Abstract
Clostridium difficile is the etiological agent of antibiotic-associated diarrhea, a potentially serious condition frequently affecting elderly hospitalized patients. While tissue damage is primarily induced by two toxins, the mechanism of gut colonization, and particularly the role of bacterial adherence to the mucosa, remains to be clarified. Previous studies have shown binding of C. difficile whole cells to cultured cell lines and suggested the existence of multiple adhesins, only one of which has been molecularly characterized. In this paper, we have investigated tissue binding of C. difficile surface layer proteins (SLPs), which are the predominant outer surface components and are encoded by the slpA gene. The adherence of C. difficile to HEp-2 cells was studied by enzyme-linked immunosorbent assay and fluorescence-activated cell sorter analysis, which showed that antibodies to the high-molecular-weight (MW) SLP inhibited adherence. Immunohistochemical analysis of human gastrointestinal tissue sections revealed strong binding both to the surface epithelium lining the digestive cavities and to the subjacent lamina propria, while glands were negative. A similar pattern was observed in the mouse. By using purified recombinant SLPs, we show that binding is largely mediated by the high-MW SLP. By Western blotting analysis, we have identified two potential ligands of the C. difficile SLPs, one of which may be specific to the gut. By using purified extracellular matrix components immobilized on nitrocellulose, we also show SLP binding to collagen I, thrombospondin, and vitronectin, but not to collagen IV, fibronectin, or laminin. These results raise the possibility that the SLPs play a role both in the initial colonization of the gut by C. difficile and in the subsequent inflammatory reaction.
Clostridium difficile, a gram-positive anaerobic bacterium, is a frequent cause of morbidity in hospitalized patients, being the etiological agent of antibiotic-associated diarrhea and pseudomembranous colitis (1, 5, 18). Two factors have been assigned main roles in the pathogenesis of these conditions, the suppression of the resident intestinal flora by the administration of antibiotics (12, 13) and the production by the bacterium of two high-molecular-weight (MW) toxins (toxins A and B) (25, 37). However, not all infections with toxigenic strains result in disease. This has prompted a search for additional virulence factors (1, 11, 29, 34, 36).
As with other enterotoxigenic pathogens, delivery of toxins follows C. difficile colonization of the gut, which requires bacterial adherence to the mucosa. In a Syrian hamster model of clindamycin-induced disease, a correlation was reported between variable efficiencies of gut colonization by different C. difficile strains and their abilities to associate with regions of the gastrointestinal tract spanning from the jejunum to the colon (2). At least part of the variation was suggested to be due to factors other than toxin production. However, the adherence of a poorly virulent strain to the small bowel mucosa was increased by the administration of a crude toxin preparation, possibly due to the unmasking of receptor sites following cell damage.
While no direct microscopic evidence of the adherence of C. difficile to the gastrointestinal mucosa has so far been produced, the phenomenon has been studied in greater detail in vitro. Binding to enterocyte-like Caco-2 cells and mucus-secreting HT29-MTX cells was reported to be increased substantially when the bacterial cells were grown in blood-containing medium and heat shocked at 60°C (11). This correlated with increased amounts of two polypeptides of 12 and 27 kDa on the bacteria. More recently, a seemingly unrelated 66-kDa protein (Cwp66) has been identified as mediating adhesion of C. difficile to Vero cells in culture (36). Both N- and C-terminal fragments of this protein, or their cognate antibodies, inhibited binding. Inhibition, however, was only partial and, surprisingly, showed an inverse correlation with the amount of blocking agent. Moreover, binding was only observed with heat-shocked bacteria, leaving open the question of the nature of the adhesins active at physiological temperatures.
Taken together, these results suggest the existence of multiple adhesins on C. difficile. In order to identify and dissect the roles of individual components, studies with whole cells have obvious limitations, since it is likely that no single factor is absolutely required for adhesion. With the sequence of the whole C. difficile genome now approaching completion, a number of novel polypeptides have been identified which may be candidates for an adhesin function. The Cwp66 protein is one of a large family of gene products encoded by the C. difficile genome with significant homology to the highly expressed surface layer proteins (SLPs), encoded by the slpA gene (4, 15). The cwp66 gene is part of a densely arranged cluster of 12 open reading frames encoding proteins related to SlpA and maps approximately 6 kb 3′ of slpA in the same transcriptional orientation. As in several other bacteria, the SLPs are the predominant surface proteins in C. difficile. However, in contrast to most other species, where the paracrystalline S-layer is composed of one protein species, the S-layer of C. difficile consists of two subunits which show interstrain variability (5, 16, 32). Both the high- and low-MW subunits are encoded by slpA and are produced from the posttranslational cleavage of a common precursor. The high-MW SLP is related both in sequence and function to amidases encoded by the cwlB and cwbA genes from Bacillus subtilis (4). Based on their localization at the outer bacterial surface, and on their abundance, it has been speculated that the SLPs may be involved in pathogen-host interactions critical to pathogenesis (27). Chemical removal of the SLPs or the presence of anti-SLP Fab fragments was shown to abolish adherence of C. difficile to human HeLa or mouse 929 cells (32a).
In the present study, we have investigated the direct binding of C. difficile SLPs to gastrointestinal tissues by both microscopic and biochemical approaches, and we present evidence that the high-MW SLP may play a role in the binding of C. difficile to host cells.
MATERIALS AND METHODS
Bacterial strains.
C. difficile strains R8366 (ribotype 001, strain 1), R7404 (ribotype 017, strain 17), and 630 were grown under anaerobic conditions as described previously (4). Lactococcus lactis strain NZ9000, a Nis− strain carrying the nisR and nisK loci at the chromosomal pepN site (18), was used for cloning and expression experiments with vector pNZ8020 (9). Escherichia coli BL21(DE3) was used for expression of genes cloned into pET28a.
Preparation of native and recombinant SLPs.
SLPs were extracted from C. difficile by treatment with 0.2 M glycine, pH 2.2, as previously described (4). For the production of recombinant SlpA precursor, the entire slpA coding sequence (CDS) was amplified by PCR from clone pCd1e4 (generated from strain 630 by the genome-sequencing project at the Sanger Centre) using primers NF163 and NF164 (Table 1). This introduced an NcoI site spanning the ATG and an XhoI site immediately 3′ of the codon for the last amino acid (Met). Amplification was carried out by Pfu polymerase, and after NcoI plus XhoI digestion, the PCR product was ligated into pET28a (Novagen, Madison, Wis.) to form pET-1e4. For expression in L. lactis, the insert in pET-1e4 was released by digestion with XbaI and Bsp1286I, blunt cloned into the EcoRV site of M13tg131 (17) to provide suitable flanking restriction sites, and recloned between the BamHI and EcoRI sites of pNZ8020 to give pNZ-1e4. A double-stranded oligonucleotide (resulting from the annealing of oligonucleotides NF238 and NF239 and encoding a hexahistidine stretch) was then inserted between the unique PvuI site (newly created in the M13tg131 clone from the ligation of the blunted Bsp1286I half site to the EcoRV half site) and the unique XhoI site at the 3′ end of pNZ-1e4 to give pNZ-1e4H.
TABLE 1.
Oligonucleotides used in the present study
| Oligonucleotide | Sequence | Description |
|---|---|---|
| NF163 | 5′-TAAGCCATGGATAAGAAAAATATAG-3′ | Forward primer for SlpA precursor |
| NF164 | 5′-AGGCTCGAGCATATCTAATAAA-3′ | Reverse primer for SlpA precursor and high-MW subunit |
| NF238 | 5′-CACCACCACCACCACCACTGAC-3′ | Sense primer; hexahistidine encoding |
| NF239 | 5′-TCGAGTCAGTGGTGGTGGTGGTGGTGAT-3′ | Antisense primer; hexahistidine encoding |
| NF244 | 5′-TAAGCCATGGCAAATGATACAA-3′ | Forward primer for high-MW subunit |
| NF245 | 5′-TAAGCCATGGCAACTACTGGAACA-3′ | Forward primer for low-MW subunit |
| NF246 | 5′-AGGCTCGAGTGATTTAGTTTCTAATC-3′ | Reverse primer for low-MW subunit |
The individual high-MW and low-MW SLPs (excluding the signal sequence) were expressed in E. coli BL21(DE3) as follows. After PCR amplification using Pfu polymerase from clone pCd1e4 with oligonucleotides NF244-NF164 and NF245-NF246 (Table 1), the PCR products were digested with NcoI and XhoI, gel purified, and ligated into pET28a. In all cases, recombinant polypeptides included a C-terminal extension of 8 amino acids, including a hexahistidine tag.
For expression in L. lactis harboring pNZ-1e4H, an exponentially growing culture in M17 medium (Merck) supplemented with 0.5% glucose at 30°C was induced with nisin (a gift from Microscience, Wokingham, United Kingdom) for 0.5 h at an optical density at 600 nm of ∼0.5. For expression in E. coli BL21(DE3) harboring pET28a constructs, cells were grown at 37°C in Luria-Bertani broth to an optical density at 550 nm of ∼0.5, induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM, and harvested after a further 3 h. The induced cells were washed once in H2O and resuspended in 10% sucrose-20 mM Tris, pH 8. After being frozen and thawed, the L. lactis cells were lysed with lysozyme (to 20 mg/ml; 10 min at room temperature) and urea (to 6 M), whereas the E. coli cells were lysed with lysozyme (to 1 mg/ml; 10 min at room temperature) followed by sonication on ice (four bursts of 30 s each at maximum power) using a Heat Systems Ultrasonic Processor XL2020 sonicator equipped with a 2-mm-wide probe. They were then subjected to ultracentrifugation at 100,000 × g for 40 min at 4°C.
Affinity chromatography was carried out on Ni-nitrilotriacetic acid (NTA) agarose (Qiagen). Bacterial lysates were brought to 0.5 M in NaCl and 5 mM in imidazole prior to being applied to the column. Bound material was washed with 500 mM NaCl-20 mM Tris (pH 8)-10 mM imidazole and eluted in 20 mM Tris (pH 8)-250 mM imidazole.
The recombinant HC domain from tetanus neurotoxin (TeNT) used as a negative control for binding was prepared and purified as previously described (31).
Anti-SLP antisera.
Four 8-week-old New Zealand White rabbits were each injected subcutaneously with 50 μg of Ni-NTA-purified recombinant high-MW or low-MW subunit in Freund's complete adjuvant (two rabbits per subunit). The rabbits were boosted after 3, 5, 7, and 11 weeks, using the same amount of antigen but in incomplete Freund's adjuvant. For the experiments described in this paper, sera collected after the third boost were used, with an estimated titer in enzyme-linked immunosorbent assays (ELISAs) against the immunogen of ∼1:100,000. Mouse antisera against SLPs from C. difficile strains 1, 17, and 630 were previously described (4).
HEp-2 cell binding.
HEp-2 cells were maintained in Dulbecco's modified Eagle's medium-10% fetal bovine serum. For binding experiments, cells were seeded in 96-well plates and grown to confluence. After they were blocked with 3% bovine serum albumin (BSA) in phosphate-buffered saline (PBS), SLPs or a control protein (recombinant HC domain from TeNT) was added at ∼1 μg/ml in PBS and incubated for 1 h at room temperature. Bound proteins were detected by the sequential addition of anti-SLPs (1:500 for mouse antisera and 1:5,000 for rabbit antisera) and horseradish peroxidase (HRP)-coupled anti-immunoglobulin (Ig) (1:2,000; Dako). For immune depletions, antisera were preabsorbed (in batch mode) onto Ni-NTA agarose coated with excess recombinant SLP.
Fluorochrome labeling of C. difficile cells and binding to HEp-2 cells were essentially as described previously (10), except that they were carried out for 1 h at room temperature instead of 37°C, and the HEp-2 cells were detached by scraping rather than by trypsinization. For blocking experiments, anti-high-MW SLP antisera, or a mixture of antisera and either acid-extracted SLPs or recombinant high-MW SLP, were preincubated with labeled bacteria for 1 h at room temperature. The bacteria were then washed once in PBS prior to their addition to HEp-2 cells.
Immunohistochemistry.
Paraffin sections (5 μm thick) of mucosal biopsy specimens of human gastrointestinal tract (stomach, proximal small intestine, and colon) were obtained from patients (age range, 3 to 16 years) who had undergone routine endoscopic examination for gastrointestinal complaints after a full diagnostic evaluation with fully informed consent and ethical approval. All were classified as histologically normal. Mouse organ samples (obtained under experimentation by license from the United Kingdom government Home Office) were removed immediately following sacrifice and fixed in buffered formalin prior to being embedded in paraffin and sectioned at 4 to 6 μm. The sections were deparaffinized, rehydrated, treated with 3% H2O2 in methanol for 10 min, and blocked in 10% horse serum in PBS (blocking solution) for 30 min. SLPs or a control protein (recombinant HC domain from TeNT) was added at ∼10 μg/ml in blocking solution and incubated for 1 h at room temperature. Anti-SLP antisera (described above) were added at 1:2,000, followed by HRP-conjugated anti-Ig (Dako) at 1:100 in blocking solution. The antisera were incubated for 30 min at room temperature. The results were visualized with diaminobenzidine (Sigma). The sections were lightly counterstained with Meyer's hematoxylin.
Western blotting and dot blot analysis.
Tissue extracts were prepared by homogenizing fresh organs in 10 volumes of 10% sodium dodecyl sulfate (SDS)-10 mM EDTA-25 mM Tris-Cl (pH 6.8), using an Ultra-Turrax homogenizer (IKA). Approximately 100 μg of total protein was fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) on 10% gels and electroblotted onto a nitrocellulose membrane (Bio-Rad) in 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid buffer-10% methanol, pH 11, for 1 h at 70 V and 4°C. Purified extracellular matrix (ECM) components (Sigma) were spotted in duplicate onto the same membrane (20 ng/spot). The membranes were blocked with 10% nonfat dry milk in PBS. SLPs or a control protein (recombinant HC domain from TeNT) was added at ∼0.25 μg/ml in 3% BSA-0.5% Tween-20-0.5% TX-100 in PBS and incubated for 1 h at room temperature. Anti-SLP antisera were used at a 1:50,000 dilution in 3% BSA-0.5% Tween-20-0.5% TX-100 in PBS and incubated for 1 h at room temperature. For blocking experiments, the antisera were preincubated with a 20-fold excess of soluble, purified, recombinant high-MW subunit. Following incubation with an HRP-conjugated goat anti-rabbit Ig (Dako; 1:2,000 dilution), the blots were developed by enhanced chemiluminescence (Amersham) in accordance with the manufacturer's instructions.
RESULTS
Expression of recombinant SLPs in E. coli and L. lactis.
The entire slpA CDS, including the signal sequence, was cloned into the E. coli expression vector pET28a, resulting in an in-frame fusion to C-terminal histidines. In the absence of IPTG, transformants were found to grow significantly more slowly than clones harboring the vector alone and appeared to stop growing altogether upon addition of the inducer. SDS-PAGE analysis of induced cell extracts showed the presence in the soluble fraction of a polypeptide corresponding to the full-length SLP precursor. However, yields were low, and upon purification on Ni-NTA agarose, a number of additional, lower-mass species were seen, suggestive of extensive proteolysis (data not shown).
To overcome this problem, we turned to an expression system based on L. lactis (18). The slpA CDS was recloned into vector pNZ8020 to give pNZ-1e4. A double-stranded oligonucleotide, encoding a hexahistidine stretch, was inserted in frame immediately 3′ of the slpA sequence in order to give a C-terminal fusion (pNZ-1e4H). The resulting plasmid was transformed into L. lactis strain NZ9000. On SDS-PAGE analysis, a decrease in total cellular protein per unit volume of culture was apparent in the strain harboring pNZ-1e4H at 1 h postinduction, reflecting a growth-inhibitory or lytic effect similar to, albeit less pronounced than, that seen in E. coli. L. lactis(pNZ-1e4H) was therefore induced for 0.5 h, and lysates were purified over Ni-NTA agarose. This yielded substantial amounts of a major band (>95% pure) corresponding to the size of the SlpA precursor (Fig. 1).
FIG. 1.
SDS-PAGE analysis of native SLPs extracted from C. difficile strains 1, 17, and 630; recombinant SlpA precursor expressed in L. lactis (pNZ-1e4H); and recombinant high- and low-MW subunits expressed in E. coli (pET28-High and pET28-Low). The last three samples were purified on Ni-NTA agarose. Molecular mass standards in kilodaltons are indicated on the left.
In contrast to the whole slpA gene, fragments encoding either the low-MW or high-MW SLP could be readily expressed in E. coli upon cloning in pET28a. In both cases, purification on Ni-NTA agarose yielded a major band (>95% pure) corresponding to the expected sizes (Fig. 1). The yield of the lower subunit was higher than that of the upper subunit.
Binding of native and recombinant C. difficile SLPs to tissue culture cells.
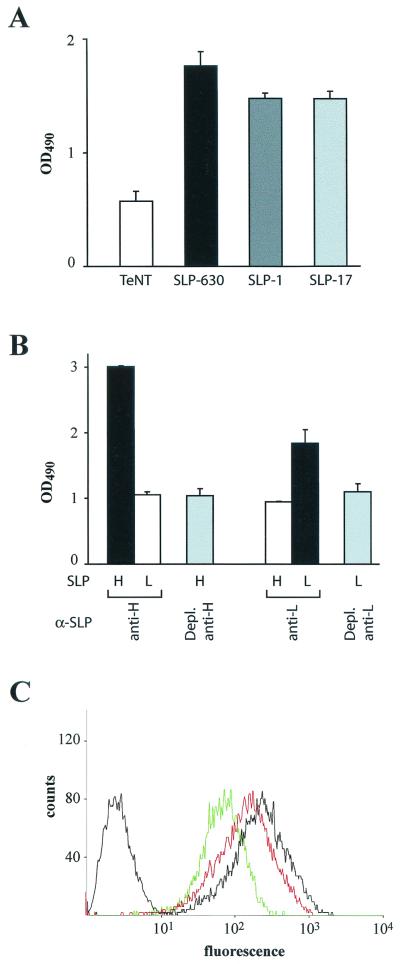
SLP binding experiments were initially performed on live cells in culture. HEp-2 cells were chosen, since although they were not of gastrointestinal origin, adherence of enterobacterial pathogens to these cells had been reported (21). As shown in Fig. 2A, acid-extracted SLPs from strains 1, 17, and 630 all showed binding to cells significantly higher than that found for a control protein (the HC domain from TeNT [31]), indicating that binding of SLPs is independent of the source strain. Further analysis of purified recombinant subunits revealed stronger binding for the high-MW SLP, although the low-MW SLP also scored positive (Fig. 2B). Anti-subunit antisera proved to be monospecific, as shown by the absence of reactivity on the heterologous subunit. Moreover, binding was abolished by removing anti-subunit activity from each antiserum by preadsorption on Ni-NTA-bound antigen.
FIG. 2.
Binding of C. difficile SLPs to cultured HEp-2 cells. (A) Binding of native SLPs from strains 630, 1, and 17 as detected by ELISA using mouse antisera raised against strain-specific SLPs. The HC domain from TeNT, followed by anti-SLP630, was used as a negative control. (B) Binding of purified recombinant high- and low-MW subunits detected by ELISA using subunit-specific rabbit antisera. Antiserum specificity is shown by the lack of reactivity on the heterologous subunit, as well as following preadsorption on immobilized homologous antigen. The bars (with standard deviations) represent the ratios of values obtained with the indicated protein to those with BSA. (C) FACS profiles of HEp-2 cells bound to fluorochrome-labeled C. difficile preincubated with rabbit preimmune serum (rightmost black line), to fluorochrome-labeled C. difficile preincubated with rabbit antiserum raised against the purified high-MW SLP (red line), or to fluorochrome-labeled C. difficile in the presence of a fivefold excess of unlabeled bacteria (green line). The leftmost black line corresponds to the profile of unlabeled HEp-2 cells. Depl., depleted.
In order to investigate whether the SLPs were involved in C. difficile binding to human epithelial cells, a blocking assay was used. Fluorochrome-labeled C. difficile was prepared by incubation with 2′,7′-bis-(carboxyethyl)-5(6′)-carboxyfluorescein acetoxymethyl ester according to a recently published method (10). Fluorescently labeled bacteria were incubated with HEp-2 cells, and bacteria bound to the cells were visualized by fluorescence-activated cell sorter (FACS) analysis (Fig. 2C). Addition of antiserum against the high-MW subunit to labeled bacteria caused an ∼20 to 30% decrease in cell binding relative to preimmune serum, based on the intensity of HEp-2-bound fluorescence. In parallel experiments, a fivefold excess of unlabeled bacteria resulted in 60 to 65% inhibition (Fig. 2C). Preincubation of the antiserum with the recombinant high-MW subunit completely abolished the blocking effect (data not shown). Thus, the high-MW SLP subunit is involved in binding of C. difficile to human epithelial cells.
Binding of native and recombinant C. difficile SLPs to gut tissue sections.
Direct binding of purified SLPs to human gastrointestinal tissues was investigated on histologically normal sections of mucosal biopsy specimens. SLP binding was revealed by subsequent incubation with antisera raised against the SLP complex or individual subunits, followed by HRP-conjugated secondary antiserum. Initial experiments tested the binding of either native SLPs, purified from C. difficile strain 630 by extraction at low pH, or purified recombinant SLPs expressed in L. lactis as an uncleaved precursor. Strong, specific binding was found both to the epithelium and the lamina propria of the mucosa at all levels of the gastrointestinal tract (Fig. 3). Anti-SLP antisera did not show any reactivity on sections preincubated with a control protein (recombinant HC domain from TeNT) (Fig. 3A, D, and G, 4, and 5A)
FIG. 3.
Low-power views of representative sections from biopsy specimens of human stomach, duodenum, and colon incubated with the HC domain from TeNT as a negative control, with native SLPs extracted from strain 630 (SLP-630), or with purified recombinant SlpA precursor (pNZ-1e4H). Positive binding is indicated by the brown color following immune detection with anti-SLP antiserum and HRP-coupled anti-Ig.
Epithelial staining was strongest in the surface epithelium lining the lumen of the digestive cavities, particularly at the apexes of gastric ridges (Fig. 3B and C) or small intestinal villi (Fig. 3E and F). The highest positivity was found in the apical cytoplasm, and particularly along the apical border, suggesting prevalent localization to the brush border. In contrast, staining was conspicuously absent in the glands, both the oxyntic glands in the stomach and the intestinal crypts of Lieberkühn in the intestine. In the lamina propria, staining was both diffuse and finely fibrillar, indicating binding to components of the ECM. In the stomach, it appeared stronger in the deeper layer, whereas in the colon it was most notable in the most superficial subepithelial layer. Recombinant SlpA precursor gave a pattern largely superimposable on that of native SLPs (Fig. 3).
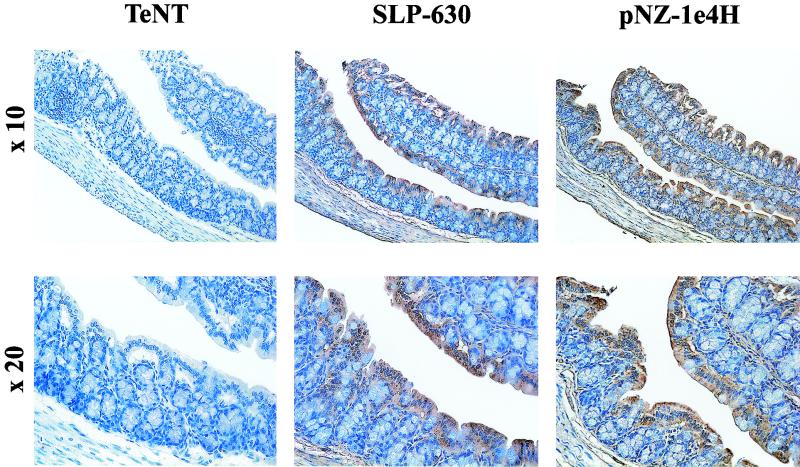
These studies were confirmed and extended with mouse tissues (Fig. 4). Examination of cross sections of the colon spanning the whole intestinal wall showed, in addition to staining of the surface epithelium and, more weakly, of the lamina propria, a distinct positivity of the serosa. Thus, binding of C. difficile SLPs to gastrointestinal tissues is conserved in the mouse, a species that is susceptible to C. difficile-induced disease, albeit to a more limited extent than the hamster (23).
FIG. 4.
Low (×10)- and high (×20)-power views of representative sections from mouse colon incubated with the HC domain from TeNT as a negative control, with native SLPs from strain 630 (SLP-630), or with purified recombinant SlpA precursor (pNZ-1e4H). Positive binding is indicated by the brown color following immune detection with anti-SLP antiserum and HRP-coupled anti-Ig.
In order to identify the SLP sequences that mediate binding, purified recombinant high- and low-MW SLPs were used on human gastrointestinal sections. Most of the intensity and pattern of staining obtained with native SLPs was replicated by the high-MW subunit (Fig. 5B and D). However, while the low-MW subunit did not give any signal on the lamina propria, weak, punctate staining was observed in a subpopulation of epithelial cells both in the surface epithelium and in the necks of the glands in the colon (Fig. 5C).
FIG. 5.
High-power (×20) views of representative sections from a biopsy of human colon, incubated with the HC domain from TeNT as a negative control, with native SLPs from strain 630 (SLP-630), and with purified recombinant low-MW SLP (pET-Low) or high-MW SLP (pET-High). Positive binding is indicated by the brown color following immune detection with antisera specific for native SLPs (TeNT and 630) or individual subunits and HRP-coupled anti-Ig.
Biochemical characterization of tissue SLP ligands.
The identities of potential protein ligands of the C. difficile SLPs were investigated by Western blotting analysis. Since binding is conserved between humans and mice, the analysis was carried out on mouse tissues, which were more readily accessible. Whole extracts were prepared from mouse small and large intestine, as well as from two structurally and functionally distinct organs: the kidney, a mainly epithelial organ, and the spleen, composed mainly of lymphoid cells of mesenchymal origin. Extracts were run on SDS-PAGE, transferred to membranes, and sequentially incubated with purified recombinant high-MW subunit or recombinant HC domain from TeNT as a negative control, followed by anti-SLP antiserum and HRP-coupled anti-Ig. In preliminary experiments, both the high-MW subunit and the native SLPs were found to give a relatively high uniform background binding to nylon or polyvinylidene difluoride membranes. This phenomenon, the basis of which has not been investigated further, was not due to the anti-SLP sera, since it was not observed when the sera were used following incubation with recombinant HC domain (Fig. 6, left blot), occurred in the absence of blotted proteins, and was not abolished by increasing the concentration of blocking agents and of detergents in the hybridization and washing fluids. The background was reduced, although still noticeable, on a nitrocellulose membrane, which was therefore used for all subsequent experiments. Representative results are shown in Fig. 6. Two bands at ∼63 and 81 kDa are visible with both the high-MW SLP and the control protein. However, two species show specific binding to the high-MW SLP: a band at ∼97 kDa is present in all four organs, whereas one at ∼107 kDa is specific to the small and large intestine. Competition experiments with excess soluble SLP confirmed the specificities of both bands (Fig. 6C).
FIG. 6.
Western blotting analysis of SLP ligands in mouse tissues. Extracts from the indicated tissues were fractionated by SDS-PAGE, blotted onto nitrocellulose, and probed with the HC domain from TeNT as a negative control (left) or with purified recombinant high-MW SLP (H) (middle and right). Bound protein was detected with anti-high-MW subunit (α-H). In the case of the right-hand panel, this had been preincubated with soluble purified recombinant high-MW SLP as a specificity control. The two arrowheads at the right of the middle panel point to an ∼97- and an ∼107-kDa species which bind the high-MW SLP specifically. Molecular mass standards in kilodaltons are indicated on the left.
The MW of the larger SLP-binding species is close to that of a collagen monomer. Binding of SLPs to collagens, as well as to other ECM components, was tested in a separate dot blot experiment, since most such components have a large mass and are not easily resolved on SDS-PAGE. The results are shown in Fig. 7. Specific binding of both the native SLPs and the high-MW subunit, but not of the low-MW subunit, was observed to collagen I, to thrombospondin, and, more weakly, to vitronectin. In contrast, there was no binding to collagen IV, fibronectin, or laminin.
FIG. 7.
Binding of C. difficile SLPs to immobilized ECM components. Aliquots (∼20 ng) of the indicated proteins were spotted in duplicate onto a nitrocellulose membrane, which was then probed with the HC domain from TeNT as a negative control, with native SLP from strain 630, or with purified recombinant high-MW (pET-High) or low-MW (pET-Low) subunit. The blots were developed with antisera specific for native SLPs (TeNT and 630) or individual subunits, followed by HRP-coupled anti-Ig and enhanced chemiluminescence.
DISCUSSION
In this paper, we show strong and specific binding of purified C. difficile SLPs both to human epithelial cells and to gastrointestinal tissues. Binding to both cells and tissues is observed with acid-extracted SLPs and with the soluble recombinant high-MW subunit. In contrast, much less or no less binding is observed with the soluble recombinant low-MW subunit, expressed and purified under the same conditions as the recombinant high-MW subunit. Finally, we have shown that the anti-high-MW subunit serum partially blocks adherence of C. difficile to HEp-2 cells, and therefore we conclude that the SLPs are involved in the adherence of whole bacteria to epithelial cells. It is unlikely that binding of SLPs to cells and tissues is an artifact of the method of preparation of the proteins. Both the native and recombinant SLP preparations retain their characteristic peptidoglycan hydrolase activities, the acid-extracted SLPs are capable of polymerizing into higher-order structures upon the addition of calcium (4), and antisera raised against both recombinant subunits react at high titers with whole C. difficile cells, suggesting that the natural conformation of the proteins is retained.
In a previous study, only partial inhibition of adherence was also reported with antibodies against a distinct but related cell surface component of C. difficile, the SlpA-like ORF3/Cwp66 (36). As is the case with many bacterial pathogens, adhesion of C. difficile to host cells is probably multifactorial, involving several distinct bacterial proteins and possibly other macromolecules.
The C. difficile SLPs bind to different components of gastrointestinal tissues, at both the morphological and biochemical levels. Morphologically, SLP binding occurs both to epithelial and connective tissue. In the intestinal epithelium, positive cells correspond to enterocytes, while goblet cells are negative. Thus, although an association of whole C. difficile cells to mucus-secreting HT29-MTX cells has been reported (11), it is unlikely that the SLP ligand is a mucus constituent. Based on the concentration of epithelial staining along the luminal border, it seems more likely that it corresponds to a component of the brush border. Binding shows an intriguing correlation with the architectural organization and hence the differentiation state of the intestinal epithelium. Thus, it is largely restricted to cells lining the main digestive cavity and is highest at the apexes of villi, which correspond to the most differentiated enterocytes. In contrast, glands and crypts, which represent the proliferative compartment, are negative. The staining obtained on the gastrointestinal connective tissue indicates binding to the ECM rather than to cells. Both a diffuse and a fibrillar pattern are observed, suggestive of binding to multiple components.
The immunohistochemistry data are supported and extended by the results of biochemical analysis. On Western blotting, the SLPs were found to bind specifically to two major polypeptides, one of which may be specific to the intestine. The nature and the precise cellular origin of these polypeptides remain to be established. Candidate ECM ligands were investigated after immobilization on a membrane. A clear signal was obtained with collagen I, thrombospondin, and vitronectin, but not with collagen IV, fibronectin, or laminin. The last results indicate that the SLPs do not bind to the basement membrane of the epithelium, consistent with the immunohistochemistry data. They also suggest that binding is mediated by specific structural domains rather than by nonspecific interactions. We cannot exclude binding to additional ECM components, such as the glycosaminoglycans that are the main constituents of the connective tissue ground substance. Binding of whole C. difficile cells has recently been reported for fibrinogen, vitronectin, fibronectin, and collagen types I, III, IV, and V but not for laminin (5a).
SLP binding to tissues or ECM molecules is mediated almost exclusively by the high-MW subunit. The sequence of this subunit is well conserved among strains, suggesting that binding is not strain dependent. This is supported by the ELISA data on HEp-2 cells using acid-extracted SLPs from three independent strains. While binding of soluble SLPs is sufficiently strong to be easily detected under the stringent conditions used in our histochemical and biochemical assays, even higher affinity is likely to exist for polymeric SLPs, such as that found within the bacterial S-layer. In contrast, the low-MW SLP does not bind to the mucosal connective tissue or to purified ECM components. Binding is observed, however, to rare gastrointestinal epithelial cells in a weak, punctate pattern, as is binding to HEp-2 cells in ELISA, although the latter are not derived from the gastrointestinal epithelium. Thus, the SLPs may be a multifunctional adhesin.
The role of SLP binding to tissue components in C. difficile-associated disease remains to be investigated. In several bacteria, surface molecules have evolved to interact with animal cells. Thus, in the case of Listeria monocytogenes, InlA and InlB binding to E-cadherins on enterocytes (8) and to gC1q-R and Met, respectively, on other epithelial cell types (3, 30), is required for internalization. Similarly, in Yersinia species, invasin binding to β1 integrins promotes entry into intestinal M cells (14). In both cases, the cellular ligands are adhesion molecules, the engagement of which activates intracellular signaling pathways and leads to rearrangements of the actin cytoskeleton. There is no evidence that C. difficile can gain entry into nonphagocytic cells. However, disruption of the actin cytoskeleton is among the most notable effects of the C. difficile toxins (25, 37). The resulting damage to the intestinal surface epithelium allows the bacteria access to the underlying mucosal connective tissue. A number of studies have shown bacterial binding to ECM components and documented its role in infection. Thus, S. aureus binding to fibronectin via FnBP-A (fibronectin binding protein) or to collagen via CNBP (collagen binding protein) has been shown to play a role in animal models of infection (20, 22, 24, 26). In Yersinia species, mutations of the YadA surface protein causing loss of collagen binding resulted in a loss of virulence in mice (33). Binding of SLPs to ECM components is not unique to pathogenic bacteria but may also have a role in gut colonization by the normal resident flora (35).
We propose that SLP-mediated binding may play a role in at least two separate stages. First, it could mediate binding of C. difficile to brush border components and therefore allow targeted delivery of toxins to enterocytes. This may be essential in order to avoid dilution of the toxins or their degradation by digestive proteolytic enzymes (7). At a later stage, following toxin-induced epithelial damage, SLP binding to ECM components is likely to contribute significantly to further tissue damage. In the intestinal mucosa, collagen I plays a major structural role, whereas thrombospondin mediates the assembly of multiprotein complexes that can modulate cell adhesion or signaling (6, 19) and vitronectin plays a key regulatory role in the integrity of the ECM (28). SLP binding may lead to the disruption of these crucial interactions. In addition, binding to tissue components may contribute to pathogenesis by masking potential antigenic determinants and allowing the bacteria to escape from the immune response. A thorough testing of this hypothesis will require the development of C. difficile slpA mutants and an analysis of their pathogenicities in an experimental animal model.
Acknowledgments
We thank the Sanger Centre, Cambridge, United Kingdom, for providing clone pCd1e4 and Christine Hale for help with FACS analysis.
E.C. was supported by a fellowship from the Blanceflor Boncompagni Ludovisi Foundation, Stockholm, Sweden.
Editor: B. B. Finlay
REFERENCES
- 1.Borriello, S. P., H. A. Davies, S. Kamiya, P. J. Reed, and S. Seddon. 1990. Virulence factors of Clostridium difficile. Rev. Infect. Dis. 12:S185-S191. [DOI] [PubMed] [Google Scholar]
- 2.Borriello, S. P., A. R. Welch, F. E. Barclay, and H. A. Davies. 1988. Mucosal association by Clostridium difficile in the hamster gastrointestinal tract. J. Med. Microbiol. 25:191-196. [DOI] [PubMed] [Google Scholar]
- 3.Braun, L., B. Ghebrehiwet, and P. Cossart. 2000. gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J. 19:1458-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calabi, E., S. Ward, B. Wren, T. Paxton, M. Panico, H. Morris, A. Dell, G. Dougan, and N. Fairweather. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40:1187-1199. [DOI] [PubMed] [Google Scholar]
- 5.Cerquetti, M., A. Molinari, A. Sebastianelli, M. Diociaiuti, R. Petruzzelli, C. Capo, and P. Mastrantonio. 2000. Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb. Pathog. 28:363-372. [DOI] [PubMed] [Google Scholar]
- 5a.Cerquetti, M., A. Serafino, A. Sebastianelli, and P. Mastrantonio. 2002. Binding of Clostridium difficile to Caco-2 epithelial cell line and to extracellular matrix proteins. FEMS Immunol. Med. Microbiol. 32:211-218. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H., M. E. Herndon, and J. Lawler. 2000. The cell biology of thrombospondin-1. Matrix Biol. 19:597-614. [DOI] [PubMed] [Google Scholar]
- 7.Corthier, G., M. C. Muller, G. W. Elmer, F. Lucas, and F. Dubos Ramare. 1989. Interrelationships between digestive proteolytic activities and production and quantitation of toxins in pseudomembranous colitis induced by Clostridium difficile in gnotobiotic mice. Infect. Immun. 57:3922-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cossart, P., and M. Lecuit. 1998. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J. 17:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.deRuyter, P., O. P. Kuipers, and W. M. deVos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drudy, D., D. P. O'Donoghue, A. Baird, L. Fenelon, and C. O'Farrelly. 2001. Flow cytometric analysis of Clostridium difficile adherence to human intestinal epithelial cells. J. Med. Microbiol. 50:526-534. [DOI] [PubMed] [Google Scholar]
- 11.Eveillard, M., V. Fourel, M. C. Barc, S. Kerneis, M. H. Coconnier, T. Karjalainen, P. Bourlioux, and A. L. Servin. 1993. Identification and characterization of adhesive factors of Clostridium difficile involved in adhesion to human colonic enterocyte-like Caco-2 and mucus-secreting HT29 cells in culture. Mol. Microbiol. 7:371-381. [DOI] [PubMed] [Google Scholar]
- 12.Freeman, J., and M. H. Wilcox. 1999. Antibiotics and Clostridium difficile. Microbes Infect. 1:377-384. [DOI] [PubMed] [Google Scholar]
- 13.George, W. L. 1984. Antimicrobial agent-associated colitis and diarrhea: historical background and clinical aspects. Rev. Infect. Dis. 6(Suppl. 1):S208-S213. [DOI] [PubMed] [Google Scholar]
- 14.Isberg, R. R., and P. Barnes. 2001. Subversion of integrins by enteropathogenic Yersinia. J. Cell Sci. 114:21-28. [DOI] [PubMed] [Google Scholar]
- 15.Karjalainen, T., A. J. Waligora-Dupriet, M. Cerquetti, P. Spigaglia, A. Maggioni, P. Mauri, and P. Mastrantonio. 2001. Molecular and genomic analysis of genes encoding surface-anchored proteins from Clostridium difficile. Infect. Immun. 69:3442-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawata, T., and K. Masuda. 1984. Electron-microscopy of a regular array in the cell-wall of Clostridium difficile. J. Electron Microsc. 33:297. [Google Scholar]
- 17.Kieny, M. P., R. Lathe, and J. P. Lecocq. 1983. New versatile cloning and sequencing vectors based on bacteriophage M13. Gene 26:91-99. [DOI] [PubMed] [Google Scholar]
- 18.Kuipers, O. P., P. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 19.Lawler, J. 2000. The functions of thrombospondin-1 and -2. Curr. Opin. Cell Biol. 12:634-640. [DOI] [PubMed] [Google Scholar]
- 20.Mamo, W., P. Jonsson, J. I. Flock, M. Lindberg, H. P. Muller, T. Wadstrom, and L. Nelson. 1994. Vaccination against Staphylococcus aureus mastitis: immunological response of mice vaccinated with fibronectin-binding protein (FnBP-A) to challenge with S. aureus. Vaccine 12:988-992. [DOI] [PubMed] [Google Scholar]
- 21.McKee, M. L., A. R. Meltoncelsa, R. A. Moxley, D. H. Francis, and A. D. Obrien. 1995. Enterohemorrhagic Escherichia coli O157-H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect. Immun. 63:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson, I. M., J. M. Patti, T. Bremell, M. Hook, and A. Tarkowski. 1998. Vaccination with a recombinant fragment of collagen adhesin provides protection against Staphylococcus aureus-mediated septic death. J. Clin. Investig. 101:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onderdonk, A. B., R. L. Cisneros, and J. G. Bartlett. 1980. Clostridium difficile in gnotobiotic mice. Infect. Immun. 28:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patti, J. M., T. Bremell, D. Krajewska Pietrasik, A. Abdelnour, A. Tarkowski, C. Ryden, and M. Hook. 1994. The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect. Immun. 62:152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pothoulakis, C., and J. T. Lamont. 2001. Microbes and microbial toxins: paradigms for microbial-mucosal interactions. II. The integrated response of the intestine to Clostridium difficile toxins. Am. J. Physiol.-Gastroint. Liver Physiol. 280:G178-G183. [DOI] [PubMed] [Google Scholar]
- 26.Rhem, M. N., E. M. Lech, J. M. Patti, D. McDevitt, M. Hook, D. B. Jones, and K. R. Wilhelmus. 2000. The collagen-binding adhesin is a virulence factor in Staphylococcus aureus keratitis. Infect. Immun. 68:3776-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sara, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schvartz, I., D. Seger, and S. Shaltiel. 1999. Vitronectin. Int. J. Biochem. Cell Biol. 31:539-544. [DOI] [PubMed] [Google Scholar]
- 29.Seddon, S. V., and S. P. Borriello. 1992. Proteolytic activity of Clostridium difficile. J. Med. Microbiol. 36:307-311. [DOI] [PubMed] [Google Scholar]
- 30.Shen, Y., M. Naujokas, M. Park, and K. Ireton. 2000. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103:501-510. [DOI] [PubMed] [Google Scholar]
- 31.Sinha, K., M. Box, G. Lalli, G. Schiavo, H. Schneider, M. Groves, I. G. Siligard, and N. Fairweather. 2000. Analysis of mutants of tetanus toxin HC fragment: ganglioside binding, cell binding and retrograde axonal transport properties. Mol. Microbiol. 37:1041-1050. [DOI] [PubMed] [Google Scholar]
- 32.Takeoka, A., K. Takumi, T. Koga, and T. Kawata. 1991. Purification and characterization of S layer proteins from Clostridium difficile GAI 0714. J. Gen. Microbiol. 137:261-267. [DOI] [PubMed] [Google Scholar]
- 32a.Takumi, K., T. Koga, T. Oka, and Y. Endo. 1991. Self-assembly, adhesion, and chemical properties of tetragonally arrayed S-layer proteins of Clostridium. J. Gen. Appl. Microbiol. 37:455-465. [Google Scholar]
- 33.Tamm, A., A. M. Tarkkanen, T. K. Korhonen, P. Kuusela, P. Toivanen, and M. Skurnik. 1993. Hydrophobic domains affect the collagen-Binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10:995-1011. [DOI] [PubMed] [Google Scholar]
- 34.Tasteyre, A., M. C. Barc, T. Karjalainen, P. Dodson, S. Hyde, P. Bourlioux, and P. Borriello. 2000. A Clostridium difficile gene encoding flagellin. Microbiology 146:957-966. [DOI] [PubMed] [Google Scholar]
- 35.Toba, T., R. Virkola, B. Westerlund, Y. Bjorkman, J. Sillanpaa, T. Vartio, N. Kalkkinen, and T. K. Korhonen. 1995. A collagen-binding S-layer protein in Lactobacillus crispatus. Appl. Environ. Microbiol. 61:2467-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waligora, A. J., C. Hennequin, P. Mullany, P. Bourlioux, A. Collignon, and T. Karjalainen. 2001. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect. Immun. 69:2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wren, B. W. 1992. Molecular characterisation of Clostridium difficile toxins A and B. Rev. Med. Microbiol. 3:21-27. [Google Scholar]