Abstract
The expression of two Enterococcus faecalis extracellular virulence-related proteins, gelatinase (GelE) and serine protease (SprE), has been shown to be positively regulated by the fsr quorum-sensing system. We recently developed a novel system for studying E. faecalis pathogenicity that involves killing of the nematode worm Caenorhabditis elegans and showed that an E. faecalis fsrB mutant (strain TX5266) exhibited attenuated killing. We explore here the role of the fsr/gelE-sprE locus in pathogenicity by comparing results obtained in the nematode system with a mouse peritonitis model of E. faecalis infection. Insertion mutants of fsrA (TX5240) and fsrC (TX5242), like fsrB (TX5266), were attenuated in their ability to kill C. elegans. A deletion mutant of gelE (TX5264) and an insertion mutant of sprE (TX5243) were also attenuated in C. elegans killing, although to a lesser extent than the fsr mutants. Complementation of fsrB (TX5266) with a 6-kb fragment containing the entire fsr locus restored virulence in both the nematode and the mouse peritonitis models. The fsr mutants were not impaired in their ability to colonize the nematode intestine. These data show that extracellular proteases and the quorum-sensing fsr system are important for E. faecalis virulence in two highly divergent hosts: nematodes and mice.
Enterococci are gram-positive bacteria that are normal inhabitants of the alimentary tract of humans and other animals. They have been recognized as a cause of infective endocarditis for more than a century (17) and have gained prominence over the last two decades as being among the most common pathogens found in hospital-acquired infections, including urinary tract infections, bloodstream infections, and surgical-site infections (27). The increasing importance of enterococci as nosocomial pathogens can be attributed in part to intrinsic and acquired antibiotic resistance (17, 26). Treatment of multidrug-resistant enterococcal infections poses a significant challenge to clinicians (4, 8), and the potential of these organisms to serve as a reservoir for antibiotic resistance genes is of great concern (6, 20, 21). Despite increasing recognition of the clinical importance of enterococcal infections, their pathogenic mechanisms are not well understood (11).
We have recently developed a novel model host-pathogen system for Enterococcus faecalis by using the nematode Caenorhabditis elegans (7). Adult worms feeding on lawns of E. faecalis die over the course of several days in a process that has the hallmarks of an active infection rather than an intoxication; that is, live bacteria colonize and proliferate within the intestine of adult C. elegans, causing a persistent and deadly infection. Cytolysin, a well-studied enterococcal virulence factor important in mammalian pathogenesis (recently reviewed by Haas and Gilmore [10]), increases the rate of nematode killing.
E. faecalis gelatinase (GelE) and serine protease (SprE) are two additional putative virulence factors thought to play a role in systemic disease in mammalian hosts (5, 9, 23, 28). GelE is a secreted 30-kDa metalloprotease that shares homology with Staphylococcus aureus aureolysin and Pseudomonas aeruginosa elastase (29). Insertion disruption of gelE significantly attenuates virulence in a mouse peritonitis model (28). The serine protease gene sprE, which lies immediately downstream of and is cotranscribed with gelE, encodes a secreted 26-kDa serine protease that shares homology with S. aureus V8 protease (22, 23). Insertion disruption of sprE also attenuates virulence in the mouse peritonitis model (23). Transcription of the gelE-sprE operon is positively regulated in a growth-phase-dependent fashion by the fsr locus, which shares many similarities with the well-studied S. aureus agr regulatory locus (23). The fsr locus is composed of three regulatory genes—fsrA, fsrB, and fsrC—located upstream of the gelE-sprE operon (23). Based on sequence homology with AgrA and AgrC, FsrA and FsrC likely constitute a classical two-component system, in which FsrC acts as a histidine kinase sensor and FsrA acts as a response regulator. Moreover, the predicted FsrB protein shares homology with AgrB (23), a membrane-bound protein thought to be involved in the production of active, thiolactone-containing, signaling peptide from the AgrD prepheromone (13). FsrB has recently been shown to have a carboxy-terminal extension of approximately 50 amino acids compared to AgrB, and this extension is processed into a lactone-containing, 11-residue peptide pheromone, termed gelatinase biosynthesis-activating pheromone (19, 22).
Previously, we demonstrated that an fsrB deletion mutant (TX5266) was attenuated in both the C. elegans and the mouse peritonitis model systems (7). In the present study, we further explore the role of fsrB, as well as that of fsrA, fsrC, gelatinase, and serine protease, in E. faecalis-mediated nematode killing and compare these findings with those obtained from a mouse peritonitis model.
MATERIALS AND METHODS
Organisms, culture conditions, and assays.
The bacterial strains and plasmids used in the present experiments are listed in Table 1. Enterococcus strains were grown by using brain heart infusion (Difco Laboratories, Detroit, Mich.) broth and agar at 37°C, supplemented, as appropriate, with rifampin (100 μg/ml), kanamycin (2,000 μg/ml), or erythromycin (10 μg/ml). Enterococcus lawns for C. elegans killing and intestinal colonization assays were prepared as previously described (7), with the following modification: ciprofloxacin (100 ng/ml) was used as a selective antibiotic against Escherichia coli strain OP50, the normal food source for C. elegans. C. elegans wild-type strain Bristol N2 was maintained at 15°C on nematode growth medium agar plates spotted with E. coli OP50 (2, 14). C. elegans killing assays (7), using L4 hermaphrodite nematodes, and the mouse peritonitis model (23), with groups of six outbred ICR female mice for each dose (mean weight of 25 g; Harlan Sprague-Dawley, Houston, Tex.), were carried out as previously described. Nematode alimentary tracts were examined by Nomarski differential interference contrast microscopy by standard techniques (30).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| OG1RF | Wild-type E. faecalis strain; Gel+ Spr+ Rifr Fusr | 18 |
| TX5128 | OG1RF gelE transposon mini-γδ insertion mutant; Gel− Spr− Rifr Fusr Kanr | 28 |
| TX5240 | OG1RF fsrA insertional mutant; Gel− Spr− Rifr Fusr Kanr | 23 |
| TX5242 | OG1RF fsrC insertional mutant; Gel− Spr− Rifr Fusr Kanr | 23 |
| TX5243 | OG1RF sprE insertional mutant; Gel+ Spr− Rifr Fusr Kanr | 23 |
| TX5264 | OG1RF gelE in-frame deletion mutant (bp 252 to 1490), Gel− Spr+ Rifr Fusr | This study |
| TX5266 | OG1RF fsrB in-frame deletion mutant (bp 79 to 684), Gel− Spr− Rifr Fusr | 29, 22 |
| TX5266.01 | TX5266 harboring plasmid pTEX5249; Gel+ Spr+ Rifr Fusr Emr | This study |
| TX5266.OS | TX5266 harboring plasmid pAT18; Gel− Spr− Rifr Fusr Emr | This study |
| E007 | E. faecium clinical isolate | 7 |
| Plasmids | ||
| pAT18 | E. coli-E. faecalis shuttle vector; Emr | 33 |
| pTEX4577 | E. faecalis suicide vector derived from pBluescript SK(−); Kanr | 28 |
| pTEX5249 | pAT18 containing 6-kb PstI/BglII fragment encoding fsrA, fsrB, and fsrC; Emr | 23 |
Abbreviations: Rif, rifampin; Fus, fusidic acid; Kan, kanamycin; Em, erythromycin; Gel, gelatinase; Spr, serine protease; r, resistance.
Deletion mutagenesis.
To make a deletion in gelE, the flanking regions of gelE were amplified by PCR with the primer pairs (i) GDF1 (AAA GAG CTC CTA AAA GTG ATT GTT GAT GTG C, from bp 639 to 618 upstream of the start codon of gelE) and GDR1 (CCG AAT TCA TCA ACA GTA ACG CCT TCC G, from bp 258 to 239 inside gelE) and (ii) GDF2 (AAG AAT TCA TTC AGG TAA ACC AAC CAA GTG, from bp 45 to 24 before the gelE stop codon) and GDR2 (TTG GTA CCG ATT ATT TGC CTT CTT TTC AGC, from bp 1047 to 1026 after the gelE stop codon) for the 5′ and 3′ regions, respectively (linker sequences are underlined). The amplicons were ligated together by the linker (EcoRI) designed in the two inner primers, GDR1 and GDF2, and inserted into the mutagenesis vector pTEX4577 by using the outer linkers (SacI and KpnI) designed in the two outer primers, GDF1 and GDR2. The resulting construct was then transformed into OG1RF by electroporation (15), and single-crossover mutants were selected on brain heart infusion agar plates supplemented with kanamycin. Single-crossover mutants were still gelatinase positive because of the duplication of the flanking regions. Since this single-crossover event creates duplicated fragments of the regions flanking the targeted gelE gene, subsequent recombination between these duplicated fragments will lead to the loss of the mutagenesis vector and one copy of the duplicated flanking sequence, giving rise to either the restored wild-type strain or the deletion mutant. After the loss of the mutagenesis vector, both the wild-type and the deletion mutant strains become kanamycin sensitive. To identify the gelE deletion mutant, we first plated the culture of the single-crossover mutants of gelE grown overnight without kanamycin on Todd-Hewitt (Difco Laboratories) agar containing 3% gelatin (Sigma, Saint Louis, Mo.) to score for the loss of gelatinase activity. Colonies that were gelatinase production negative were then scored for continued serine protease production and for the loss of kanamycin resistance. A selected gelatinase-negative, serine protease-positive, kanamycin-sensitive mutant was further confirmed as being a gelE deletion mutant by PCR with the two outer primers, GDF1 and GDR2, and by pulsed field gel electrophoresis (18).
Detection of protease activity.
The production of gelatinase in E. faecalis was detected by using Todd-Hewitt agar containing 3% gelatin, as previously described (23). The production of serine protease activity in E. faecalis was detected by 0.05% casein zymogram gel (Novex, San Diego, Calif.) analysis, as previously described (23).
Statistical analysis.
Kaplan-Meier survival estimates determined by using log-rank analysis were performed as previously described for both nematode and mouse survival by using STATA 6.0 for Windows. The data were plotted with GraphPad Prism 3.02 for Windows.
RESULTS AND DISCUSSION
Mutants of the fsr quorum-sensing system.
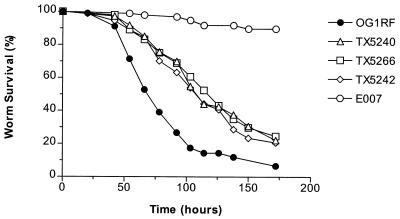
To determine whether the previously observed effect of an fsrB mutation on E. faecalis virulence in the C. elegans model was specific to fsrB or representative of the entire fsr locus, we evaluated the ability of fsrA (TX5240) and fsrC (TX5242) insertion mutants to kill C. elegans. Compared with the wild-type strain OG1RF, strains TX5240 (P ≤ 0.0001) and TX5242 (P ≤ 0.0001) were highly defective in their ability to kill C. elegans, similar to strain TX5266 (ΔfsrB) (P ≤ 0.0001) (Fig. 1). There were no significant differences in killing among the three fsr mutants (P > 0.05). As a control, the clinical E. faecium isolate E007 demonstrated little nematocidal activity, as has been shown previously (7). These data suggest that enterococci employ quorum-sensing during the pathogenic process in C. elegans. We had previously obtained similar data showing that the gram-negative human pathogen, P. aeruginosa, also employs quorum-sensing during pathogenic processes in both nonvertebrate and vertebrate hosts. Mutants of the quorum-sensing-associated regulators lasR, gacA, and mvfR of P. aeruginosa PA14 exhibit reduced virulence in C. elegans (16, 31, 32), Arabidopsis thaliana (3, 24, 25), and Galleria mellonella (lasR and gacA mutants) (12), as well as in a mouse full-skin-thickness burn model of P. aeruginosa infection (24, 25, 32).
FIG. 1.
Survival of C. elegans on fsr mutants. Kaplan-Meier survival plots of worms feeding on OG1RF and the fsr mutants TX5240 (fsrA), TX5266 (ΔfsrB), and TX5242 (fsrC), as well as the E. faecium clinical isolate, E007 (negative control). Similar data were obtained in three independent experiments.
Mutants of the protease genes gelE and sprE.
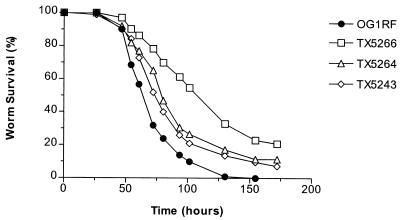
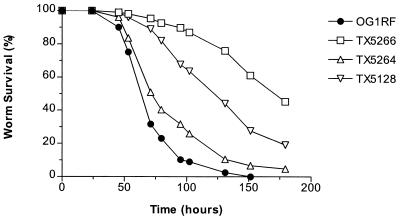
Because neither gelatinase nor serine protease activity are detectable in the fsr mutants TX5240, TX5242, and TX5266 (22, 23), we evaluated the contribution of E. faecalis gelatinase and serine protease separately to nematocidal activity. A gelE in-frame deletion mutant (TX5264) was constructed by allelic replacement (22) and, as shown in Fig. 2, TX5264 (P ≤ 0.0001) and the previously constructed sprE insertion mutant TX5243 (23) (P ≤ 0.0001) were each found to be attenuated in nematode killing, although to a lesser degree than TX5266. Compared to each other, TX5264 and TX5243 were not significantly different in their ability to kill C. elegans (P > 0.05) (Fig. 2). Nematode killing by the mini-γδ transposon insertion mutant of gelE, TX5128 (28), was more attenuated than the gelE in-frame deletion mutant TX5264 (P ≤ 0.0001) but less attenuated than the fsrB deletion mutant TX5266 (P ≤ 0.0001) (Fig. 3). Since the transposon mutation in gelE has a polar effect on the downstream sprE gene (as demonstrated by Northern blot, reverse transcription-PCR, and zymographic analyses [22, 23]), these results suggest that GelE and SprE may have additive effects on virulence. However, because TX5266 is even more attenuated than TX5128, these results also raise the possibility that the fsr system regulates genes involved in virulence for C. elegans in addition to gelE and sprE.
FIG. 2.
Survival of C. elegans on gelE and sprE mutants. Kaplan-Meier survival plots of worms feeding on OG1RF, TX5266 (ΔfsrB), TX5264 (ΔgelE), and TX5243 (sprE). Similar data were obtained in three independent experiments.
FIG. 3.
Survival of C. elegans on insertion and deletion gelE mutants. Kaplan-Meier survival plots of worms feeding on OG1RF, TX5266 (ΔfsrB), TX5264 (ΔgelE), and TX5128 (gelE::mγδ). Similar data were obtained in three independent experiments.
Complementation of the fsrB mutant.
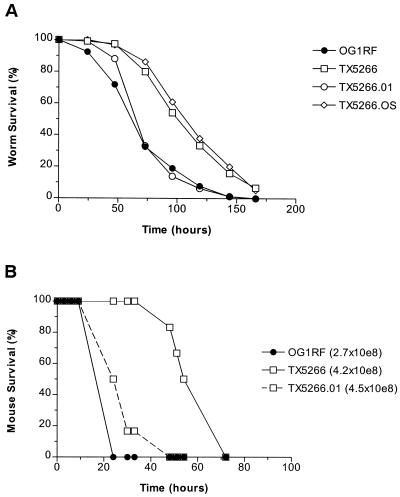
To confirm that the effect on E. faecalis virulence in TX5266 is due to the loss of a functional fsr system, we complemented the fsrB mutant TX5266 with the plasmid pTEX5249, which contains a 6-kb PstI/BglII fragment encoding fsrA, fsrB, and fsrC. This construct, TX5266.01, demonstrated restored gelatinase production (data not shown). As shown in Fig. 4, TX5266.01 had pathogenicity restored to near-wild-type levels in both the C. elegans killing assay and the mouse peritonitis model, indicating that virulence can be complemented with fsr function provided in trans. As a control, the fsrB mutant TX5266 transformed with the pAT18 “empty” vector alone (TX5266.OS) had no significant difference in C. elegans virulence compared to plasmid-free TX5266.
FIG. 4.
Complementation of fsrB in trans restores virulence in both the nematode and the mouse peritonitis models. (A) Survival plots of worms on OG1RF, TX5266, TX5266 carrying the fsrABC+ vector pTEX5249 (TX5266.01), and TX5266 carrying the “empty” vector pAT18 (TX5266.OS). Similar data were obtained in three independent experiments. (B) Kaplan-Meier survival plots of mice infected peritoneally with OG1RF, TX5266 (ΔfsrB), and TX5266 carrying the fsrABC+ vector pTEX5249 (TX5266.01). TX5266 was significantly attenuated compared to OG1RF at a smaller inoculum (P = 0.0009; closed circles versus open squares). TX5266.01 was significantly more virulent than TX5266 at similar inocula (P = 0.0012; open dashed squares versus open squares). Similar delays in lethality of TX5266 compared to OG1RF and TX5266.01 were also observed in a separate experiment with one-half of these inocula (data not shown).
Microscopy.
We previously found that live bacteria colonize the alimentary tract of adult C. elegans feeding on E. faecalis, similar to worms fed P. aeruginosa and Salmonella enterica (7). Such colonization is not seen when worms are fed E. coli or Bacillus subtilis (1, 7, 31). A small inoculum of E. faecalis is sufficient to colonize the nematode digestive tract and is followed by bacterial proliferation within the gut, leading to marked distention of the intestinal lumen (7). Mutants of the PA14 virulence regulators gacA and lasR fail to accumulate to a significant degree in the nematode alimentary tract after 24 to 48 h of feeding, suggesting that the establishment and/or proliferation of bacteria within the nematode may be dependent on the quorum-sensing mechanisms of P. aeruginosa (31, 32). However, in contrast to the results obtained with quorum-sensing mutants of P. aeruginosa, microscopic examination of adult nematodes feeding on lawns of either OG1RF or the fsrB mutant TX5266 showed almost identical gut distention with intact bacteria throughout the life span of the worms (data not shown). These data suggest that the fsr system does not play a role in E. faecalis colonization of the nematode intestine.
Conclusion.
The E. faecalis fsr system is the second example of a quorum-sensing system that regulates virulence gene expression in bacterial infection of both simple model organisms and mammalian hosts. Quorum sensing may be an important mechanism used by many prokaryotes to adapt to different environments encountered during pathogenesis. Our results also raise the possibility that the fsr system in E. faecalis regulates virulence genes in addition to gelE and sprE in this pathogen.
Acknowledgments
This research was supported by Postdoctoral Research Fellowships for Physicians from the Howard Hughes Medical Institute (C.D.S. and E.M.), by a postdoctoral fellowship from the Irvington Institute for Immunological Research (D.A.G.), by grant AI47923 from the National Institute of Allergy and Infectious Diseases (B.E.M.), and by a grant from Aventis SA (F.M.A. and S.B.C).
Editor: B. B. Finlay
REFERENCES
- 1.Aballay, A., P. Yorgey, and F. M. Ausubel. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539-1542. [DOI] [PubMed] [Google Scholar]
- 2.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, H., G. Krishnan, B. Goumnerov, J. Tsongalis, R. Tompkins, and L. G. Rahme. 2001. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self- regulatory mechanism. Proc. Natl. Acad. Sci. USA 98:14613-14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupont, H., P. Montravers, J. Mohler, and C. Carbon. 1998. Disparate findings on the role of virulence factors of Enterococcus faecalis in mouse and rat models of peritonitis. Infect. Immun. 66:2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.French, G. L. 1998. Enterococci and vancomycin resistance. Clin. Infect. Dis. 27(Suppl. 1):S75-S83. [DOI] [PubMed] [Google Scholar]
- 7.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold, H. S. 2001. Vancomycin-resistant enterococci: mechanisms and clinical observations. Clin. Infect. Dis. 33:210-219. [DOI] [PubMed] [Google Scholar]
- 9.Gold, O. G., H. V. Jordan, and J. van Houte. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch. Oral Biol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 10.Haas, W., and M. S. Gilmore. 1999. Molecular nature of a novel bacterial toxin: the cytolysin of Enterococcus faecalis. Med. Microbiol. Immunol. 187:183-190. [DOI] [PubMed] [Google Scholar]
- 11.Hancock, L. E., and M. S. Gilmore. 2000. Pathogenicity of enterococci, p. 251-258. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. A. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 12.Jander, G., L. G. Rahme, and F. M. Ausubel. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182:3843-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 14.Lewis, J. A., and J. T. Fleming. 1995. Basic culture methods, p. 3-29. In H. F. Epstein and D. C. Shakes (ed.), Caenorhabditis elegans: modern biological analysis of an organism. Academic Press, San Diego, Calif.
- 15.Li, X., G. M. Weinstock, and B. E. Murray. 1995. Generation of auxotrophic mutants of Enterococcus faecalis. J. Bacteriol. 177:6866-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 17.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. Akkermans, W. M. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 20.Noble, W. C., Z. Virani, and R. G. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72:195-198. [DOI] [PubMed] [Google Scholar]
- 21.Poyart, C., C. Pierre, G. Quesne, B. Pron, P. Berche, and P. Trieu-Cuot. 1997. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob. Agents Chemother. 41:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 25.Rahme, L. G., M. W. Tan, L. Le, S. M. Wong, R. G. Tompkins, S. B. Calderwood, and F. M. Ausubel. 1997. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 94:13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice, L. B. 2001. Emergence of vancomycin-resistant enterococci. Emerg. Infect. Dis. 7:183-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 28.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 29.Su, Y. A., M. C. Sulavik, P. He, K. K. Makinen, P. L. Makinen, S. Fiedler, R. Wirth, and D. B. Clewell. 1991. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect. Immun. 59:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulston, J., and J. Hodgkin. 1988. Methods, p. 587-606. In W. B. Wood (ed.), The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 31.Tan, M. W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]