Abstract
We report that the archetypal Escherichia coli strain C5 causing neonatal meningitis harbors a pathogenicity island (PAI) designated PAI IC5 that is similar to the PAI IIJ96 of uropathogenic E. coli J96 inserted in the leuX-tRNA gene. PAI-negative C5 mutants had a lower capacity than C5 to induce high-level bacteremia in a neonatal rat model. However, no change in their resistance to the bactericidal effect of serum and their capacity to cross the blood-brain barrier was observed.
Escherichia coli K1 is the leading gram-negative bacterial cause of neonatal meningitis. The rates of mortality and neurologic sequelae remain high despite advances in intensive care (11, 27). The two major pathophysiological steps in E. coli neonatal meningitis consist of bacteremia with intravascular growth (14, 26) and passage of bacteria across the blood-brain barrier (BBB). Clinical and experimental studies have demonstrated a relationship between the magnitude of bacteremia and the development of meningitis (9, 12, 20). Although the K1 capsule, the adhesin Sfa, the invasin IbeA, and the recently described genes traJ, cnf1, cigA, and nilA-C have been incriminated in the virulence of E. coli causing neonatal meningitis (ECNM) (1, 2, 17-19, 25), these factors have not been shown to be sufficient for the development of the disease (1, 2). This suggests that other genes may be involved in the two major pathophysiological steps of E. coli meningitis.
We recently reported that genes encoding P-related fimbriae (prs), hemolysin (hly), cytotoxic necrotizing factor (cnf1), and a heat-resistant agglutinin (hra) were found to be clustered together on the bacterial chromosome of the archetypal ECNM strain C5 (8). The combination of these four genes in association with the insertion element IS100 has previously been described in uropathogenic E. coli strain J96, and they are characteristic of a pathogenicity island (PAI) designated PAI IIJ96 (16, 29).
We now provide evidence that strain C5 harbors a PAI similar to PAI IIJ96. In accordance with the nomenclature described by Hacker et al. (16), we propose to name it PAI IC5. Furthermore, by using a neonatal rat model, we also show that this PAI contributes to the pathogenicity of ECNM strains by inducing high-level bacteremia.
Bacterial strains.
Strain C5 (O18:K1:H7), kindly provided by C. Bortolussi (9), harbors the specific virulence genes encoding the adhesin Sfa and the invasin IbeA (5, 19). The C5 chromosome also contains prs, hly, cnf1, hra, and IS100 genes (8). Strain ECOR 4, which lacks known virulence factors, was used as a nonpathogenic reference (8, 24).
Selection of C5 mutant strains.
Nonhemolytic C5 mutants with spontaneous deletion of the putative uropathogenicity island were selected on sheep blood agar.
Molecular methods.
DNA from strain C5 and its mutants digested by NotI were subjected to pulsed-field gel electrophoresis (PFGE) and then to Southern blotting and hybridization as previously described (8). The primers for PCR probe synthesis were as follows: papG.1, 5′-GACTCTTTCTGTGTCTTGCG-3′; papG.2, 5′-GAACCAGATAGTACTCCTGG-3′; hly.1, 5′-AGGTTCTTGGGCATGTATCCT-3′; hly.2, 5′-TTGCTTTGCAGACTGCAGTGT-3′; cnf1.1, 5′-CAGTGACCGGATCTCCGTTAT-3′; cnf1.2, 5′-CGTGTAATTCTTCTGTACTTCC-3′; hra.1, 5′-CAGAAAACAACCGGTATCAG-3′; hra.2, 5′-ACCAAGCATGATGTCATGAC-3′; IS100.1, 5′-TGCCGTTCGGTTCGAAACTG-3′; and IS100.2, 5′-TGTACTGCACCATCCGTTCC-3′. Detection of probes by chemiluminescence was performed according to the manufacturer's instructions (DIG luminescent detection kit for nucleic acid; Roche Boehringer Mannheim). The primers for PCR that were used for localization of the PAI and for determination of the sequence at the junction of the PAI and chromosome were as follows: pheR.1, 5′-GCCGCAATCTTAAGCAGTTG-3′; pheR.2, 5′-GCACGACATTTCACGTCAGT-3′; leuX.1, 5′-GTGCGACAGGTATAATCCAC-3′; leuX.2, 5′-GACTCGATTTGCATACGGTG-3′; and TspE4.D3, 5′-CTTAACCACCGATAAGGAGC-3′. Long-range PCR was performed as previously described (10). Sequence analyses were performed by ESGS (Evry, France).
Animal model of meningitis.
E. coli bacteremia and meningitis were induced in 107 rats as described by Bortolussi et al. (9) and Kim et al. (20). Briefly, pathogen-free Sprague-Dawley rats were obtained from Janvier Laboratories (Le Genest St Isle, France) at 4 days of age, together with the mothers. At 5 days of age all pups were inoculated intraperitoneally with 200 ± 50 CFU in physiological saline of the C5 or mutant strains. Three and 18 h after inoculation, 5 μl of blood was obtained by tail incision. At 18 h, animals were sacrificed and 5 μl of cerebrospinal fluid (CSF) was immediately obtained by cisternal puncture. Quantitative cultures of all samples was performed. Neonates with negative blood cultures were considered nonbacteremic. The detection limit of bacteria in blood was 4 × 102 CFU/ml. During cisternal puncture, contamination of CSF by small amounts of infected blood can occur. As bacterial counts in blood may be very high (up to 108 CFU/ml), even slight contamination by blood can lead to a wrong diagnosis of meningitis. To prevent this, all CSF samples were examined microscopically and red cells were counted. When red cells were present, the probable number of bacteria derived from infected blood was calculated using the following formula: [level of bacteremia (in CFU per milliliter)] × [number of red cells in CSF (per millimeter3)/number of red cells in blood (per millimeter3) (determined in a pilot experiment to be approximately 5 × 106 per mm3)]. Thus, only CSF samples in which bacterial counts were 10 times higher than expected were considered truly infected.
Serum bactericidal activity.
Serum was obtained from three informed healthy volunteers. Blood was allowed to coagulate for 2 h at 4°C, and the serum was then decanted and immediately aliquoted and stored at −80°C until use. To measure serum bactericidal activity, 20 μl of a bacterial inoculum of 108 CFU/ml in physiological serum was added to 180 μl of freshly thawed pure serum. Quantitative cultures were done 5 h later. Experiments were repeated five times.
Statistical analysis.
Comparison of proportion between two groups was performed using the Pearson chi-square test or the Fisher exact test, when appropriate. Comparisons of bacterial counts in the animal model and in the serum bactericidal experiment were performed using a two-sample unpaired and a paired t test, respectively. Data are expressed as mean ± standard errors. A P value of less than 0.05 was considered statistically significant.
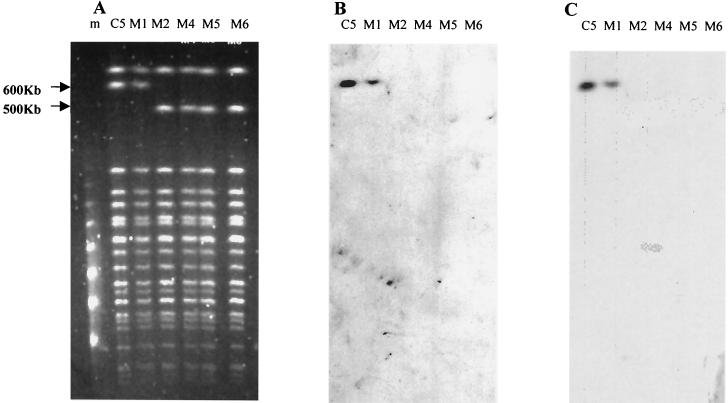
Nonhemolytic C5 mutants were obtained at a rate of 10−5. The five mutants obtained from five different experiments (C5M1, C5M2, C5M4, C5M5, and C5M6) were further characterized. PFGE of DNA digested by NotI yielded a DNA fragment 100 kb shorter than that of the wild-type strain (600 versus 500 kb) with all the mutants except C5M1 (Fig. 1A). PFGE Southern blot hybridization with probes specific for prs, hly, cnf1, hra, and IS100 showed that these genes were physically linked on the wild-type 600-kb NotI DNA restriction fragment and that they were absent from the 500-kb DNA fragment of mutants C5M2, C5M4, C5M5, and C5M6 (Fig. 1B and C, hly and cnf1 probes, respectively). These results showed that the genes characteristic of PAI IIJ96 are clustered on a DNA fragment of approximately 100 kb on the C5 chromosome, and they can be spontaneously deleted en bloc, suggesting that strain C5 harbored a PAI similar to PAI IIJ96 and designated PAI IC5.
FIG. 1.
Characterization of strain C5 and five nonhemolytic mutants by PFGE (A) and hybridization with hly-specific (B) and cnf1-specific (C) probes on Southern blots. Comparison of the macrorestriction profiles showed a 100-kb deletion in mutants C5M2, C5M4, C5M5, and C5M6. Southern blotting showed that the deletion corresponded to the loss of the PAI IC5. Lane C5, C5; Lane M1, C5M1; lane M2, C5M2; lane M4, C5M4; lane M5, C5M5; lane M6, C5M6; lane m: molecular mass marker (concatemers of lambda DNA).
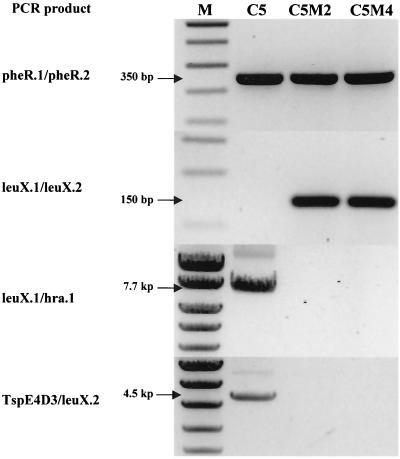
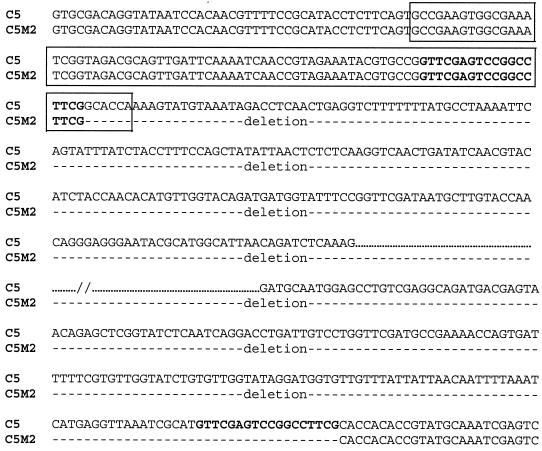
To determine whether PAI IC5 was inserted within the pheR-tRNA gene as described for strain J96 (29), PCR was performed with primers homologous to the flanking sequence of the pheR-tRNA gene (6). This yielded a fragment of 350 bp in strains C5, C5M2, and C5M4, suggesting that PAI IC5 was not inserted in this tRNA (Fig. 2). Based on work by Hacker et al., who described a PAI in strain 536 inserted in the leuX-tRNA gene (7), and on our previous mapping of the clustered genes near this region (8), we postulated that leuX-tRNA was the PAI IC5 insertion site. PCR was then performed with primers homologous to the flanking sequence of this tRNA gene (7). Amplification was negative with strain C5 but yielded a fragment of 150 bp with strains C5M2 and C5M4 (Fig. 2), suggesting that the putative PAI was inserted in the vicinity of the leuX-tRNA gene. Finally, to demonstrate the physical linkage between the PAI and the tRNA gene, we attempted long-range PCR between hra (a gene located at the end of the PAI [29]) and the leuX-tRNA region. This yielded a fragment of 7.7 kb, demonstrating that PAI IC5 was inserted in the leuX-tRNA region (Fig. 2). This DNA fragment was used to determine one side of the sequence of the PAI-chromosome junction. To determine the sequence of the other side of the junction, we attempted long-range PCR between our different subtractive clones mapped in the region of the PAI (8) and the flanking leuX-tRNA region. One of these, TspE4.D3 (accession number AF222192), yielded a fragment of 4.5 kb. The sequence of the junction site (Fig. 3) revealed that the PAI was flanked by 18-nucleotide direct repeats, as described for PAI II536 (7). After a deletion event, one of the repeats remained at the excision site, and six nucleotides of the leuX-tRNA gene were deleted.
FIG. 2.
Agarose gel showing PCR products from strain C5 and mutant strains C5M2 and C5M4. pheR.1/pheR.2, amplification of the pheR-tRNA region, leuX.1/leuX.2, amplification of the leuX-tRNA region; leuX.1/hra.1, amplification between leuX-tRNA and hra; TspE4D3/leuX.2, amplification between TspE4D3 and leuX-tRNA. Amplifications of leuX.1/hra.1 and TspED3/leuX.2 were obtained by long-range PCR. M, molecular mass marker.
FIG. 3.
Nucleotide sequence at the deletion site of the PAI. The tRNA coding sequence leuX is shown in the box. Boldface indicates the 18-nucleotide direct repeats.
Two mutants, C5M2 and C5M4, were used in the experimental meningitis model to assess the role of the uropathogenicity island in the pathogenesis of neonatal meningitis. The results were similar with the two mutants and are thus combined here. Three hours after inoculation, the proportion of bacteremic neonates was similar among the mutants (58 of 61 [95%]) and the wild-type strain, C5 (41 of 46 [90%]), and so was the average bacterial blood count (2.95 ± 0.37 versus 2.82 ± 0.44 log CFU/ml) (results not shown). However, 18 h after inoculation, the proportion of bacteremic neonates was significantly lower among the mutants (46 of 61 [76%]) than with the wild-type strain (46 of 46 [100%]) (P < 0.001) (Fig. 2), and the average bacterial blood count of bacteremic neonates was also significantly lower among the mutants (5.2 ± 1.3 versus 5.7 ± 0.86 log CFU/ml; P < 0.018). Despite this significant difference in level of bacteremia, the rate of meningitis among bacteremic neonates was not significantly lower in the mutants (35%) than in strain C5 (52%) (P = 0.09) (Table 1). Given the known importance of the magnitude of bacteremia in the onset of meningitis, the rates of meningitis among mutants and strain C5 were also compared according to the degree of bacteremia at 18 h postinfection; no significant difference was observed. All these results suggested that PAI IC5 in strain C5 is not involved in the passage across the BBB (Table 1).
TABLE 1.
Development of meningitis (defined as positive CSF culture) in newborn rats with varying degrees of bacteremia
| Level of bacteremia (CFU/ml of blood) | No. of animals with positive CSF culture/no. of bacteremic animals (%)
|
|
|---|---|---|
| Strain C5 | Strains C5M2 and C5M4 | |
| 4 × 102 to <104 | 0/2 | 0/7 |
| 104 to <105 | 3/5 (60) | 2/11 (18)a |
| 105 to <106 | 10/20 (50) | 5/15 (33)a |
| 106 to <107 | 10/16 (62) | 6/10 (60)a |
| 107 to <108 | 1/3 (33) | 3/3 (100)a |
| All bacteremic neonates | ||
| 4 × 102 to <108 | 24/46 (52) | 16/46 (35)a |
P, nonsignificant versus strain C5.
To investigate how the PAI IC5 increases the level of bacteremia, we assessed bactericidal activity of human serum. Strain C5 was resistant relative to the nonpathogenic strain ECOR 4 (Table 2). No difference was observed between the two mutants and C5, suggesting that this PAI is not involved in resistance to complement activity.
TABLE 2.
Determination of serum resistance of strain C5, two mutants (C5M2 and C5M4), and strain ECOR 4
| Serum sample no. | Mean bacterial count in log CFU/ml (standard error) for straina
|
|||
|---|---|---|---|---|
| C5 | C5M2 | C5M4 | ECOR 4 | |
| 1 | 6.7 (0.7) | 5.7 (0.8)b | 5.8 (0.9)b | 2.8 (0.9)c |
| 2 | 5.7 (1.1) | 5.8 (0.6)b | 5.37 (0.8)b | 2.3 (0.6)c |
| 3 | 5.3 (0.8) | 4.91 (0.4)b | 4.95 (0.5)b | 2.5 (0.8)c |
Bacterial count was obtained 5 h after exposure to a 107 CFU per ml standardized inoculum in serum.
P, nonsignificant versus C5 bacterial count.
P< 0.05 versus C5 bacterial count.
In this study we obtained strong evidence that strain C5 harbors a PAI similar to PAI IIJ96. Indeed, prs, hly, cnf1, hra, and IS100 genes colocalized on a DNA fragment with a length (approximately 100 kb) compatible with that of PAI IIJ96 (110 kb). Furthermore, this fragment was spontaneously deleted, en bloc, at a frequency of about 10−5, by recombinational events involving nucleotide repeats as previously described for PAI (7, 15, 21). However, contrary to strain J96, the insertion site of this PAI was in the leuX-tRNA region but not in the PheR-tRNA (29).
One characteristic of ECMN isolates is their ability to induce high-level bacteremia, which is required for passage across the BBB. The capsular polysaccharide K1 plays a key role in this latter process (9, 20), but it is not sufficient to explain the intravascular growth of ECNM, as naturally occurring K1 strains do not all exhibit identical pathogenicities in an experimental model (26). Other extraintestinal virulence factors that might be involved in intravascular growth (4, 5, 10) include hemolysin and cytotoxic necrotizing factor. Indeed, hemolysin gene transfer can convert avirulent fecal E. coli strains into virulent organisms in an experimental model (30), and both cytotoxic necrotizing factor and hemolysin are associated with resistance to phagocytic cells (3, 22, 28). In contrast, previous work suggests that neither factor is involved in extraintestinal pathogenicity (13, 23), and the roles of hly and cnf1 are therefore still not clearly established in extraintestinal pathogenicity. These conflicting results prompted us to investigate the contribution of PAI IC5, which harbors cnf1 and hly, in the virulence of ECNM, focusing on the bacteremic step.
We used a well-documented and relevant experimental model (9, 14, 20). With strain C5, we obtained results very similar to those of Bortolussi et al. and Kim et al. (9, 20). Indeed, we observed that the development of E. coli meningitis was associated with a threshold level of bacteremia (approximately 104 CFU/ml) and that a relationship exists between the magnitude of bacteremia and the rate of meningitis when the number of rats (>10) was sufficient for comparison (Table 1) (9, 20). We also used a method of spontaneous deletion to investigate the influence of the PAI IC5 on virulence. It must be pointed out that the length of this PAI (>100 kb) did not allow a step of complementation of mutants in the aim to control the restitution of virulence. However, the observed change in virulence was unlikely to be due to another concomitant deletion or mutation, as two different mutants from two different cultures were studied and gave similar results.
The mutants displayed the same capacity as the wild-type strain C5 to induce bacteremia at 3 h postinfection. In contrast, at 18 h, 24% of the mutant-infected rats had cleared the bacterium from the blood, compared to 0% of C5-infected rats. Moreover, the average bacterial count in blood of bacteremic neonates was significantly lower at 18 h in pups infected by the mutants than in those infected by strain C5. These results suggested that the PAI IC5 contributed to bacterial survival in blood. To our knowledge, this is the first time that a putative PAI has been directly incriminated in the bacteremic phase of neonatal meningitis. However, the PAI did not influence resistance to the bactericidal activity of serum. Although it can be excluded that different activities of the complement may exist between human and rat species, these results suggest that PAI IC5 may be associated with resistance to other innate defenses, such as neutrophil phagocytosis (3, 22, 28).
Comparing the rate of meningitis between mutants and C5, according to the degree of bacteremia, we found no effect of the PAI IC5 on the second pathophysiological step of E. coli meningitis represented by the BBB passage. This result is conflicting with the recent report of Badger et al. (2) identifying the cnf1 gene in strain RS218 as a contributor to E. coli K1 passage across the BBB. It must be pointed out that the 50% reduction in invasiveness observed in cell cultures was not confirmed by an animal model in the study of Badger et al. (2). On the other hand, it cannot be ruled out that the number of infected pups we used was not sufficient to detect a slight effect of the loss of the cnf1 gene on the traversal of the BBB.
In conclusion, we have demonstrated the presence in strain C5 of a PAI, designated PAI IC5, and its contribution to extraintestinal virulence in a setting other than urinary tract infection, specifically in the bacteremic phase of neonatal meningitis. Further studies are needed to determine which genes of this PAI are specifically contributing to the high sustained bacteremia of strain C5.
Acknowledgments
We thank Etienne Carbonelle and Claire Boumaila for their help.
This work was supported in part by the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires (Appel d'offre 1998), Recherche de déterminants génétiques de pathogénicité chez E. coli K1 responsable de méningite néonatale, and by the Programme Hospitalier de Recherche Clinique (grant AOM 96069).
Editor: V. J. DiRita
REFERENCES
- 1.Badger, J. L., C. A. Wass, and K. S. Kim. 2000. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol. Microbiol. 36:174-182. [DOI] [PubMed] [Google Scholar]
- 2.Badger, J. L., C. A. Wass, S. J. Weissman, and K. S. Kim. 2000. Application of signature-tagged mutagenesis for identification of Escherichia coli K1 genes that contribute to invasion of human brain microvascular endothelial cells. Infect. Immun. 68:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhakdi, S., M. Muhly, S. Korom, and G. Schmidt. 1990. Effects of Escherichia coli hemolysin on human monocytes. Cytocidal action and stimulation of interleukin 1 release. J. Clin. Investig. 85:1746-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingen, E., S. Bonacorsi, N. Brahimi, E. Denamur, and J. Elion. 1997. Virulence patterns of Escherichia coli K1 strains associated with neonatal meningitis. J. Clin. Microbiol. 35:2981-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bingen, E., B. Picard, N. Brahimi, S. Mathy, P. Desjardins, J. Elion, and E. Denamur. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642-650. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschape, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonacorsi, S. P., O. Clermont, C. Tinsley, I. Le Gall, J. C. Beaudoin, J. Elion, X. Nassif, and E. Bingen. 2000. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 68:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bortolussi, R., P. Ferrieri, and L. W. Wannamaker. 1978. Dynamics of Escherichia coli infection and meningitis in infant rats. Infect. Immun. 22:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clermont, O., S. Bonacorsi, and E. Bingen. 2001. The Yersinia high-pathogenicity island is highly predominant in virulence-associated phylogenetic groups of Escherichia coli. FEMS Microbiol. Lett. 196:153-157. [DOI] [PubMed] [Google Scholar]
- 11.de Louvois, J. 1994. Acute bacterial meningitis in the newborn. J. Antimicrob. Chemother. 34(Suppl. A):61-73. [DOI] [PubMed] [Google Scholar]
- 12.Dietzman, D. E., G. W. Fischer, and F. D. Schoenknecht. 1974. Neonatal Escherichia coli septicemia-bacterial counts in blood. J. Pediatr. 85:128-130. [DOI] [PubMed] [Google Scholar]
- 13.Fournout, S., C. M. Dozois, M. Odin, C. Desautels, S. Peres, F. Herault, F. Daigle, C. Segafredo, J. Laffitte, E. Oswald, J. M. Fairbrother, and I. P. Oswald. 2000. Lack of a role of cytotoxic necrotizing factor 1 toxin from Escherichia coli in bacterial pathogenicity and host cytokine response in infected germ-free piglets. Infect. Immun. 68:839-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glode, M. P., A. Sutton, E. R. Moxon, and J. B. Robbins. 1977. Pathogenesis of neonatal Escherichia coli meningitis: induction of bacteremia and meningitis in infant rats fed E. coli K1. Infect. Immun. 16:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacker, J., L. Bender, M. Ott, J. Wingender, B. Lund, R. Marre, and W. Goebel. 1990. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb. Pathog. 8:213-225. [DOI] [PubMed] [Google Scholar]
- 16.Hacker, J., J. Blum-Oehler, B. Janke, G. Nagy, and W. Hoebel. 1999. Pathogenicity islands of extraintestinal Escherichia coli, p. 59-76. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 17.Hoffman, J. A., C. Wass, M. F. Stins, and K. S. Kim. 1999. The capsule supports survival but not traversal of Escherichia coli K1 across the blood-brain barrier. Infect. Immun. 67:3566-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, S. H., Z. S. Wan, Y. H. Chen, A. Y. Jong, and K. S. Kim. 2001. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J. Infect. Dis. 183:1071-1078. [DOI] [PubMed] [Google Scholar]
- 19.Huang, S. H., C. Wass, Q. Fu, N. V. Prasadarao, M. Stins, and K. S. Kim. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 63:4470-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, K. S., H. Itabashi, P. Gemski, J. Sadoff, R. L. Warren, and A. S. Cross. 1992. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J. Clin. Investig. 90:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knapp, S., J. Hacker, T. Jarchau, and W. Goebel. 1986. Large, unstable inserts in the chromosome affect virulence properties of uropathogenic Escherichia coli O6 strain 536. J. Bacteriol. 168:22-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May, A. K., T. G. Gleason, R. G. Sawyer, and T. L. Pruett. 2000. Contribution of Escherichia coli alpha-hemolysin to bacterial virulence and to intraperitoneal alterations in peritonitis. Infect. Immun. 68:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moxley, R. A., E. M. Berberov, D. H. Francis, J. Xing, M. Moayeri, R. A. Welch, D. R. Baker, and R. G. Barletta. 1998. Pathogenicity of an enterotoxigenic Escherichia coli hemolysin (hlyA) mutant in gnotobiotic piglets. Infect. Immun. 66:5031-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkkinen, J., T. K. Korhonen, A. Pere, J. Hacker, and S. Soinila. 1988. Binding sites in the rat brain for Escherichia coli S fimbriae associated with neonatal meningitis. J. Clin. Investig. 81:860-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pluschke, G., A. Mercer, B. Kusecek, A. Pohl, and M. Achtman. 1983. Induction of bacteremia in newborn rats by Escherichia coli K1 is correlated with only certain O (lipopolysaccharide) antigen types. Infect. Immun. 39:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pong, A., and J. S. Bradley. 1999. Bacterial meningitis and the newborn infant. Infect. Dis. Clin. North Am. 13:711-733. [DOI] [PubMed] [Google Scholar]
- 28.Rippere-Lampe, K. E., A. D. O'Brien, R. Conran, and H. A. Lockman. 2001. Mutation of the gene encoding cytotoxic necrotizing factor type 1 (cnf1) attenuates the virulence of uropathogenic Escherichia coli. Infect. Immun. 69:3954-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swenson, D. L., N. O. Bukanov, D. E. Berg, and R. A. Welch. 1996. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect. Immun. 64:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch, R. A., E. P. Dellinger, B. Minshew, and S. Falkow. 1981. Haemolysin contributes to virulence of extra-intestinal Escherichia coli infections. Nature 294:665-667. [DOI] [PubMed] [Google Scholar]