promega), upstream of the luciferase gene. The predicted initiation site (CTT) for transcription (to be determined in future) was designated as +1 whereas the start codon (ATG) for translation was located at +287 (Figure 4A). The sequence analysis for the approximately 1.9 kb LO promoter indicated that it contains 286 bp downstream of the predicted transcriptional initiation site, the 5′-untranslation region (5′UTR), with 1,579 bp upstream of the predicted transcription initiation site, the putative cis-element containing region. In addition to TATA-like (TF II), GC (SP1) and GGG(A/C)GGGG (AP2) boxes, the cloned rat LO promoter also contains the metal response element (MRE, core sequence = TGCRCNC; where R = purine, N = any nucleotide) (Stuart et al., 1984; Lichtlen et al., 2001), the hypoxia response element (HRE, core sequence = RCGTG) (Semenza, 2000) and the antioxidant response element (ARE, core sequence =RTGACNNNGC) (Chen et al., 2003; Hayes et al., 2000). As shown here, CSC inhibited LO promoter-directed luciferase expression in treated cells such that 120 μg/ml of CSC reduced the reporter gene expression to 33% of the control (Figure 4B). This could result from perturbation by CSC of the interaction of cis-elements with their corresponding transcription factors. Note that upregulation of metallothionein (MT) as a major phenotypic change was accompanied with downregulation of LO in CSC treated cells (Chen et al., 2005). Many MRE-related transcription factors would be recruited to bind to MREs in the MT promoters for gene activation thus competing with their binding to the LO promoter. As reported, MT-I and MT-II genes contain 6 MREs in their promoter regions, respectively. In addition, inactivation of HRE-related transcription factors by oxidant-containing CSC may also inhibit LO promoter activity. These possibilities are under study. Results presented suggest that CSC inhibition of LO promoter activity may be a critical mechanism leading to decreased levels in LO relative transcriptional rates as shown by the nuclear run-on assay (Figure 2) and in LO steady-state mRNAs revealed by Northern blot analysis (Figure 1) in cells treated with CSC under the same conditions.
The lung ECM is a dynamic structure composed of a number of functionally diverse elements which were integrated mainly by interstitial cells, e.g., fibroblasts (Davidson, 1990; Campa et al., 1993). The overall pattern of the lung ECM results from an intricate balance between the synthesis and degradation of its major structural components such as collagen and elastin. Emphysema is currently defined as “a condition of the lung characterized by abnormal enlargement of respiratory airspaces with destruction of alveolar walls and the loss of elasticity of the lung (Snider et al., 1986). The importance of LO in emphysema pathogenesis was demonstrated by the fact that chicks, rats and hamsters fed with a copper-deficient diet developed lung lesions in these animals similar to panlobular emphysema in humans (Harris, 1986; O’Dell et al., 1978; Soskel et al., 1984). β-Aminoproprionitrile (BAPN), an irreversible inhibitor of LO, can limit the fibrogenic response in certain surgical procedures and decrease collagen/elastin deposition in models of lung fibrosis. Feeding of BAPN markedly enhanced elastase-induced emphysema in hamsters (Kuhn and Starcher, 1980). Moreover, LO regulation of the substrate gene expression is further supported by the observation that treatment of rat neonatal aortic smooth muscle cells with BAPN induced downregulation of tropoelastin mRNAs (Jackson et al., 1991) and that overexpression of LO in COS7 cells by gene transfection enhanced collagen type III promoter activity (Giampuzzi et al., 2000). In this study, we indicated inhibition of LO mRNA expression by CSC associated with downregulation of collagen type I and tropoelastin transcripts, consistent with our previous findings that CSC suppressed production of collagen and elastin proteins (Chen et al., 2005). In this case, the solubilized collagen and elastin as a result of LO deficiency may inhibit their own gene expression by activation of the feedback regulation mechanism (Ivarsson et al., 1993; Riikonen et al., 1995; Jackson et al., 1991). Thus, LO may play a critical role in regulation of its substrate expression. Loss of ECM components, which may induce emphysema development, may result from inhibition of synthesis of ECM precursors important for the repair process. Briefly, our findings as shown in this study indicate perturbation of LO transcription and posttranscriptional modification by CSC coupled with downregulation of transcripts of collagen type I and tropoelastin, substrates of LO, enhancing our understanding of the molecular mechanism of CS damage to the lung ECM relevant to emphysema pathogenesis.
Acknowledgments
This work was supported by research grants from the Philip Morris External Research Program and NIH R01-ES 11340.
Footnotes
Conflict of Interest Statement There are no conflicts of interest.
References
- American Lung Association of Washington. (2004). Lung Disease. http://www.alaw.org/lung_disease
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K., Albright, L.M., Coen, D.M., Varki, A., and Janssen, K. (1995). Current Protocols in Molecular Biology pp. 9.4.1–9.4.3, John Wiley & Sons, Inc., New York.
- Campa, J.S., Harrison, N.K., and Laurent, G. (1993). Regulation of matrix production in the airways. In T-Lymphocyte and Inflammatory Cell Research in Asthma (G. Jolles, J-A. Karlsson, and J. Taylor, Eds.), pp.221–235, Academic Press, London.
- Chen L-J, Zhao Y, Gao S, Chou IN, Toselli P, Stone P, Li W. Downregulation of lysyl oxidase and upregulation of cellular thiols in rat fetal lung fibroblasts treated with cigarette smoke condensate. Toxicol Sci. 2005;83:372–379. doi: 10.1093/toxsci/kfi019. [DOI] [PubMed] [Google Scholar]
- Chen XL, Varner SE, Rao AS, Grey JY, Thomas S, Cook CK, Wasserman MA, Medford RM, Jaiswal AK, Kunsch C. Laminar flow induction of antioxidant response element-mediated genes in endothelial cells. J Biol Chem. 2003;278:703–711. doi: 10.1074/jbc.M203161200. [DOI] [PubMed] [Google Scholar]
- Dani C, Blanchard JM, Piechaczyk M, Sabouty SE, Marty L, Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci USA. 1984;81:7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Armiento JL, Dalal SS, Okada Y, Berg RA, Chada K. Collagenase expression in the lungs of transgenic mice causes pulmonary emphysema. Cell. 1992;71:955–961. doi: 10.1016/0092-8674(92)90391-o. [DOI] [PubMed] [Google Scholar]
- Davidson JM. Biochemistry and turnover of lung interstitium. Eur Respir J. 1990;3:1048–1063. [PubMed] [Google Scholar]
- Dyer RB, Herzog NK. Isolation of intact nuclei for nuclear extract preparation from a fragile B-lymphocyte cell line. BioTechniques. 1995;19:192–195. [PubMed] [Google Scholar]
- Elia MC, Storer RD, Harmon LS, Kraynak AR, McKelvey TW, Hertzog PR, Keenan KP, DeLuca JG, Nichols WW. Cytotoxicity as measured by trypan blue as a potentially confounding variable in the in vitro alkaline elution/rat hepatocyte assay. Mutat Res. 1993;291:193–205. doi: 10.1016/0165-1161(93)90159-w. [DOI] [PubMed] [Google Scholar]
- Giampuzzi M, Botti G, Duca MD, Arata L, Ghiggeri G, Gusmano R, Ravazzolo R, Donato AD. Lysyl oxidase activates the transcription activity of human collagene III promoter. J Biol Chem. 2000;275:36341–36349. doi: 10.1074/jbc.M003362200. [DOI] [PubMed] [Google Scholar]
- Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- Harris ED. Biochemical defect in chick lung resulting from copper deficiency. J Nutr. 1986;116:252–258. doi: 10.1093/jn/116.2.252. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Chanas SA, Henderson CJ, McMahon MM, Sun C, Moffact GJ, Wolf CR, Yamamoto M. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Chemoprotection. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- Hoffmann D, Hoffmann I, El-Bayoumy K. The less harmful cigarette: a controversial issue. A tribute to Ernst L Wynder. Chem Res Toxicol. 2001;14:767–790. doi: 10.1021/tx000260u. [DOI] [PubMed] [Google Scholar]
- Hoffmann, D., and Wynder, E.L. (1999). Active and passive smoking. In Toxicology (H. Marquardt, S. G. Schaefer, R. McClellan, and F. Welsch, Eds.). pp. 879–898, Academic press, New York.
- Howe KJ. RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochem Biophys Acta. 2002;1577:308–324. doi: 10.1016/s0167-4781(02)00460-8. [DOI] [PubMed] [Google Scholar]
- IARC. (1985). Tobacco smoking. IARC Monographs on the Evaluation of Carcinogenic Risk to Humans. Vol. 38, pp163–198. IARC, Lyon.
- Ivarsson M, McWhirter A, Black CM, Rubin K. Impaired regulation of collagen pro-α1 (I) mRNA and change in pattern of collagen-binding integrins on scleroderma fibroblasts. J Invest Dermatol. 1993;101:216–221. doi: 10.1111/1523-1747.ep12364810. [DOI] [PubMed] [Google Scholar]
- Jackson LE, Faris B, Martin BM, Jones HV, Rich CB, Foster JA, Franzblau C. The effect of β-aminopropionitrile on elastin gene expression in smooth muscle cell cultures. Biochem Biophys Res Commun. 1991;179:939–944. doi: 10.1016/0006-291x(91)91909-v. [DOI] [PubMed] [Google Scholar]
- Kagan HM, Li W. Lysyl oxidase: Properties, specificity and biological roles inside of the cell. J Cell Biochem. 2003;88:660–672. doi: 10.1002/jcb.10413. [DOI] [PubMed] [Google Scholar]
- Kimbel, P. (1985). Proteolytic damage and emphysema pathogenesis. In Chronic Obstructive Pulmonary Disease (T. L. Petty, Ed.), pp. 105–127, Marcel Dekker, Inc., New York.
- Kuhn C, III, Starcher BC. The effects of lathyrogens on the evolution of elastase-induced emphysema. Am Rev Respir Dis. 1980;122:453–460. doi: 10.1164/arrd.1980.122.3.453. [DOI] [PubMed] [Google Scholar]
- Li W, Chou IN, Boak A, Kagan HM. Downregulation of lysyl oxidase in cadmium-resistant fibroblasts. Am J Respir Cell Mol Biol. 1995;13:418–425. doi: 10.1165/ajrcmb.13.4.7546771. [DOI] [PubMed] [Google Scholar]
- Lichtlen P, Wang Y, Belser T, Georgiev O, Certa U, Sack R, Schaffner W. Target gene search for the metal-responsive transcription factor MTF1. Nuc Aci Res. 2001;29:1514–1523. doi: 10.1093/nar/29.7.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’ Dell BL, Kilburn KH, Mckenzie WN, Thurston RJ. The lung of the copper-deficient rat. A model for developmental pulmonary emphysema. Am J Pathol. 1978;91:413–432. [PMC free article] [PubMed] [Google Scholar]
- Parent A, Benzaghou I, Bougie I, Bisaillon M. Transcription and mRNA processing events: the importance of coordination. J Biol Sci. 2004;4:624–627. [Google Scholar]
- Rat Genome Sequencing Consortium (2003). Rattus norvegicus chromosome 18 WGS supercontig. NW_047513. http://www.ncbi.nlm.nih.gov/entrez/viewer
- Riikonen T, Westermarck J, Koivisto L, Kahari VM, Heino J. Integrin α2β1 is a positive regulator of collagenase (MMP-1) and collagen α1 (I) gene expression. J Biol Chem. 1995;270:13548–13552. doi: 10.1074/jbc.270.22.13548. [DOI] [PubMed] [Google Scholar]
- Schubeler, D., and Bode, J. (1997). A sensitive transcription assay based on a simplified nuclear runon protocol. Elsevier Trends J. Tech Tips Online, http://tto.trends.com/T01178
- Semenza GL. Expression of hypoxia-inducible factor 1: mechanisms and consequences. Biochem Pharmacol. 2000;59:47–53. doi: 10.1016/s0006-2952(99)00292-0. [DOI] [PubMed] [Google Scholar]
- Snider GL, Lucey EC, Stone PJ. Animal models of emphysema. Am Rev Respir Dis. 1986;133:149–169. doi: 10.1164/arrd.1986.133.1.149. [DOI] [PubMed] [Google Scholar]
- Soskel NT, Watanabe S, Sandberg LB. Mechanisms of lung injury in the copper-deficient hamster model of emphysema. Chest. 1984;85:70s–72s. doi: 10.1378/chest.85.6_supplement.70s. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Searle PF, Chen HY, Brinster R, palmiter RD. A 12 –base pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc Natl Acad Sci USA. 1984;81:7318–7322. doi: 10.1073/pnas.81.23.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trackman PC, Bedell-Hogan D, Tang J, Kagan HM. Post-translational glycosylation and proteolytic processing of a lysyl oxidase precursor. J Biol Chem. 1992;267:8666–8671. [PubMed] [Google Scholar]
- Wright JL, Churg A. Smoke-induced emphysema in guinea pigs is associated with morphometric evidence of collagen breakdown and repair. Am J Physiol. 1995;268:L17–20. doi: 10.1152/ajplung.1995.268.1.L17. [DOI] [PubMed] [Google Scholar]


