Abstract
Although T-cell responses are known to be critical for effective host defenses against the fungal pathogen Cryptococcus neoformans, the antigens that stimulate protective responses are poorly characterized but are thought to be comprised, at least in part, of mannoproteins. Recently, we created a panel of murine CD4+-T-cell hybridomas that react with C. neoformans antigens. A mannoprotein antigen, MP98, that stimulated one of the hybridomas was purified, and the gene encoding MP98 was cloned. In the present study, the cryptococcal antigen, MP88, that stimulated a second T-cell hybridoma, X5A3, to secrete interleukin-2 was characterized. MP88 was purified from supernatants of glass bead-disrupted C. neoformans by anion-exchange and hydrophobic interaction chromatography. A single band with an apparent molecular mass of 88 kDa was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to partial internal amino acid sequencing. The gene encoding MP88 was cloned and sequenced. MP88 features a C-terminal serine/threonine-rich region, which presumably serves as a site for extensive O glycosylation, followed by a putative glycosylphosphatidylinositol anchor site. A search of C. neoformans genomic databases revealed that MP88 shares this feature with at least 11 other genes, including MP98. The mannoprotein nature of MP88 was established based upon the capacity of (i) the mannoprotein fraction of C. neoformans supernatants to stimulate X5A3 and (ii) mannosylated ligands to competitively inhibit this stimulation. Thus, a second cryptococcal mannoprotein has been identified which stimulates T-cell responses and is a vaccine candidate.
The encapsulated fungus Cryptococcus neoformans is a major cause of morbidity and mortality in individuals with impaired cell-mediated immunity, particularly in those with quantitative or qualitative defects in CD4+-T-cell function (2, 7). In patients with AIDS, cryptococcosis ranks as one of the five most common life-threatening opportunistic infections. In contrast, clinically apparent cryptococcosis is rare in individuals with intact immune systems, with an annual incidence estimated at <0.001%. Meningoencephalitis is the most frequent clinical presentation, although any organ system can be affected, and disseminated disease is common in severely immunocompromised patients. In animal models of cryptococcosis, impairment of either CD4+- or CD8+-T-cell function is deleterious (5, 14, 15). The mechanisms by which T cells contribute to host defenses appear to be multifactorial and include activation of macrophages via production of cytokines, as well as possibly direct antifungal activity (2, 8).
The requirement for T cells to effectively defend against cryptococcosis has led investigators to search for immunoreactive cryptococcal antigens that could serve as vaccine candidates. Murphy isolated a crude culture supernatant, designated C. neoformans culture filtrate antigen (CneF), which stimulated delayed-type hypersensitivity responses and cytokine production in immunized mice (17). Fractionation of CneF and similar preparations revealed that it was the mannoprotein fraction, defined by the ability to adhere to a concanavalin A (ConA) affinity column, that was predominantly responsible for the delayed-type hypersensitivity responses (18). Mannoprotein has also been shown to stimulate lymphoproliferative responses and cytokine production from patients recovered from cryptococcosis (6, 9). Fractionation of mannoprotein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblotting and staining with a biotinylated ConA probe, demonstrates at least five distinct bands (13). Several of the bands negatively stain when silver stained, suggesting heavy glycosylation (10, 13). Further evidence for extensive glycosylation comes from biochemical analysis of unfractionated mannoprotein, which reveals a carbohydrate/protein ratio of greater than 5:1 (13).
T cells obtained from mice immunized with C. neoformans mannoprotein proliferate in vitro when stimulated with mannoprotein in the presence of antigen-presenting cells (APC). When separated by SDS-PAGE, the fraction with an apparent molecular mass of >60 kDa contains the majority of the stimulatory activity (13). Recently, we created a panel of CD4+-T-cell hybridomas by fusing splenic T cells from mice immunized with crude C. neoformans antigens with the thymoma cell line BW5147 (10). The T-cell receptor on each hybridoma recognizes a C. neoformans antigen fragment presented by class II major histocompatibility complex molecules on an APC. In the presence of an APC, the antigen of interest will stimulate the hybridoma to make interleukin-2 (IL-2) or IL-4. By using this system, we reported on one such hybridoma, P1D6, with specificity for a mannoprotein, MP98, with an apparent molecular mass of 98 kDa. In the present study, we describe a second mannoprotein, MP88, which stimulates the T-cell hybridoma X5A3. MP88 was purified to homogeneity, and the gene encoding the protein was cloned and sequenced. MP88 and MP98 share structural features, including signal sequences, serine/threonine-rich C-terminal regions (which likely serve as sites of extensive O-glycosylation), and glycosylphosphatidylinositol anchor (GPI-anchor) motifs.
MATERIALS AND METHODS
Reagents.
All chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise noted. High-pressure liquid chromatography (HPLC)-grade reagents were purchased from Fisher Scientific (Fairlawn, N.J.). Restriction endonucleases were purchased from New England BioLabs, Inc. (Beverly, Mass.). Phosphate-buffered saline (PBS), RPMI 1640, and fetal bovine serum (FBS) were purchased from Life Technologies (Rockville, Md.). Complete media is defined as RPMI 1640 with 10% FBS supplemented with penicillin, streptomycin, ciprofloxacin, amphotericin B, and l-glutamine. All incubations were at 37°C in humidified air supplemented with 5% CO2, except where otherwise noted.
Mice.
Male C57BL/6J mice were obtained from The Jackson Laboratory (Bar Harbor, Maine) and housed under pathogen-free conditions at the Boston University Laboratory Animal Science Center.
Fungi.
Acapsular C. neoformans strain cap67 (ATCC 52817) was used for all experiments unless otherwise indicated. Other cryptococcal strains utilized were serotype D strain B3501 (the encapsulated parent strain of cap67); serotype A strains 6 (ATCC 62066), 145 (ATCC 62070), and H99 (ATCC 208821); serotype B strain 444 (ATCC 32609); and serotype C strain 18 (ATCC 24066). Fleischmann's RapidRise baker's yeast was purchased at Donelan's Supermarket (Wayland, Mass.) and used as the source of Saccharomyces cerevisiae. Strain SC5314 served as the source of Candida albicans.
T-cell hybridomas.
The procedure utilized for generating T-cell hybridomas reactive with C. neoformans antigens was exactly as described previously (10). Briefly, supernatants of glass bead-disrupted cap67 were suspended in the RIBI adjuvant system, and mice were given two intraperitoneal injections on days 0 and 21. Mice were sacrificed on day 28, and splenocytes were isolated. After a 3-day in vitro stimulation with C. neoformans supernatants, T cells were fused with BW5147 TCR-α−β− thymoma cells (a gift of George Deepe, University of Cincinnati School of Medicine) by using 50% polyethylene glycol (molecular mass, 15 kDa; Roche Molecular Biochemicals, Mannheim, Germany). Unfused BW5147 cells were killed by the addition of hypoxanthine-aminopterin-thymidine, and growing hybridomas were identified by inverted microscopy. The hybridoma utilized in the studies, X5A3, was cloned by limiting dilution and expanded in complete media.
To determine which C. neoformans antigen reacted with X5A3, hybridoma cells were incubated for 18 h with irradiated (3,000 rads) splenocytes in the presence of C. neoformans antigens. Supernatants were collected, freeze-thawed, and incubated with the IL-2/IL-4-sensitive CTLL-2 cell line (16). After 24 h, 10 μl of alamarBlue (Biosource International, Camarillo, Calif.) was added to each well. Plates were incubated an additional 18 h, and the A570 to A600 values were determined on a plate reader. The results were then compared with a standard curve generated with known amounts of IL-2. Results were interpreted semiquantitatively. Thus, detectable levels of IL indicated the presence of the antigen of interest. To determine whether X5A3 produced IL-2 or IL-4, proliferation was determined in the presence of neutralizing rat anti-mouse monoclonal antibody to IL-2 (R&D Systems, Minneapolis, Minn.).
Purification of the antigen of interest.
The purification scheme was similar to prior studies (10). cap67 was disrupted with glass beads and ultracentrifuged. Supernatants were collected, passed through a 0.22-μm (pore-size) filter and diluted in 20 mM Tris-HCl buffer. Anion-exchange chromatography was performed with a Mono-Q column (Amersham Pharmacia, Piscataway, N.J.) by fast-performance liquid chromatography by using an ÄKTA-FPLC machine (Amersham Pharmacia) equipped with UV, pH, and conductivity detectors. Buffer A consisted of 20 mM Tris-HCl, whereas buffer B consisted of 1 M NaCl in 20 mM Tris HCl. The sample was applied to the column by using a 50-ml Superloop and then eluted with an increasing gradient of buffer B. Then, 1-ml fractions were collected. Sequential runs at pH 6.0 and 7.0 were performed. Separation on the basis of hydrophobicity was performed by using reversed-phase HPLC (RP-HPLC; 1090 series II; Hewlett-Packard, Burlington, Mass.). Antigens were applied to C18 and C4 columns (catalog numbers 218TP54 and 214TP54, respectively; Vydac, Heperia, Calif.) equilibrated in solvent A (aqueous 0.1% trifluoroacetic acid [TFA]) and eluted with a linear gradient of solvent B (0.09% TFA in acetonitrile) at a flow rate of 1 ml/min. The percentage of buffer B was 0% for the first 5 min and then increased to 40% at 20 min, 60% at 50 min, and 100% at 60 min. Fractions (0.5 ml) were dried by using a speed vacuum apparatus and dissolved in 100 μl of PBS. For both fast-performance liquid chromatography and the RP-HPLC runs, fractions were collected and screened for their capacity to stimulate X5A3 to produce IL-2 by using the CTLL-2 assay described above. The active fraction obtained after the final RP-HPLC run was resolved on a 1-mm-thick 7.5% SDS-PAGE gel, lightly stained with Coomassie brilliant blue to identify the protein of interest, and destained, and the heart of the band was excised. The gel fragment was washed with HPLC-grade 50% acetonitrile-50% water and sent to the Harvard Microchemistry Facility (Cambridge, Mass.) for internal sequencing, as described in Results.
PCR.
PCR primers were designed based upon the results of a TBLASTN search of the Stanford University Technology Center C. neoformans genome project (http://www-sequence.stanford.edu/group/C.neoformans/index.html) and a BLASTN search of the University of Oklahoma's Advanced Center for Genome Technology C. neoformans strain H99 database (http://www.genome.ou.edu/cneo.html). The forward primers included CF0 (5′-ATCTCGGCTAGCTACCTAGT), CF1 (5′-CTCATGGCCGGTGTGGTAT), CF3 (5′-AACGTCAACGCTCAGGTCAC), CF6 (5′-GTCCTTACCGCCGTCCACTT), CF7 (5′-CAACGGCTCCACCTCTACCT), and T7 (5′-GTAATACGACTCGACTCACTATAGGGC). The reverse primers included CR2 (5′-CGGCGGTAAGGACACCATC), CR3 (5′-GATGGGTGAAGTCACCGAAAC), CR5 (5′-CCGACAGAGGTGTAGGTCTT), CR7 (5′-CACATCCATGCAGCAACAAG), and T3 (5′-AATTAACCCTCACTAAAGGG). The primers were custom synthesized by Life Technologies, Inc. (Rockville, Md.). A total of 25 cycles of PCR were run by using Taq polymerase (Promega Corp., Madison, Wis.). Each cycle consisted of 45 s of denaturation at 95°C, 30 s of annealing at 55°C, and 45 s of extension at 72°C. For the first cycle, the denaturing step was extended to 5 min, whereas for the last cycle the extension time was increased to 6 min. PCR amplimers were passed through a purifying column (Promega) and sequenced on an ABI 377-96 automated DNA sequencer (Applied Biosystems, Inc., Foster City, Calif.) at the Boston University Medical Center Molecular Genetics Core Facility. PCR amplimers were labeled with [32P]dCTP (New England Nuclear Life Science Products, Inc., Boston, Mass.) by using the RTS RadPrime DNA labeling system (Life Technologies).
C. neoformans cDNA, genomic DNA ( gDNA), and RNA.
A cDNA library of C. neoformans B3501 constructed in the Uni-Zap XR vector (Stratagene) has been previously described (10). Mass excision of the pBluescript phagemid from the Uni-Zap XR vector was performed according to the manufacturer's instructions (Stratagene). Phagemid DNA was purified by using the Qiagen plasmid Midi-kit (Qiagen, Valencia, Calif.).
Cloning of MP88.
C. neoformans cDNA library was mixed with XL-Blue MRF′ Escherichia coli host cells and plated at 2 × 104 to 5 × 104 phage/plate. After overnight growth at 37°C, plaques were transferred to nylon membranes (Amersham International plc, Amersham, United Kingdom). The lifted membrane was dried and cross-linked. Prehybridization and hybridization with the 32P-labeled amplimer were performed at 65°C, and positive colonies were identified by autoradiography. Confirmation of positive colonies was performed with a second round of screening and by PCR. The pBluescript SK plasmid containing the insert of interest was excised from positive phage by using the ExAssist helper phage and transformed into E.coli SOLR cells according to the protocol supplied by Stratagene. Plasmid DNA was isolated from transformed SOLR cells by using Qiagen Minikit and was digested with the restriction enzymes BseRI and NaeI to check the fragment size. Sequencing of the plasmid was performed at a minimum of twofold redundancy with the primers T7, CF3, CF6, CF7, T3, CR2, CR3, CR5, and CR7.
To clone MP88 gDNA, total gDNA was extracted from glass bead-disrupted C. neoformans B3501 by phenol-chloroform extraction as described previously (2). The PCR primers CF0, CF1, CF3, CF6, CF7, CR2, CR3, CR5, and CR7 were used to amplify the gDNA. The amplimers were purified and sequenced.
Secreted C. neoformans antigens.
For the experiments described in Fig. 4, crude mannoprotein and flowthrough fractions of cap67 culture supernatants were collected exactly as described previously (13). Briefly, filter-sterilized C. neoformans culture supernatants were dialyzed against PBS and concentrated by tangential flow filtration by using a 10-kDa cassette (Millipore, Bedford, Mass.). This material, referred to as the “crude” fraction, was separated by ConA affinity chromatography into a nonbinding “flowthrough” fraction and a “mannoprotein” fraction eluted with 0.2 M methyl α-d-mannopyranoside. Flowthrough and eluted fractions were boiled for 5 min to inactivate any ConA that might have leached from the column (3) and tested for the capacity to stimulate IL-2 production from the hybridomas. After reconstitution with PBS, the protein concentration was assessed by using the bicinchoninic acid assay (Pierce, Rockford, Ill.), whereas the total carbohydrate was measured by using the phenol-sulfuric acid assay (4). Chemical O-linked deglycosylation (β-elimination) was performed as in our previous studies (13) by incubation of the mannoprotein fraction in 0.1 M NaOH for 24 h at 37°C. NaOH was neutralized with glacial acetic acid. Mock β-eliminated protein was treated in an identical manner except that it was neutralized immediately after the addition of NaOH.
FIG. 4.
Gene structure and encoded domains of MP88. (A) Combined sequences of genomic DNA, cDNA, and the deduced protein of MP88. The long horizontal arrows depict the span of genomic DNA that was sequenced. The shaded amino acids indicate peptide sequences that were directly determined for purified MP88 protein. A vertical arrow demarks the predicted cleavage of the signal peptide. Double underlined amino acids span the Ser/Thr-rich region, which is also schematically depicted in panel B.
Accession numbers.
The cDNA and amino acid sequence of MP88 was deposited in GenBank under accession number AF395487. The gDNA was deposited in GenBank under accession number AF480842.
RESULTS
Purification of the antigen that stimulates X5A3.
In previous studies, a panel of T-cell hybridomas reactive with C. neoformans antigens was created (10). The present study focused on determining the immunoreactive antigen that stimulated one hybridoma, X5A3, from this panel. By using neutralizing monoclonal antibodies, it was determined that X5A3 secretes IL-2. Expression of CD4 and the T-cell receptor was demonstrated by flow cytometry by using fluorescence-tagged antibodies (data not shown). X5A3 secreted IL-2 when stimulated with splenocytes pulsed with a 10-μg/ml portion of glass bead-disrupted C. neoformans strains cap67, B3501, 6, 145, H99, 444, and 18, but not when stimulated with S. cerevisiae or C. albicans (data not shown).
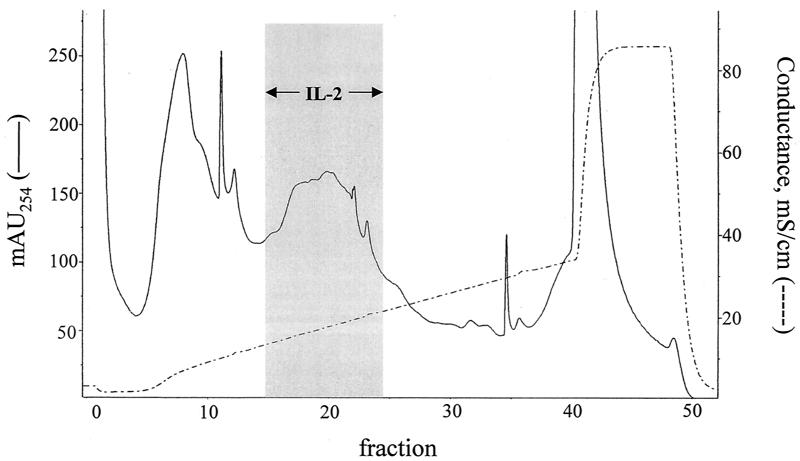
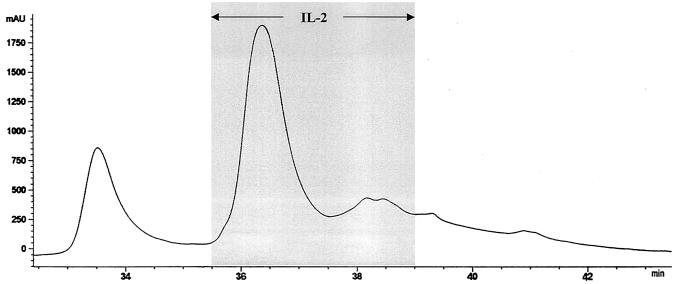
Supernatants from glass-bead disrupted C. neoformans cap67 were applied to an anion-exchange column at pH 6.0 and eluted with a sodium chloride gradient (Fig. 1). As previously demonstrated (10), three broad peaks were obtained. Fractions were incubated with X5A3 in the presence of APC. Based upon the capacity to stimulate IL-2 production from X5A3, the antigen of interest was located in fractions 15 through 23. Fractions 18, 19, and 20, which contained peak activity, were pooled and again placed on the anion-exchange column, but this time at pH 7.0. Fractions 20, 21, and 22, representing the three most active fractions (data not shown), were pooled and then resolved by RP-HPLC with a C4 column. At least 15 distinct peaks were obtained, including one large peak that contained the antigen of interest (data not shown). As a final polishing step, this peak was applied to a C18 column and eluted with a gradient of acetonitrile in TFA. Activity was located in the large peak, although the smaller two peaks, which eluted immediately after the first peak, also had activity (Fig. 2).
FIG. 1.
Separation of cryptococcal antigens by anion-exchange chromatography. Supernatants from glass bead-disrupted C. neoformans were separated on a Mono-Q column at pH 6 by using an NaCl gradient. One-milliliter fractions were collected and tested for their capacity to stimulate hybridoma X5A3 to produce IL-2 in the presence of APC. The gray box indicates the fractions that stimulated IL-2 production. The chromatogram is representative of 10.
FIG. 2.
Purification of the cryptococcal protein that stimulates X5A3 by RP-HPLC. The active fractions from an C18 column were applied to a C4 column and eluted with a gradient of acetonitrile in 0.1% TFA (buffer B). Bioactivity (as measured by the capacity to stimulate X5A3 to produce IL-2 in the presence of APC) was associated with the large peak seen at the 36-min elution time point. The two smaller peaks, eluted immediately after the large peak, also had bioactivity. The results are representative of four column runs.
Internal sequencing of the antigen of interest.
The fractions containing the large peak with the antigen of interest isolated after elution on the C4 column were combined, concentrated, and subjected to SDS-7.5% PAGE. A single band with an apparent molecular mass of ca. 88 kDa was seen after Coomassie blue staining. The antigen of interest was named MP88 on the basis of its predicted mannosylation (see below) and apparent molecular size. The heart of the band was excised, the protein was eluted from the gel, and the sample was sent to the Harvard Microchemistry Facility. There, it was subjected to trypsin digestion, and the fragments were resolved by HPLC. One peak was sequenced by Edman degradation, yielding the amino acid sequence VIPDGVLTAVHFVK. The protein purification procedure was repeated, and the gel slice was again sent to the Harvard Microchemistry Facility. Trypsin-digested samples this time were analyzed by microcapillary RP-HPLC nanoelectrospray tandem mass spectrometry on a Finnigan LCQ quadrupole ion trap mass spectrometer. A 30-amino-acid (aa) sequence, TPIYWQIQGFGDFTHLNIQDGDEGGELDPH, was obtained. In addition, a 14-aa sequence, VIPDGVLTAVHFVK, again was obtained, as well as the sequences MAYDQFCLR and TEAEEVAWCVQPR. Searches of the Stanford University Technology Center C. neoformans genome project and the University of Oklahoma's Advanced Center for Genome Technology C. neoformans strain H99 and JEC21 databases revealed expressed sequence tags (ESTs) with 100% predicted identity for all of the amino acid sequences.
Cloning and sequencing of mp88.
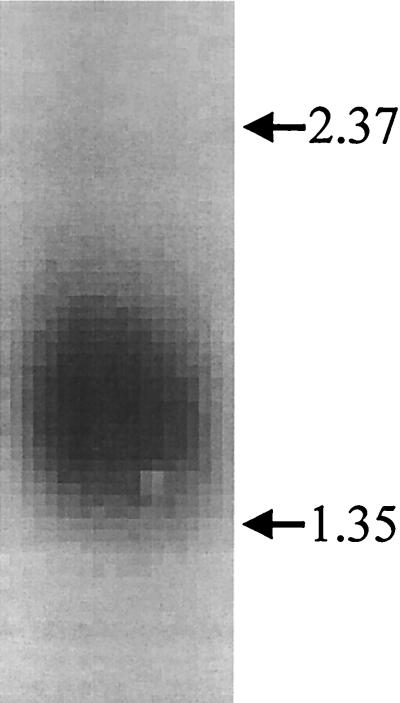
Primers were designed based upon the DNA sequences obtained from the two C. neoformans genome databases. Primers CF3 and CR3 were used to amplify cDNA that was mass excised from the C. neoformans phagemid library. A PCR amplimer of the expected size of 310 bp was obtained, and its identity was confirmed by DNA sequencing. The amplimer was 32P labeled. Northern blotting of C. neoformans RNA revealed a single band migrating at 1.5 kb (Fig. 3). The probe was then used to screen the C. neoformans cDNA library. After two rounds of screening, 15 positive plaques were obtained. Phagemids from three of these positive plaques were excised and transformed into E. coli, and the inserts were sequenced with the primer sets described in Materials and Methods. The cDNA was sequenced in both directions with a minimum of twofold redundancy (Fig. 4).
FIG. 3.
Northern blot demonstrating MP88 gene expression. RNA extracted from C. neoformans was probed for MP88. The arrows indicate the locations of the 2.37- and 1.35-kb RNA molecular weight markers.
The primer sets described in Materials and Methods were then used to sequence gDNA from C. neoformans B3501. The sequence obtained was identical to the gDNA sequence data posted on the Stanford University Technology Center C. neoformans genome project database, with the exception of a single base pair difference in intron 4. Figure 4 shows the cDNA/gDNA sequence of the full-length insert and the predicted 388 amino acid sequence of MP88. The open reading frame of MP88 contains 1,167 bp. There is a 243-bp intron in the 5′ untranslated region of the gene, as well as four introns ranging from 50 to 74 bp in the open reading frame. The protein contains a signal sequence at the N-terminal end. There are seven potential sites for N glycosylation containing the Asn-X-Ser/Thr glycosylation sequon. There is a serine/threonine-rich (30 of 56 aa [54%]) region toward the C-terminal which could serve as a site for O-linked glycosylation. Immediately downstream of the serine/threonine rich region is a hydrophobic tail with a GPI-anchor site predicted by two web-based software programs (Research Institute of Molecular Pathology, http://mendel.imp.univie.ac.at/; Detection or prediction of GPI cleavage site, http://129.194.18 6.123/GPI-anchor/index_en.html).
X5A3 is stimulated by C. neoformans mannoproteins.
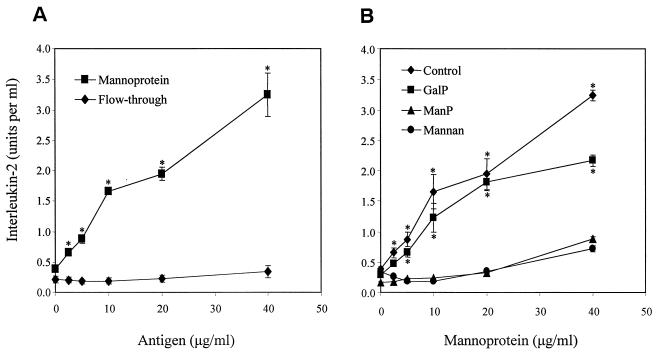
The presence of numerous sites for O-linked and N-linked glycosylation suggests that MP88, like MP98, exhibits terminal mannosylation. To support this proposition experimentally, culture supernatants from cap67 were collected and filter sterilized. This crude material was then separated into MP and flowthrough fractions based upon its capacity to bind to ConA (13). X5A3 was stimulated by the MP fraction but not the flowthrough fraction (Fig. 5, left panel).
FIG. 5.
Effect of C. neoformans supernatant fractions and mannosylated inhibitors on activation of the T-cell hybridoma cell line X5A3. (A) Irradiated naive splenocytes (2 × 105/well) and X5A3 hybridoma cells (105/well) were incubated for 24 h in the presence of the indicated protein concentrations of C. neoformans mannoprotein and flowthrough culture supernatant fractions. (B) The contribution of mannose receptors to the X5A3 response to MP was assessed by using the following inhibitors: no inhibitor (control), 50 mM methyl α-d-galactopyranoside (GalP), 50 mM methyl α-d-mannopyranoside (ManP), or 1 mg of S. cerevisiae mannans/ml (mannan). For both panels, supernatants were collected, and the activation of X5A3 was measured by assaying for IL-2 production by using the CTLL-2 bioassay, as described in Materials and Methods. Data are the means ± the standard error of the means of three representative independent experiments, each performed in duplicate or triplicate. (A: *, P < 0.03, by Student t test, comparing MP versus FT; B: *, P < 0.03 comparing control or GalP versus ManP or mannan).
Previously, we demonstrated that blocking mannose receptors on APC profoundly inhibited the capacity of these cells to take up and present C. neoformans mannoprotein to T cells (13). To examine whether this is also the case with the presentation of MP88 to X5A3, mannose receptors on irradiated splenocytes were blocked by pretreatment with the ligands mannan (derived from S. cerevisiae) and methyl α-d-mannopyranoside. Splenocytes were then exposed to MP in the presence of X5A3, and IL-2 production by the hybridoma cells was determined. Both mannan and methyl α-d-mannopyranoside inhibited stimulated IL-2 production. Methyl α-d-galactopyranoside, which served as an osmotic control, did not significantly inhibit IL-2 secretion (Fig. 5, right panel).
The final experiments examined the effect of removal of O linkages by β-elimination on the capacity of mannoproteins to stimulate X5A3. X5A3 cells failed to make significant amounts of IL-2 after incubation with irradiated splenocytes and up to 47 μg of β-eliminated mannoprotein/ml. In contrast, mock β-eliminated mannoprotein stimulated X5A3 to secrete IL-2 at concentrations as low as 6 μg/ml (data not shown).
DISCUSSION
The data presented here describe the purification to homogeneity and cloning of a cryptococcal glycoprotein, MP88, that stimulates T-cell responses in immunized mice. The genes encoding three other C. neoformans antigens with T-cell-stimulating capacity have been cloned. One gene, DHA1, encodes for an estimated 20-kDa glycoprotein of unknown function that elicits delayed-type hypersensitivity reactions in mice immunized against C. neoformans (12). The second gene, MP98, encodes for a T-cell-stimulating mannoprotein with an apparent molecular mass of 98 kDa (10). MP98 exhibits strong homology to chitin deacetylases. Recently, another putative deacetylase with T-cell-stimulating properties was identified (1).
Several pieces of evidence, taken together, strongly suggest that MP88, like MP98, is a mannoprotein. First, the mannoprotein fraction, as defined by ConA binding, stimulated X5A3. In contrast, the flowthrough fraction did not. Second, blockade of mannose receptors with mannans or methyl α-d-mannopyranoside inhibited the capacity of irradiated splenocytes to present MP88 to X5A3. A similar effect was seen after β-elimination of mannoprotein. Third, analysis of the gene structure of MP88 reveals numerous potential sites for O-linked and N-linked glycosylation.
MP98 and MP88 share several structural features, including a signal sequence, a serine/threonine-rich C-terminal region, and a putative GPI-anchor. Whereas MP98 has a chitin deacetylase domain (10), MP88 has sequence similarity to only one protein in GenBank, Mal f 8 (GenBank accession no. AJ011958), which is an allergen secreted by the fungal dermatophyte Malassezia furfur (21). The region of similarity (45%) is between the signal peptide and Ser/Thr-rich region of MP88 (aa 55 to 207). Although there are no discernible domains in this region that would suggest the function of MP88 or Mal f 8, searches of the Stanford Genomic DNA (update 10/15/01) and Oklahoma cDNA (release 11/20/01) databases identified three other C. neoformans proteins with the same homologous region. Each C. neoformans protein sequence was identified by TBLASTN with the full-length MP88 protein sequence as query. The three deduced proteins, which we have called MP-A (401 aa), MP-B (444 aa), and MP-C (421 aa), are predicted to be about the same size as MP88 (Table 1). They also are predicted to each have an N-terminal signal peptide and C-terminal 100-aa region that are initially Ser/Thr-rich and then terminate with GPI-anchor motifs. Pairwise alignments by the NCBI BLAST 2 Sequences Program of full-length sequences of MP88 with each of the other three gave identities ranging from 31 to 58% and similarities ranging from 45 to 70%. All three, likewise, show similarity with Mal f 8.
TABLE 1.
Cryptococcus mannoproteins with Ser/Thr-rich and GPI-anchor domains similar to the C-terminal 88 aa of mp88
| Protein | Size (aa) | cDNA contigb
|
Ser/Thr-rich regiond (%Ser, Thr) | Other conserved domains, patterns, and commentse | |
|---|---|---|---|---|---|
| JEC21 | H99 | ||||
| MP88 | 388 | 1623 | ESTs | 286-361 (46) | None |
| MP-A | 401 | 1283 | ESTs | 298-377 (55) | None, 58% identity with MP88 (aa 23-284) |
| MP-B | 444 | None | None | 324-410 (61) | None, 34% identity with MP88 (aa 45-287) |
| MP-C | 421 | None | EST | 276-398 (35) | None, 31% identity with MP88 (aa 50-274) |
| MP98 | 458a | 1494c | ESTs | 380-432 (64) | Chitin deacetylase catalytic |
| MP-D | 309 | None | 428 | 216-281 (53) | Phosphatidylethanolamine-binding protein; Antifreeze-Type I |
| MP-E | 221 | 1628 | 434 | 123-204 (61) | Antifreeze-Type I |
| MP-F | 213 | 1151 | None | 52-176 (40) | Antifreeze-Type I |
| MP-G | 226 | 223c | 373 | 141-200 (63) | Antifreeze-Type I |
| MP-H | 251 | 1126 | 426 | 93-227 (47) | Antifreeze-Type I, 57% identity with MP-G (aa 19-107) |
| MP-I | 197 | 1579 | 438 | 96-171 (59) | Antifreeze-Type I, 46% identity with MP-G (aa 1-103) |
| MP-J | 197 | 1570 | ESTs | 106-170 (72) | Antifreeze-Type I, 57% identity with MP-G (aa 9-103) |
Based on GenBank accession no. AF361369.
Contig numbers are from data release 11/20/01 for strain JEC21 and data release 12/3/99 for strain H99 by the Oklahoma cDNA Sequencing Project (http://www.genome.ou.edu/cneo.html). Where contigs have not been assembled, cDNA may have been partially sequenced and listed as “ESTs” or else not sequenced by the data release date (indicated by “none”).
Sequence is not full length.
Ser/Thr-rich regions were identified as Ser-rich by InterProScan (http://www.ebi.ac.uk/interpro/scan.html) and reevaluated to include flanking Thrs. Each region is indicated by amino acid positions; the percent amino acids that are Ser or Thr in a defined region is given in parentheses.
Domains and patterns were identified by InterProScan. The percent identities were the results of BLAST 2 Sequence comparisons (National Center for Biotechnology Information).
C. neoformans mannoproteins that have in common a Ser/Thr-rich region, followed by a putative GPI-anchor motif, as identified for MP88 and its homologs were also identified in the Oklahoma databases of cDNA sequences. A sequence composed of the last 88 aa of the MP88 protein was used to query the B3501/JEC21 and H99 databases by TBLASTN. The most significant similarities found were with cDNAs which, when translated, had both motifs of the query. Sequences with only a Ser/Thr-rich region showed less similarity and were not analyzed further.
In addition to identifying MP98 (10) in the search, several new sequences that encode mannoproteins were found. DNA sequences that had been assembled into contigs in the Oklahoma databases were downloaded and scrutinized individually. The longest open reading frame encoded by each contig was found to have a Ser/Thr-rich region and GPI-anchor motifs at the C terminus. The deduced protein sequence of each cDNA was also predicted to have a signal peptide at the N terminus by the SignalP V2.0 program (http://www.cbs.dtu.dk/services/SignalP-2.0/). A summary of the features of each new MP is given in Table 1.
By BLASTP searches of GenBank, MP-D had the greatest similarity in a search of GenBank with the antigenic glycoprotein OV-16 of the parasite Onchocerca volvulus (GenBank accession #P31729 [11]). As with the MP88 family of proteins identified above, wherein a C. neoformans mannoprotein shows sequence similarity to another antigenic protein from a different species, the region of sequence similarity of MP-D is between the signal peptide and Ser/Thr-rich domain. Of the C. neoformans mannoproteins that have been identified, MP98 was found to have a polysaccharide (chitin) deacetylase catalytic domain (Pfam 01522), whereas MP-D has a phosphatidylethanolamine-binding domain (Pfam 01161). Several of the mannoproteins (Table 1) were also predicted by InterProScan to have an “Antifreeze-Type I” motif, which consists of three blocks 10 to 15 aa in length (22) that help protect the organisms from freezing. It will be interesting to determine whether C. neoformans mannoproteins with this motif serve a similar role in environmental protection.
Like MP98, MP88 was identified based upon its capacity to stimulate a single T-cell hybridomas. One potential pitfall with this approach is that it is unknown what percentage of T cells from immunized mice respond to MP88. We have previously shown that primary T cells from the lymph nodes of mice immunized with C. neoformans mannoproteins respond predominantly to the >60-kDa mannoprotein fraction (13). It may be no coincidence that both MP88 and MP98 have serine/threonine-rich C-terminal regions. Extensive terminal mannosylation of these regions should promote avid recognition by mannose receptors on APC (13).
Final proof that MP88 is indeed the antigen that stimulates X5A3 to produce IL-2 will come once MP88 is expressed recombinantly. Nevertheless, several lines of evidence, taken together, strongly point to MP88 as the correct immunoreactive glycoprotein. First, after two rounds of anion-exchange chromatography, followed by two RP-HPLC column runs, the antigen that stimulated X5A3 eluted as a single peak. This peak was further resolved by SDS-PAGE before sequencing was performed. Second, MP88 was independently purified and sequenced twice. Both times, the obtained sequences were from the same protein. Third, based on its affinity for ConA, the immunoreactive antigen is likely a mannoprotein. As noted above, MP88 has features typically found in mannoproteins.
Cryptococcal mannoproteins account for a large percentage of the secreted and cell-associated material from C. neoformans and are thus likely to be encountered frequently during the course of a cryptococcal infection (13). In preliminary experiments, C57BL/6 mice immunized with cryptococcal mannoproteins are partially protected from a subsequent intravenous challenge with live C. neoformans (M. K. Mansour and S. M. Levitz, unpublished data). Efforts are currently under way to express recombinant mannoproteins, including MP88, in yeast vectors in sufficient quantities so that the individual protective components of the mannoprotein fraction can be determined. It is likely that some mannoproteins will be immunodominant, whereas others will elicit a poor T-cell response. Moreover, even among antigens that elicit a strong T-cell response, only some may be able to elicit protective responses. Support for this concept comes from studies demonstrating that activated T cells are elicited after immunization of mice with either a cryptococcal culture filtrate or heat-killed C. neoformans (19, 20). However, protection (albeit partial) to subsequent cryptococcal challenge is only seen in the mice vaccinated with the culture filtrate.
Acknowledgments
This work was supported by National Institutes of Health grants RO1 AI37532, RO1 AI25780, and T32 AI07309. S.M.L. is a recipient of a Burroughs-Wellcome Fund Scholar Award in Pathogenic Mycology.
We thank Xiulin Liu for technical assistance in making the hybridoma. We also thank George Deepe for helpful discussions.
Editor: T. R. Kozel
REFERENCES
- 1.Biondo, C., C. Beninati, D. Delfino, et al. 2002. Identification and cloning of a cryptococcal deacetylase that produces protective immune responses. Infect. Immun. 70:2383-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, D.C.
- 3.Chaka, W., A. F. Verheul, V. V. Vaishnav, et al. 1997. Cryptococcus neoformans and cryptococcal glucuronoxylomannan, galactoxylomannan, and mannoprotein induce different levels of tumor necrosis factor alpha in human peripheral blood mononuclear cells. Infect. Immun. 65:272-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 5.Hill, J. O. 1992. CD4+ T cells cause multinucleated giant cells to form around Cryptococcus neoformans and confine the yeast within the primary site of infection in the respiratory tract. J. Exp. Med. 175:1685-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoy, J. F., J. W. Murphy, and G. G. Miller. 1989. T-cell response to soluble cryptococcal antigens after recovery from cryptococcal infection. J. Infect. Dis. 159:116-119. [DOI] [PubMed] [Google Scholar]
- 7.Levitz, S. M. 1991. The ecology of Cryptococcus neoformans and the epidemiology of cryptococcosis. Rev. Infect. Dis. 13:1163-1169. [DOI] [PubMed] [Google Scholar]
- 8.Levitz, S. M., H. L. Mathews, and J. W. Murphy. 1995. Direct antimicrobial activity of T cells. Immunol. Today 16:387-391. [DOI] [PubMed] [Google Scholar]
- 9.Levitz, S. M., and E. A. North. 1997. Lymphoproliferation and cytokines profiles in human peripheral blood mononuclear cells stimulated by Cryptococcus neoformans. J. Med. Vet. Mycol. 35:229-236. [DOI] [PubMed] [Google Scholar]
- 10.Levitz, S. M., S. Nong, M. K. Mansour, C. Huang, and C. A. Specht. 2001. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T-cell responses to Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 98:10422-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lobos, E., M. Altmann, G. Mengod, N. Weiss, W. Rudin, and M. Karam. 1990. Identification of an Onchocerca volvulus cDNA encoding a low-molecular-weight antigen uniquely recognized by onchocerciasis patient sera. Mol. Biochem. Parasitol. 39:135-145. [DOI] [PubMed] [Google Scholar]
- 12.Mandel, M. A., G. G. Grace, K. I. Orsborn, et al. 2000. The Cryptococcus neoformans gene DHA1 encodes an antigen that elicits a delayed-type hypersensitivity reaction in immune mice. Infect. Immun. 68:6196-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansour, M. K., L. S. Schlesinger, and S. M. Levitz. 2002. Optimal T-cell responses to Cryptococcus neoformans mannoprotein are dependent on recognition of conjugated carbohydrates by mannose receptors. J. Immunol. 168:2872-2879. [DOI] [PubMed] [Google Scholar]
- 14.Mody, C. H., M. F. Lipscomb, N. E. Street, and G. B. Toews. 1990. Depletion of CD4+ (L3T4+) lymphocytes in vivo impairs murine host defense to Cryptococcus neoformans. J. Immunol. 144:1472-1477. [PubMed] [Google Scholar]
- 15.Mody, C. H., G. H. Chen, C. Jackson, J. L. Curtis, and G. B. Toews. 1994. In vivo depletion of murine CD8-positive T cells impairs survival during infection with a highly virulent strain of Cryptococcus neoformans. Mycopathologia 125:7-17. [DOI] [PubMed] [Google Scholar]
- 16.Mosmann, T. R., H. Cherwinski, M. W. Bond, M. A. Giedlin, and R. L. Coffman. 1986. Two types of murine helper T-cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 136:2348-2357. [PubMed] [Google Scholar]
- 17.Murphy, J. W. 1988. Influence of cryptococcal antigens on cell-mediated immunity. Rev. Infect. Dis. 10:S432-S435. [DOI] [PubMed] [Google Scholar]
- 18.Murphy, J. W., R. L. Mosley, R. Cherniak, G. H. Reyes, T. R. Kozel, and E. Reiss. 1988. Serological, electrophoretic, and biological properties of Cryptococcus neoformans antigens. Infect. Immun. 56:424-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy, J. W., F. Schafer, A. Casadevall, and A. Adesina. 1998. Antigen-induced protective and nonprotective cell-mediated immune components against Cryptococcus neoformans. Infect. Immun. 66:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichols, K. L., S. K. Bauman, F. B. Schafer, and J. W. Murphy. 2002. Differences in components at delayed-type hypersensitivity reaction sites in mice immunized with either a protective or a nonprotective immunogen of Cryptococcus neoformans. Infect. Immun. 70:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasool, O., A. Zargari, J. Almqvist, H. Eshaghi, P. Whitley, and A. Scheynius. 2000. Cloning, characterization, and expression of complete coding sequences of three IgE binding Malassezia furfur allergens, Mal f 7, Mal f 8, and Mal f 9. Eur. J. Biochem. 267:4355-4361. [DOI] [PubMed] [Google Scholar]
- 22.Wen, D., and R. A. Laursen. 1993. Structure-function relationships in an antifreeze polypeptide: the role of charged amino acids. J. Biol. Chem. 268:16396-16400. [PubMed] [Google Scholar]