Abstract
Mycobacterium microti is a member of the Mycobacterium tuberculosis complex that causes tuberculosis in voles. Most strains of M. microti are harmless for humans, and some have been successfully used as live tuberculosis vaccines. In an attempt to identify putative virulence factors of the tubercle bacilli, genes that are absent from the avirulent M. microti but present in human pathogen M. tuberculosis or Mycobacterium bovis were searched for. A minimal set of 50 bacterial artificial chromosome (BAC) clones that covers almost all of the genome of M. microti OV254 was constructed, and individual BACs were compared to the corresponding BACs from M. bovis AF2122/97 and M. tuberculosis H37Rv. Comparison of pulsed-field gel-separated DNA digests of BAC clones led to the identification of 10 regions of difference (RD) between M. microti OV254 and M. tuberculosis. A 14-kb chromosomal region (RD1mic) that partly overlaps the RD1 deletion in the BCG vaccine strain was missing from the genomes of all nine tested M. microti strains. This region covers 13 genes, Rv3864 to Rv3876, in M. tuberculosis, including those encoding the potent ESAT-6 and CFP-10 antigens. In contrast, RD5mic, a region that contains three phospholipase C genes (plcA to -C), was missing from only the vole isolates and was present in M. microti strains isolated from humans. Apart from RD1mic and RD5mic other M. microti-specific deleted regions have been identified (MiD1 to MiD3). Deletion of MiD1 has removed parts of the direct repeat region in M. microti and thus contributes to the characteristic spoligotype of M. microti strains.
Mycobacterium microti, a member of the Mycobacterium tuberculosis complex, was originally described as the infective agent of a tuberculosis-like disease in voles (Microtus agrestis) in the 1930s (47, 48). Until recently, M. microti strains were thought to be pathogenic only for voles, but not for humans, and some were even used as live vaccines. In fact, the vole bacillus proved to be safe and effective in preventing clinical tuberculosis in a trial involving roughly 10,000 adolescents in the United Kingdom in the 1950s (16). At about the same time, another strain, OV166, was successfully administered to half a million newborns in Prague, Czech Republic, without any serious complications (39). M. microti vaccination has since been discontinued because it was no more effective than the frequently employed Mycobacterium bovis BCG vaccine.
Apart from very slow growth on solid media, where pyruvate is required, and an S-shaped-curved-rod morphology, it is difficult to distinguish M. microti from the other members of the M. tuberculosis complex by classical methods. Biochemical tests reveal that most M. microti strains yield positive reactions for urease, nicotinamidase, pyrazinamidase, and alpha-esterase activities. However, M. microti strains give variable results concerning niacin accumulation and nitrate reduction (27). Like Mycobacterium africanum and M. bovis, M. microti is sensitive to thiophen-2-carboxylic acid hydrazide.
The use of new genotyping protocols for members of the M. tuberculosis complex has provided powerful tools to identify organisms that resemble the vole bacillus, and thus those strains have been classified as M. microti (32, 44). The most frequently used methods employ interstrain variation of repetitive sequences as the typing criteria, such as the direct repeat (DR) locus (spoligotyping), restriction fragment length polymorphism (RFLP) of IS6110, the polymorphic GC-rich sequence, and the mycobacterial interspersed repetitive units. Based on these genetic markers, M. microti strains that showed spoligotype hybridization patterns that were identical to those of the vole bacilli but that exhibited different IS6110 profiles have been isolated from immunocompromised patients (44). Furthermore, M. microti has been detected in the lungs of human immunodeficiency virus (HIV)-positive (19) and HIV-negative immunocompetent adults with pulmonary tuberculosis (32). For these cases, M. microti was considered the causative agent of the disease since no other mycobacterium was found in the organs of these patients.
Thus, are there any genetic differences between human and vole isolates that might explain their different degrees of virulence and host preferences? And what makes the vole isolates harmless for humans?
To address these questions, comparative-genomics methods were employed in the present study to identify major differences that may exist between M. microti reference strain OV254 and the entirely sequenced strains of M. tuberculosis H37Rv (10) and M. bovis AF2122/97 (14). An ordered bacterial artificial chromosome (BAC) library of M. microti OV254 was constructed, and individual BAC-to-BAC comparisons of a minimal set of these clones with BAC clones from previously constructed libraries of M. tuberculosis H37Rv and M. bovis AF2122/97 were undertaken.
Ten regions in M. microti that were different from the corresponding genomic regions in M. tuberculosis and M. bovis were detected. To investigate if these regions were associated with the ability of M. microti strains to infect humans, their genetic organizations in eight additional M. microti strains, including those isolated recently from patients with pulmonary tuberculosis, were studied. This analysis identified some regions that were specifically absent from all tested M. microti strains but present in all other members of the M. tuberculosis complex and other regions that were absent only from vole isolates of M. microti.
MATERIALS AND METHODS
Bacterial strains and plasmids.
M. microti OV254, which was originally isolated from voles in the United Kingdom in the 1930s (47), was kindly supplied by M. J. Colston. DNA from M. microti OV216 and OV183 was included in a set of strains used during a multicenter study (26). M. microti Myc 94-2272 was isolated in 1988 from the perfusion fluid of a 41-year-old dialysis patient (44) and was kindly provided by L. M. Parsons. M. microti ATCC 35782 was purchased from the American Type Culture Collection (designation TMC 1608 [M.P. Prague]). Human isolates B1 type llama, B2 type llama, B3 type vole, and B4 type vole were obtained from the collection of the National Reference Center for Mycobacteria, Forschungszentrum Borstel, Borstel, Germany. M. bovis strain AF2122/97, spoligotype 9, was responsible for a herd outbreak in Devon in the United Kingdom and has been isolated from lesions in both cattle and badgers. Typically, mycobacteria were grown on 7H9 Middlebrook liquid medium (Difco) containing 10% oleic acid-dextrose-catalase (Difco), 0.2% pyruvic acid, and 0.05% Tween 80.
Library construction, preparation of BAC DNA, and sequencing reactions.
Preparation of agarose-embedded genomic DNA from M. microti strain OV254, M. tuberculosis H37Rv, and M. bovis BCG was performed as described by Brosch et al. (5). The M. microti library was constructed by ligation of partially digested HindIII fragments (50 to 125 kb) into pBeloBAC11. From the 10,000 clones that were obtained, 2,000 were picked into 96-well plates and stored at −80°C. Plasmid preparations of recombinant clones for sequencing reactions were obtained by pooling a total of eight copies from 96-well plates, with each well containing an overnight culture in 250 μl of 2× yeast-tryptone medium with 12.5 μg of chloramphenicol · ml−1. After 5 min of centrifugation of each plate at 3,000 rpm in a swing rotor (Eppendorf A4-62), the bacterial pellets were resuspended in 25 μl of solution A (25 mM Tris [pH 8.0], 50 mM glucose, 10 mM EDTA) and cells were lysed by adding 25 μl of buffer B (0.2 M NaOH, 0.2% sodium dodecyl sulfate). Then 20 μl of cold 3 M sodium acetate, pH 4.8, was added, and the suspension was kept on ice for 30 min. After centrifugation at 3,000 rpm for 30 min, the pooled supernatants (140 μl) were transferred to four new plates. Isopropanol (130 μl) was added, and, after 30 min on ice, DNA was pelleted by centrifugation at 3,500 rpm for 15 min. The supernatant was discarded, the pellet was resuspended in 50 μl of a 10-μg/ml RNase A solution (in 10 mM Tris [pH 7.5]-1 mM EDTA) and pooled again, and the suspension (100 μl) was incubated at 64°C for 15 min. After precipitation (2.5 μl of 3 M sodium acetate [pH 7], 200 μl of absolute ethanol) pellets from the two remaining plates were rinsed with 200 μl of 70% ethanol, air dried, and finally suspended in 20 μl (each) of Tris-EDTA buffer.
End-sequencing reactions were performed with a Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems) by using a mixture of 13 μl of DNA solution, 2 μl of primer (2 μM; SP6-BAC1, AGTTAGCTCACTCATTAGGCA, or T7-BAC1, GGATGTGCTGCAAGGCGATTA), 2.5 μl of Big Dye, and 2.5 μl of a 5× buffer (50 mM MgCl2, 50 mM Tris). Thermal cycling was performed on a PTC-100 amplifier (MJ Inc.) with an initial denaturation step of 60 s at 95°C, followed by 90 cycles of 15 s at 95°C, 15 s at 56°C, and 4 min at 60°C. DNA was then precipitated with 80 μl of 76% ethanol and centrifuged at 3,000 rpm for 30 min. After the supernatant was discarded, DNA was rinsed with 80 μl of 70% ethanol and resuspended in the appropriate buffers depending on the type of automated sequencer used (ABI 377 or ABI 3700). Sequence data were transferred to Digital workstations and edited with TED software from the Staden package (38). Edited sequences were compared against the M. tuberculosis H37Rv database (http://genolist.pasteur.fr/TubercuList/), the M. bovis BLAST server (http://www.sanger.ac.uk/Projects/M_bovis/blast_server.shtml), and in-house databases to determine the relative positions of the M. microti OV254 BAC end sequences.
Preparation of BAC DNA from recombinants and BAC digestion profile comparison.
DNA for digestion was prepared as previously described (4). DNA (1 μg) was digested with HindIII (Boehringer), and restriction products were separated by pulsed-field gel electrophoresis (PFGE) on a Bio-Rad CHEF-DR III system by using a 1% (wt/vol) agarose gel and a pulse of 3.5 s for 17 h at 6 V · cm−1. Low-range PFGE markers (New England Biolabs [NEB]) were used as size standards. Insert sizes were estimated after ethidium bromide staining and visualization with UV light. Different comparisons were made with overlapping clones from the M. microti OV254, M. bovis AF2122/97, and M. tuberculosis H37Rv pBeloBAC11 libraries.
PCR analysis to determine presence of genes in different M. microti strains.
Reaction mixtures contained 1.25 μl of 10× PCR buffer (600 mM Tris-HCl [pH 8.8], 20 mM MgCl2, 170 mM [NH4]2SO4, 100 mM β-mercaptoethanol), 1.25 μl of 20 mM nucleotide deoxynucleoside triphosphate mixture, 2.5 μl of each primer at 2 μM, 10 ng of template DNA, 10% dimethyl sulfoxide, 0.5 U of Taq polymerase (Gibco-BRL), and sterile distilled water in a final volume of 12.5 μl. Thermal cycling was performed on a PTC-100 amplifier (MJ Inc.) with an initial denaturation step of 90 s at 95°C, followed by 35 cycles of 45 s at 95°C, 1 min at 60°C, and 2 min at 72°C.
RFLP analysis.
In brief, agarose plugs of genomic DNA prepared as previously described (5) were digested with either AseI, DraI, or XbaI (NEB) and then electrophoresed on a 1% agarose gel and finally transferred to Hybond-C extra-nitrocellulose membranes (Amersham). Different probes were amplified by PCR from M. microti strain OV254 or M. tuberculosis H37Rv with primers for esat-6 (esat-6F, GTCACGTCCATTCATTCCCT; esat-6R, ATCCCAGTGACGTTGCCTT), the RD1mic flanking region (4340,209F, GCAGTGCAAAGGTGCAGATA; 4354,701R, GATTGAGACACTTGCCACGA), or plcA (plcA.int.F, CAAGTTGGGTCTGGTCGAAT; plcA.int.R, GCTACCCAAGGTCTCCTGGT). Amplification products were radiolabeled by using the Stratagene Prime-It II kit. Hybridizations were performed at 65°C in a solution containing 0.8 M NaCl, 5 mM EDTA, pH 8, 50 mM sodium phosphate, pH 8, 2% sodium dodecyl sulfate, 1× Denhardt's reagent, and 100 μg of salmon sperm DNA (Genaxis)/ml. Membranes were exposed to phosphorimager screens, and images were digitalized by using a STORM phosphorimager. IS6110 RFLP was performed as previously described (26) with the exception that radiolabeling and hybridization conditions described above were employed.
Nucleotide sequence accession numbers.
The nucleotide sequences that flank MiD1, MiD2, and MiD3 and the junction sequence of RD1mic have been deposited in the EMBL database. Accession numbers are AJ345005, AJ345006, AJ315556, and AJ315557, respectively.
RESULTS
Establishment of a complete ordered BAC library of M. microti OV254.
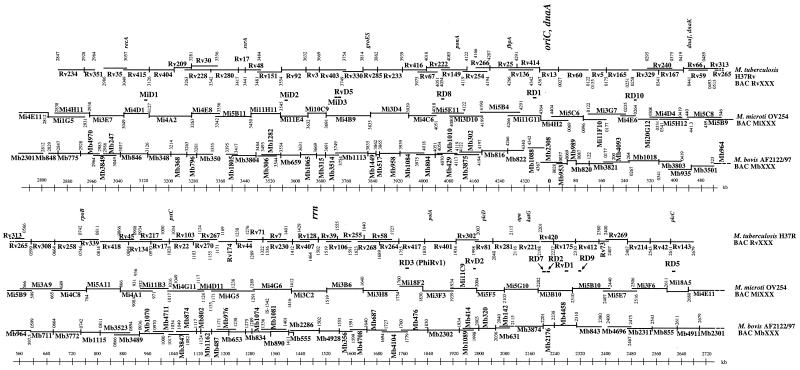
Electroporation of pBeloBAC11 containing partial HindIII digests of M. microti OV254 DNA into Escherichia coli DH10B yielded about 10,000 recombinant clones, from which 2,000 were isolated and stored in 96-well plates. With the complete sequence of the M. tuberculosis H37Rv genome as a scaffold, the end sequencing of 384 randomly chosen M. microti BAC clones allowed us to select enough clones to cover almost all of the 4.4-Mb chromosome. A few rare clones that spanned regions that were not covered by this approach were identified by PCR screening of pools as previously described (4). This resulted in a minimal set of 50 BACs, covering over 99.9% of the M. microti OV254 genome, whose positions relative to M. tuberculosis H37Rv are shown in Fig. 1. The insert size ranged between 50 and 125 kb, and the recombinant clones were stable. Compared with other BAC libraries from tubercle bacilli (4, 13) the M. microti OV254 BAC library contained clones that were generally larger than those obtained previously, which facilitated the comparative-genomics approach described below.
FIG. 1.
M. microti strain OV254 BAC map (MiXXX) overlaid on those of M. tuberculosis H37Rv (RvXXX) and M. bovis AF2122/97 (MbXXX). The scale bars indicate the positions on the M. tuberculosis genome.
Identification of DNA deletions in M. microti OV254 relative to M. tuberculosis H37Rv by comparative genomics.
The minimal overlapping set of 50 BAC clones, together with the availability of three other ordered BAC libraries from M. tuberculosis H37Rv, M. bovis BCG Pasteur 1173P2 (5, 13), and M. bovis AF2122/97 (14), allowed us to carry out direct BAC-to-BAC comparisons of clones spanning the same genomic regions. Size differences between PFGE-separated HindIII restriction fragments from M. microti OV254 BACs and restriction fragments from M. bovis and/or M. tuberculosis BAC clones identified loci that differed among the tested strains. Size variations of at least 2 kb were easily detectable, and 10 deleted regions, evenly distributed around the genome and containing more than 60 open reading frames (ORFs), were identified. These regions represent over 60 kb missing from M. microti OV254 that are present in M. tuberculosis H37Rv. First, it was found that phiRv2 (RD11), one of the two M. tuberculosis H37Rv prophages, was present in M. microti OV254, whereas phiRv1, also referred to as RD3 (29), was absent. Second, it was found that M. microti lacks four of the genomic regions that were also absent from M. bovis BCG. In fact, these four regions of difference (RD), named RD7, RD8, RD9, and RD10, are absent from all members of the M. tuberculosis complex with the exception of M. tuberculosis and Mycobacterium canettii and seem to have been lost from a common progenitor strain of M. africanum, M. microti, and M. bovis (3). As such, our results, obtained from individual BAC-to-BAC comparisons, show that M. microti is part of this non-M. tuberculosis lineage of the tubercle bacilli, and this assumption was further confirmed by sequencing the junction regions of RD7 to RD10 in M. microti OV254. The sequences obtained were identical to those from M. africanum, M. bovis, and M. bovis BCG strains. Apart from these four conserved RD and phiRv1 (RD3), M. microti OV254 did not show any RD with junction regions identical to those of M. bovis BCG Pasteur, which lacks at least 17 RD relative to M. tuberculosis H37Rv (1, 13, 36). However, five other regions missing from the genome of M. microti OV254 relative to that of M. tuberculosis H37Rv were identified (RD1mic, RD5mic, MiD1, MiD2, and MiD3). Such regions are specific either for strain OV254 or for M. microti strains in general. Interestingly, two of these regions, RD1mic and RD5mic, partially overlap RD from M. bovis BCG.
Antigens ESAT-6 and CFP-10 are absent from M. microti.
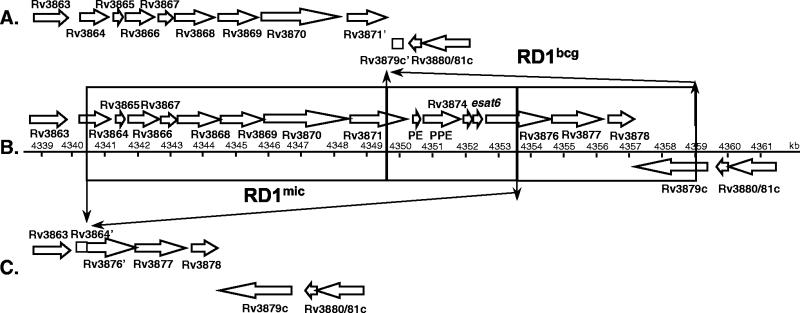
One of the most interesting findings of the BAC-to-BAC comparison was a novel deletion in a genomic region close to the origin of replication (Fig. 1). Detailed PCR and sequence analysis of this region in M. microti OV254 showed a segment of 14 kb (equivalent to M. tuberculosis H37Rv from kb 4340.4 to 4354.5) that partly overlapped RD1bcg, which is absent from M. bovis BCG, to be missing. More precisely, ORFs Rv3864 and Rv3876 are truncated in M. microti OV254 and ORFs Rv3865 to Rv3875 are absent (Fig. 2). This observation is particularly interesting as previous comparative genomic analyses identified RD1bcg as the only RD that is specifically absent from all BCG substrains but present in all other members of the M. tuberculosis complex (1, 4, 13, 29, 36). RD1bcg was therefore assumed to be directly involved in the attenuation of BCG. As shown in Fig. 2, in M. microti OV254 the RD1mic deletion is responsible for the loss of a large portion of the conserved ESAT-6 family core region (41) including the genes coding for major T-cell antigens ESAT-6 and CFP-10 (2, 15). The fact that previous deletion screening protocols employed primer sequences that were designed for the right portion of the RD1bcg region (i.e., gene Rv3878) (6, 40) explains why the RD1mic deletion was not detected earlier by these investigations. Figure 2 shows that RD1mic does not affect genes Rv3877, Rv3878, and Rv3879, which are part of the RD1bcg deletion.
FIG. 2.
Differences in the region comprising kb 4340 to 4360 between the deletions in BCG RD1bcg (A) and M. microti RD1mic (C) relative to M. tuberculosis H37Rv (B).
Deletion of phospholipase C (PLC) genes in M. microti OV254.
RD5mic, the other region absent from M. microti OV254, which partially overlapped an RD from BCG, was revealed by comparison of BAC clone Mi18A5 with BAC Rv143 (Fig. 1). PCR analysis and sequencing of the junction region revealed that the RD5mic deletion was smaller than the RD5 deletion in BCG (Tables 1 and 2). In fact, M. microti OV254 lacks genes plcA, plcB, and plcC and two specific genes (Rv2352 and Rv2353) encoding proteins from the PPE family (motif Pro-Pro-Glu). This was confirmed by the absence of a clear band on a Southern blot of AseI-digested genomic DNA from M. microti OV254 hybridized with a plcA probe. However, genes Rv2346c and Rv2347c, members of the esat-6 family, and Rv2348c, which are missing from M. bovis and BCG strains (3), are still present in M. microti OV254. The presence of an IS6110 element in this segment suggests that recombination between two IS6110 elements could have been involved in the loss of RD5mic, and this is supported by the finding that the remaining copy of IS6110 does not show a 3-bp direct repeat in strain OV254 (Table 2).
TABLE 1.
Description of the putative functions of the deleted and truncated ORFs in M. microti OV254 relative to M. tuberculosis H37Rv
| Region | Start-end (kb) | Overlapping ORFs | Gene name(s) and/or putative function or family(ies) of product |
|---|---|---|---|
| RD10 | 264.5-266.5 | Rv0221-Rv0223 | echA1 |
| RD3 | 1779.5-1788.5 | Rv1573-Rv1586 | Bacteriophage proteins |
| RD7 | 2207.5-2220.5 | Rv1964-Rv1977 | yrbE3A and -3B; mce3A to -3F; unknown |
| RD9 | 2330-2332 | Rv2072-Rv2075 | cobL; probable oxidoreductase; unknown |
| RD5mic | 2627.6-2633.4 | Rv2348-Rv2352 | plcA to -C; member of PPE family |
| MiD1 | 3121.8-3126.6 | Rv2816-Rv2819 | IS6110 transposase; unknown |
| MiD2 | 3554.0-3755.2 | Rv3187-Rv3190 | IS6110 transposase; unknown |
| MiD3 | 3741.1-3755.7 | Rv3345-Rv3349 | Members of the PE-PGRS and PPE families; insertion elements |
| RD8 | 4056.8-4062.7 | Rv3617-Rv3618 | ephA; lpqG; member of the PE-PGRS family |
| RD1mic | 4340.4-4354.5 | Rv3864-Rv3876 | Member of the CBXX/CFQX family; member of the PE and PPE families; ESAT-6; CFP10; unknown |
TABLE 2.
Sequences at the junctions of the deleted regions in M. microti OV254 relative to M. tuberculosis H37Rv
| Junction | Position (kb) | ORFs | Sequence at the junctiona | Flanking primers (5′-3′ sequences) |
|---|---|---|---|---|
| RD1mic | 4340.421-4354.533 | Rv3864-Rv3876 | CAAGACGAGGTTGTAAAACCTCGACGCAGGATCGGCGATGAAATGCCAGTCGGCGTCGCTGAGCGCGCGCTGCGCCGAGTCCCATTTTGTCGCTGATTTGTTTGAACAGCGACGAACCGGTGTTGAAAATGTCGCCTGGGTCGGGGATTCCCT | 4340, 209F (GCAGTGCAAAGGTGCAGATA), 4354,701R (GATTGAGACACTTGCCACGA) |
| RD5mic | 2627.831-2635.581 | Rv2349-Rv2355 | CCTCGATGAACCACCTGACATGACCCCATCCTTTCCAAGAACTGGAGTCTCCGGACATGCCGGGGCGGTTCACTGCCCCAGGTGTCCTGGGTCGTTCCGTTGACCGTCGAGTCCGAACATCCGTCATTCCCGGTGGCAGTCGGTGCGGTGAC | 2627, 370F (GAATGCCGACGTCATATCG), 2633, 692R (CGGCCACTGAGTTCGATTAT) |
| MiD1 | 3121.880-3126.684 | Rv2815c-Rv2818c | CACCTGACATGACCCCATCCTTTCCAAGAACTGGAGTCTCCGGACATGCCGGGGCGGTTCAGGGACATTCATGTCCATCTTCTGGCAGATCAGCAGATCGCTTGTTCTCAGTGCAGGTGAGTC | 3121, 690F (CAGCCAACACCAAGTAGACG), 3126, 924R (TCTACCTGCAGTCGCTGTG) |
| MiD2 | 3554.066-3555.259 | Rv3188-Rv3189 | GCTGCCTACTACGCTCAACGCCAGAGACCAGCCGCCGGCTGAGGTCTCAGATCAGAGAGTCTCCGGACTCACCGGGGCGGTTCATAAAGGCTTCGAGACCGGACGGGCTGTAGGTTCCTCAACTGTGTGGCGGATGGTCTGAGCACTTAAC | 3553, 880F (GTCCATCGAGGATGTCGAGT), 3555, 385R (CTAGGCCATTCCGTTGTCTG) |
| MiD3 | 3741.139-3755.777 | Rv3345c-Rv3349c | TGGCGCCGGCACCTCCGTTGCCACCGTTGCCGCCGCTGGTGGGCGCGGTGCCGTTCGCCCCGGCCGAACCGTTCAGGGCCGGGTTCGCCCTCAGCCGCTAAACACGCCGACCAAGATCAACGAGCTACCTGCCCGGTCAAGGTTGAAGAGCCCCCATATCAGCAAGGGCCCGGTGTCGGCG | 3740, 950F (GGCGACGCCATTTCC), 3755, 988R (AACTGTCGGGCTTGCTCTT) |
Sequences in bold, lefthand portion of junction sequence; underlined sequences, short repeats in M. tuberculosis H37Rv.
Lack of MiD1 provides a genomic clue for the M. microti OV254 characteristic spoligotype.
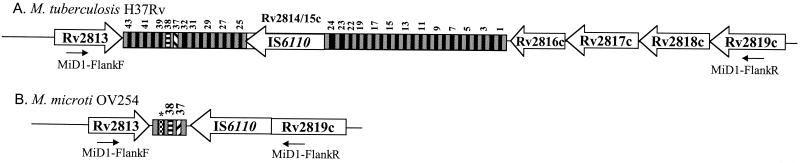
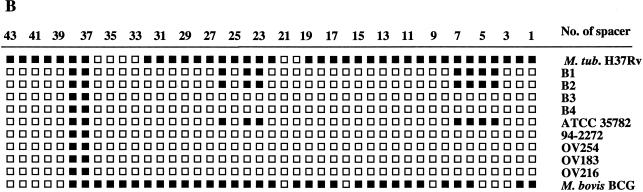
MiD1 encompasses ORFs Rv2816, Rv2817, and Rv2818, which encode putative proteins whose functions are yet unknown, and is in the direct repeat region (DR), a polymorphic locus in the genomes of the tubercle bacilli that contains a cluster of direct repeats of 36 bp separated by unique spacer sequences of 36 to 41 bp (17) (Fig. 3). The presence or absence of 43 unique spacer sequences that intercalate the DR sequences is the basis of spacer oligotyping, a powerful typing method for strains from the M. tuberculosis complex (23). M. microti isolates exhibit a characteristic spoligotype with an unusually small DR cluster due to the presence of only spacers 37 and 38 (44). In M. microti OV254, the absence of spacers 1 to 36, which are present in many other M. tuberculosis complex strains, appears to result from an IS6110-mediated deletion of 636 bp of the DR. Amplification and PvuII restriction analysis of a 2.8-kb fragment obtained with primers located in the genes that flank the DR (Rv2813c and Rv2819) showed that there is only one copy of IS6110 remaining in this region (Fig. 3). This IS6110 element is inserted into ORF Rv2819 at position 3119932 relative to the M. tuberculosis H37Rv genome. As for other IS6110 elements that result from homologous recombination between two copies (7), no 3-bp direct repeat was found for this copy of IS6110 in the DR. Concerning the absence of spacers 39 to 43, it was found that M. microti showed a slightly different organization of this locus than M. bovis strains, which also characteristically lack spacers 39 to 43 (Fig. 3). In M. microti OV254 an extra spacer of 36 bp that was not present in M. bovis or in M. tuberculosis H37Rv was found. The sequence of this specific spacer was identical to that of spacer 58, reported by van Embden and colleagues (43). In their study of the DRs in many strains from the M. tuberculosis complex this spacer was found only in M. microti strain NLA000016240 (AF189828) and in some ancestral M. tuberculosis strains (3, 43). Like MiD1, MiD2 most probably results from an IS6110-mediated deletion of two genes (Rv3188 and Rv3189) that encode putative proteins whose function is unknown (Tables 2 and 3).
FIG. 3.
Difference in the region comprising kb 3121 to 3127 between M. tuberculosis H37Rv (A) and M. microti OV254 (B). Gray boxes, DRs; black boxes, unique numbered spacer sequences; ∗, spacer sequence identical to that of spacer 58 reported by van Embden et al. (43). Note that spacers 33 to 36 and 20 to 22 are not shown because H37Rv lacks these spacers.
TABLE 3.
Presence of the RD and MiD region in different M. microti strains
| Region | Presence in straina:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| OV254 | OV183 | OV216 | ATCC 35782 | Myc 94-2272 | B3 type vole | B4 type vole | B1 type llama | B2 type llama | |
| RD1mic | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| RD3 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| RD7 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| RD8 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| RD9 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| RD10 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| MiD3 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| MiD1 | Absent | Absent | Absent | Present | Partial | Partial | Partial | Present | Present |
| RD5mic | Absent | Absent | Absent | Present | Present | Present | Present | Present | Present |
| MiD2 | Absent | Absent | Absent | Present | Present | Present | Present | Present | Present |
Strains OV254, OV183, and OV216 are from voles; strains Myc 94-2272, B3 type vole, B4 type vole, B1 type llama, and B2 type llama are from humans; the origin of strain ATCC 35782 is unknown.
Absence of some members of the PPE family in M. microti.
MiD3 was identified by the absence of two HindIII sites in BAC Mi4B9 that exist at kb 3749 and 3754 in the M. tuberculosis H37Rv chromosome. By PCR and sequence analysis, it was determined that the MiD3 deletion corresponds to a 12-kb deletion that truncates or removes five genes orthologous to Rv3345c to Rv3349c. Rv3347c encodes a protein of 3,157 amino acids that belongs to the PPE family, and Rv3346c encodes a conserved protein that is also present in Mycobacterium leprae. The function of both these putative proteins is unknown, while Rv3348 and Rv3349 are part of an insertion element (Table 1). At present, the consequences of the MiD3 deletions for the biology of M. microti remain entirely unknown.
Extra DNA in M. microti OV254 relative to M. tuberculosis H37Rv.
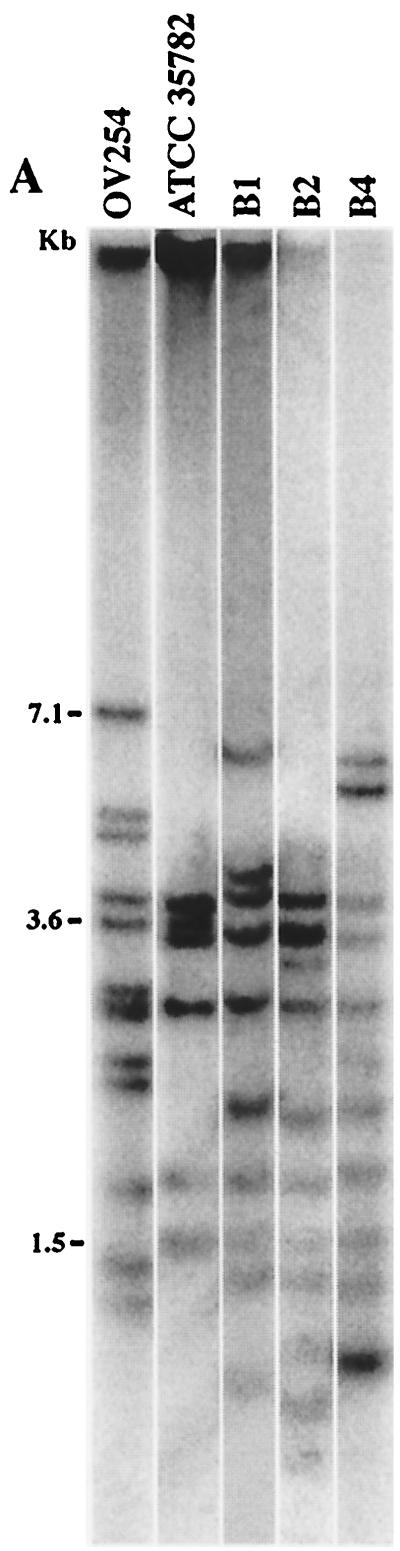
M. microti OV254 possesses regions RvD1 to RvD5 and TBD1, which are absent from sequenced strain M. tuberculosis H37Rv but which have been shown to be present in other members of the M. tuberculosis complex, such as M. canettii, M. africanum, M. bovis, and M. bovis BCG (3, 7, 13). In M. tuberculosis H37Rv, four of these regions (RvD2 to -5) contain a copy of IS6110 which is not flanked by a direct repeat, suggesting that recombination of two IS6110 elements was involved in the deletion of the intervening genomic regions (7). In consequence, it seems plausible that these regions were deleted from the M. tuberculosis H37Rv genome rather than specifically acquired by M. microti. Three other small insertions have also been found, and they are due to the presence of an IS6110 element in a location different from those in M. tuberculosis H37Rv and M. bovis AF2122/97. Indeed, PvuII RFLP analysis of M. microti OV254 reveals 13 IS6110 elements (Fig. 4A) (44).
FIG. 4.
(A) IS6110 RFLP after PvuII digestion from various M. microti strains. (B) Spoligotypes of various M. microti and control strains.
Genomic diversity of M. microti strains.
To obtain a more global picture of the genetic organization of the taxon M. microti, we evaluated the presence or absence of the variable regions found in strain OV254 in eight other M. microti strains. These strains, which were isolated from humans and voles, have been designated M. microti mainly on the basis of their specific spoligotypes (26, 32, 44) and can be further divided into subgroups according to the host such as voles, llamas, and humans (Table 3). As stated in the introduction, M. microti is rarely found in humans, unlike M. tuberculosis. So the availability of nine strains from variable sources for genetic characterization is an exceptional resource. Among them was one strain (Myc 94-2272) from a severely immunocompromised individual (44), and four strains were isolated from HIV-positive or HIV-negative humans with spoligotypes typical of llama and vole isolates. For strain ATCC 35782 (TMC 1608 [M.P. Prague]), we could not identify with certainty the original host from which the strain was isolated or if this strain corresponds to M. microti OV166, which was received by L. Sula from A. Q. Wells and used thereafter for the vaccination program in Prague in the 1960s (39).
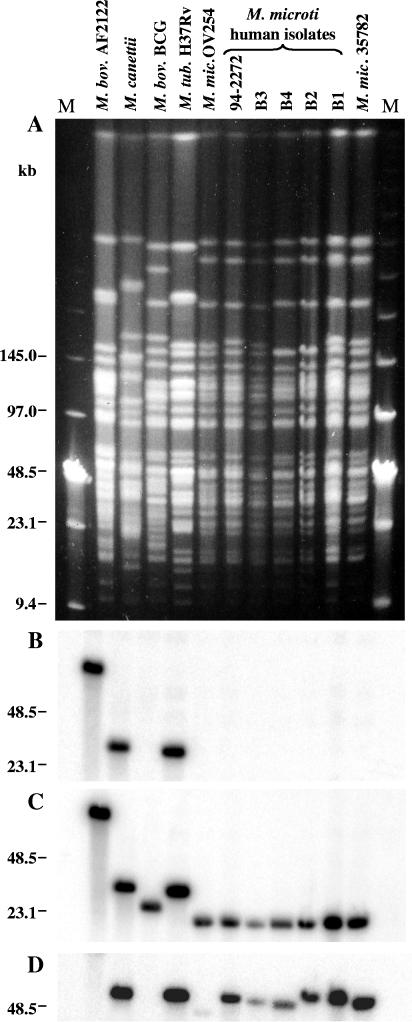
First, we were interested if these nine strains, designated M. microti on the basis of their spoligotypes, also resembled each other by other molecular typing criteria. As RFLP of pulsed-field gel-separated chromosomal DNA represents probably the most accurate molecular typing strategy for bacterial isolates, we determined the AseI profiles of the available M. microti strains and found that the profiles resembled each other closely but differed significantly from the macrorestriction patterns of M. tuberculosis, M. bovis, and M. bovis BCG strains used as controls. However, as depicted in Fig. 5A, the patterns were not identical to each other and each M. microti strain showed subtle differences, suggesting that they were not epidemiologically related. A similar observation was made with other rare-cutting restriction enzymes, such as DraI and XbaI (data not shown).
FIG. 5.
(A) AseI restriction fragment PFGE profiles of various M. microti strains; (B) hybridization with a radiolabeled esat-6 probe; (C) hybridization with a probe of the RD1mic flanking region; (D) hybridization with a plcA probe. Lanes: 1 (second lane from left), M. bovis AF2122/97; 2, M. canetti; 3, M. bovis BCG Pasteur; 4, M. tuberculosis H37Rv; 5, M. microti OV254; 6, M. microti Myc 94-2272; 7, M. microti B3 type vole; 8, M. microti B4 type vole; 9, M. microti B2 type llama; 10, M. microti B1 type llama; 11, M. microti ATCC 35782; M, low-range PFGE marker (NEB).
Common and diverging features of M. microti strains.
Two strategies were used to test for the presence or absence of variable regions in these strains, for which we do not have ordered BAC libraries. First, PCRs using internal and flanking primers of the variable regions were employed and amplification products of the junction regions were sequenced. Second, probes from the internal portions of variable regions absent from M. microti OV254 were obtained by amplification of M. tuberculosis H37Rv DNA with specific primers. Hybridization with these radiolabeled probes was carried out on blots from PFGE-separated AseI restriction digests of the M. microti strains. In addition, we confirmed the findings obtained by these two techniques by using a focused macroarray containing some of the genes identified in variable regions of the tubercle bacilli to date (data not shown).
This led to the finding that the RD1mic deletion is specific for all M. microti strains tested. Indeed, none of the M. microti DNA digests hybridized with the radiolabeled esat-6 probe (Fig. 5B) but rather with the RD1mic flanking region (Fig. 5C). In addition, PCR amplification with primers flanking the RD1mic region (Table 2) yielded fragments of the same size for all M. microti strains whereas no products were obtained for M. tuberculosis, M. bovis, and M. bovis BCG strains. Furthermore, the sequences of the junction regions for the strains were found to be identical, which confirms that the genomic organizations of the RD1mic loci in all tested M. microti strains were the same (Table 3). This clearly demonstrates that M. microti lacks the conserved ESAT-6 family core region, stretching in other members of the M. tuberculosis complex from Rv3864 to Rv3876, and, as such, represents a taxon of naturally occurring ESAT-6 and CFP-10 deletion mutants.
Like RD1mic, MiD3 was found to be absent from all nine M. microti strains tested and, therefore, appears to be a specific genetic marker that is restricted to M. microti strains (Table 3). However, PCR amplification showed that RD5mic is absent only from vole isolates OV254, OV216, and OV183 and is present in M. microti strains isolated from human and other sources (Table 3). This was confirmed by the presence of single bands of differing sizes on a Southern blot hybridized with a plcA probe for all M. microti tested strains except OV254 (Fig. 5D). The observed size differences of the corresponding AsnI fragments are probably due to the presence of a variable numbers of insertion elements in these fragments. Interestingly, the presence or absence of RD5mic correlated with the similarity of IS6110 RFLP profiles. The profiles of the three M. microti strains isolated from voles in the United Kingdom differed considerably from the IS6110 RFLP patterns of humans isolates (Fig. 4) (44). Taken together, these results underline the proposed involvement of IS6110-mediated deletion of the RD5 region and further suggest that RD5 may be involved in the variable potential of M. microti strains to cause disease in humans. Similarly, it was found that MiD1 was missing from vole isolates OV254, OV216, and OV183 whereas for strains B1 type llama, B2 type llama, and ATCC 35782, all displaying the llama spoligotype (Fig. 4B), the MiD1 region was present. PCR analysis further revealed that, in human isolates B3 type vole, B4 type vole, and Myc 94-2272, gene Rv2818c was present and the MiD1 region was only partially deleted. These findings correlate with the described spoligotypes of the different isolates. Strains that had region MiD1 present contained more spacers than strains with partial or full deletion of MiD1, suggesting that the strains characterized by the llama spoligotype are ancestral to the ones showing only spacers 37 and 38.
As regards region MiD2, we observed that it was absent only from vole isolates OV254, OV216, and OV183 but was present in all other tested strains (Table 3). Further studies are needed to investigate if proteins encoded by genes Rv3188 and Rv3189 may have an impact on the virulence of the strains.
DISCUSSION
In this study we have searched for major genomic variations, due to insertion-deletion events, between the vole pathogen M. microti and the human pathogen M. tuberculosis. BAC-based comparative genomics led to the identification of 10 regions absent from the genome of the vole bacillus M. microti OV254 and several insertions due to IS6110. Seven of these deleted regions were also absent from eight other M. microti strains isolated from voles or humans, and they account for more than 60 kb of genomic DNA. Of these regions, RD1mic is of particular interest, because absence of part of this region has been found to be restricted to the BCG vaccine strains to date. As M. microti was originally described as nonpathogenic for humans, it is thus tempting to speculate that RD1 genes may be involved in the pathogenicity for humans. Preliminary results with M. bovis ESAT-6 knockout mutants support this hypothesis (46). This is reinforced by the fact that RD1bcg (29) has lost putative ORFs belonging to the esat-6 gene cluster, including the genes encoding ESAT-6 and CFP-10 (Fig. 2) (41). Both polypeptides have been shown to act as potent stimulators of the immune system and are antigens recognized during the early stages of infection (8, 12, 20, 35). Moreover, the biological importance of this region for mycobacteria is underlined by the fact that it is also conserved in M. leprae, where genes ML0047 to ML0056 show high similarities in their sequences and operon organization to the genes in the esat-6 core region of the tubercle bacilli (11). In spite of the radical gene decay observed in M. leprae, the esat-6 operon apparently has kept its functionality in this organism.
However, the RD1 deletion may not be the only reason why the vole bacillus is attenuated for humans. Indeed, it remains unclear why certain M. microti strains included in the present study that show exactly the same RD1mic deletion as vole isolates have been found to be causative agents of human tuberculosis. As human M. microti cases are extremely rare, the most plausible explanation for this phenomenon is that the infected people were particularly susceptible for mycobacterial infections in general. This could have been due to an immunodeficiency (32, 44) or to a rare genetic host predisposition such as gamma interferon or interleukin-12 receptor modification (22).
In addition, the finding that human M. microti isolates differed from vole isolates by the presence of region RD5mic may also have an impact on the increased potential of human M. microti isolates to cause disease. Intriguingly, BCG and the vole bacillus lack overlapping portions of this chromosomal region, which encompasses three (plcA, plcB, and plcC) of the four genes encoding PLC in M. tuberculosis. PLC has been recognized as an important virulence factor in numerous bacteria, including Clostridium perfringens, Listeria monocytogenes, and Pseudomonas aeruginosa, where it plays a role in cell-to-cell spread of bacteria, intracellular survival, and cytolysis (37, 42). To date, the exact role of PLC for the tubercle bacilli remains unclear. However, a recent study suggests that PLC is involved in the virulence of M. tuberculosis (34a). plcA encodes antigen mtp40, which has previously been shown to be absent from seven tested vole and hyrax isolates (28). PLC activity in M. tuberculosis, M. microti, and M. bovis, but not in M. bovis BCG, has been reported (21, 49). The levels of PLC activity detected in M. bovis were much lower than those seen in M. tuberculosis, consistent with the loss of plcABC. It is likely that plcD is responsible for the residual PLC activity in strains lacking RD5, such as M. bovis and M. microti OV254. Indeed, the plcD gene is located in region RvD2, which is present in some but not all tubercle bacilli (13, 18, 24). PLC-encoding genes have been recognized as hot spots for integration of IS6110, and it appears that regions RD5 and RvD2 undergo independent deletion processes more frequently than any other genomic regions (45). Thus, the number of functional plc genes necessary for the synthesis of an appropriate amount of PLC may have an impact on the virulence of individual M. microti strains and possibly also influences the natural host spectrum of certain other tubercle bacilli, such as M. bovis strains, which lack the plcABC genes.
Another detail revealed by this study is that, among the deleted genes, seven code for members of the PPE family of Gly-, Ala-, Asn-rich proteins. A closer look at the sequences of these genes showed that in some cases such as Rv3873, located in the RD1mic region, and Rv2352c and Rv2353c, located in the RD5mic region, their products were small proteins with unique sequences. Others, such as Rv3347c, located in the MiD3 region, code for a much larger PPE protein (3,157 amino acids). In this case a neighboring gene (Rv3345c), belonging to another multigene family, the PE-polymorphic GC-rich sequence family, was partly affected by the MiD3 deletion. While the function of the PE/PPE proteins is currently unknown, their predicted abundances in the proteome of M. tuberculosis suggest that they may play an important role in the life cycle of the tubercle bacilli. Indeed, recently some of them were shown to be involved in the pathogenicity of M. tuberculosis (9) and Mycobacterium marinum (34). Complementation of such genomic regions in M. microti OV254 should enable us to carry out proteomics and virulence studies of animals in order to understand the role of such ORFs in pathogenesis.
In conclusion, this study has shown that M. microti, a taxon originally named after its major host, Microtus agrestis, the common vole, represents a relatively homogenous group of tubercle bacilli. Although all tested strains showed unique PFGE macrorestriction patterns that differed slightly, deletions that were common to all M. microti isolates (RD7 to RD10, MiD3, and RD1mic deletions) have been identified. The conserved nature of these deletions suggests that these strains are derived from a common precursor that has lost these regions, and their loss may account for some of the observed common phenotypic properties of M. microti, such as the very slow growth on solid media and the formation of tiny colonies. This finding is consistent with results from a recent study that showed that M. microti strains carry a particular mutation in the gyrB gene (31). Of particular interest, some of these common features (e.g., the flanking regions of RD1mic and MiD3) could be exploited for an easy-to-perform PCR identification test, similar to the one proposed for a range of tubercle bacilli (33). This test should enable unambiguous and rapid identification of M. microti isolates in order to obtain a better estimate of the overall rate of M. microti infections in humans and other mammalian species. However, our study also shows that, in spite of an extensive homogeneity, there are important genomic differences among some of the strains designated M. microti, which may be implicated in the different degrees of pathogenicity of M. microti strains in certain hosts. While this work was in progress, Manabe et al. confirmed the exquisite specificity of ATCC 19422 for the vole (30). However, some other M. microti strains may never have been associated with voles but, instead, may have their reservoirs in other mammals (25). Now that these strains have been subdivided by using deletion typing methods, it would be of great interest to determine their relative levels of virulence in the vole or mouse models of disease. Complementation of the vole bacillus with the individual RD should help to uncover the function of the genes located in these regions and should further allow evaluations of the potential of recombinant M. microti strains as improved live vaccines. The present study represents a first step toward understanding why certain strains cause human disease. In a continuation of this work, determination of the genome sequence of the vole bacillus and in depth analysis, as well as proteomics studies, have been started recently.
Acknowledgments
We are grateful to M. J. Colston and L. M. Parsons for supplying M. microti strains OV254 and Myc 94-2272 and for fruitful discussions. We also thank C. Lacroix, S. Duthoy, and N. Barilone for their help.
Financial support was received from the European Union (grant QLK2-CT-1999-01093), the Institut Pasteur (PTR35/2000), the Génopole Programme, and the Association Française Raoul Follereau.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 2.Berthet, F.-X., P. B. Rasmusse, I. Rosenkrands, P. Andersen, and B. Gicquel. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144:3195-3203. [DOI] [PubMed] [Google Scholar]
- 3.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito, B. G. Barrell, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosch, R., S. V. Gordon, C. Buchrieser, A. S. Pym, T. Garnier, and S. T. Cole. 2000. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast 17:111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosch, R., S. V. Gordon, A. Pym, K. Eiglmeier, T. Garnier, and S. T. Cole. 2000. Comparative genomics of the mycobacteria. Int. J. Med. Microbiol. 290:143-152. [DOI] [PubMed] [Google Scholar]
- 7.Brosch, R., W. J. Philipp, E. Stavropoulos, M. J. Colston, S. T. Cole, and S. V. Gordon. 1999. Genomic analysis reveals variation between Mycobacterium tuberculosis H37Rv and the attenuated M. tuberculosis H37Ra strain. Infect. Immun. 67:5768-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brusasca, P. N., R. Colangeli, K. P. Lyashchenko, X. Zhao, M. Vogelstein, J. S. Spencer, D. N. McMurray, and M. L. Gennaro. 2001. Immunological characterization of antigens encoded by the RD1 region of the Mycobacterium tuberculosis genome. Scand. J. Immunol. 54:448-452. [DOI] [PubMed] [Google Scholar]
- 9.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 10.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLeah, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Soeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 11.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 12.Elhay, M. J., T. Oettinger, and P. Andersen. 1998. Delayed-type hypersensitivity responses to ESAT-6 and MPT64 from Mycobacterium tuberculosis in the guinea pig. Infect. Immun. 66:3454-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, S. V., K. Eiglmeier, T. Garnier, R. Brosch, J. Parkhill, B. Barrell, S. T. Cole, and R. G. Hewinson. 2001. Genomics of Mycobacterium bovis. Tuberculosis (Edinburgh) 81:157-163. [DOI] [PubMed] [Google Scholar]
- 15.Harboe, M., A. S. Malin, H. S. Dockrell, H. G. Wiker, G. Ulvund, A. Holm, M. C. Jorgensen, and P. Andersen. 1998. B-cell epitopes and quantification of the ESAT-6 protein of Mycobacterium tuberculosis. Infect. Immun. 66:717-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hart, P. D. A., and I. Sutherland. 1977. BCG and vole bacillus vaccines in the prevention of tuberculosis in adolescence and early adult life. Br. Med. J. 2:293-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermans, P. W. M., D. van Soolingen, E. M. Bik, P. E. W. De Haas, J. W. Dale, and J. D. A. van Embden. 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, T. B., B. D. Robertson, G. M. Taylor, R. J. Shaw, and D. B. Young. 2000. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates. Yeast 17:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horstkotte, M. A., I. Sobottka, C. K. Schewe, P. Schaefer, R. Laufs, S. Ruesch-Gerdes, and S. Niemann. 2001. Mycobacterium microti llama-type infection presenting as pulmonary tuberculosis in a human immunodeficiency virus-positive patient. J. Clin. Microbiol. 39:406-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwitz, M. A., B. W. Lee, B. J. Dillon, and G. Harth. 1995. Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 92:1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansen, K. A., R. E. Gill, and M. L. Vasin. 1996. Biochemical and molecular analysis of phospholipase C and phospholipase D activity in mycobacteria. Infect. Immun. 64:3259-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jouanguy, E., R. Doffinger, S. Dupuis, A. Pallier, F. Altare, and J. L. Casanova. 1999. IL-12 and IFN-gamma in host defense against mycobacteria and salmonella in mice and men. Curr. Opin. Immunol. 11:346-351. [DOI] [PubMed] [Google Scholar]
- 23.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kremer, K., D. van Soolingen, J. van Embden, S. Hughes, J. Inwald, and G. Hewinson. 1998. Mycobacterium microti: more widespread than previously thought. J. Clin. Microbiol. 36:2793-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy-Frebault, V., and F. Portaels. 1992. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium spp. Int. J. Syst. Bacteriol. 42:315-323. [DOI] [PubMed] [Google Scholar]
- 28.Liebana, E., A. Aranaz, B. Francis, and D. Cousins. 1996. Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 34:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manabe, Y. C., C. P. Scott, and W. R. Bishai. 2002. Naturally attenuated, orally administered Mycobacterium microti as a tuberculosis vaccine is better than subcutaneous Mycobacterium bovis BCG. Infect. Immun. 70:1566-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niemann, S., D. Harmsen, S. Rusch-Gerdes, and E. Richter. 2000. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemann, S., E. Richter, H. Daluegge-Tamm, H. Schlesinger, D. Graupner, B. Koenigstein, G. Gurath, U. Greinert, and S. Ruesch-Gerdes. 2000. Two cases of Mycobacterium microti-derived tuberculosis in HIV-negative immunocompetent patients. Emerg. Infect. Dis. 6:539-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Britzel, D. van Soolingen, Y. Hale, and M. Salfinger. 2002. Rapid and easy-to-perform identification of Mycobacterium tuberculosis complex isolates using PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 34a.Raynaud, C., C. Guilhot, J. Rauzier, Y. Bordat, V. Pelicic, R. Manganelli, I. Smith, B. Gicquel, and M. Jackson. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 45:203-217. [DOI] [PubMed] [Google Scholar]
- 35.Rosenkrands, I., P. B. Rasmussen, M. Carnio, S. Jacobsen, M. Theisen, and P. Andersen. 1998. Identification and characterization of a 29-kilodalton protein from Mycobacterium tuberculosis culture filtrate recognized by mouse memory effector cells. Infect. Immun. 66:2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salamon, H., M. Kato-Maeda, P. M. Small, J. Drenkow, and T. R. Gingeras. 2000. Detection of deleted genomic DNA using a semiautomated computational analysis of GeneChip data. Genome Res. 10:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Songer, J. G. 1997. Bacterial phospholipases and their role in virulence. Trends Microbiol. 5:156-161. [DOI] [PubMed] [Google Scholar]
- 38.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 39.Sula, L., and I. Radkovsky. 1976. Protective effects of M. microti vaccine against tuberculosis. J. Hyg. Epidemiol. Microbiol. Immunol. 20:1-6. [PubMed] [Google Scholar]
- 40.Talbot, E. A., D. L. Williams, and R. Frothingham. 1997. PCR identification of Mycobacterium bovis BCG. J. Clin. Microbiol. 35:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tekaia, F., S. V. Gordon, T. Garnier, R. Brosch, B. G. Barrell, and S. T. Cole. 1999. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 79:329-342. [DOI] [PubMed] [Google Scholar]
- 42.Titball, R. W. 1998. Bacterial phospholipases. Soc. Appl. Bacteriol. Symp. Ser. 27:127-137. [PubMed] [Google Scholar]
- 43.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. van der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Soolingen, D., A. G. M. van der Zanden, P. E. W. De Haas, G. T. Noordhoek, A. Kiers, N. A. Foudraine, F. Portaels, A. H. J. Kolk, K. Kremer, and J. D. A. van Embden. 1998. Diagnosis of Mycobacterium microti infections among humans by using novel genetic markers. J. Clin. Microbiol. 36:1840-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vera-Cabrera, L., M. A. Hernandez-Vera, O. Welsh, W. M. Johnson, and J. Castro-Garza. 2001. Phospholipase region of Mycobacterium tuberculosis is a preferential locus for IS6110 transposition. J. Clin. Microbiol. 39:3499-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wards, B. J., G. W. de Lisle, and D. M. Collins. 2000. An esat6 knockout mutant of Mycobacterium bovis produced by homologous recombination will contribute to the development of a live tuberculosis vaccine. Tuber. Lung Dis. 80:185-189. [DOI] [PubMed] [Google Scholar]
- 47.Wells, A. Q. 1937. Tuberculosis in wild voles. Lancet i:1221.
- 48.Wells, A. Q. 1946. The murine type of tubercle bacillus. Med. Res. Counc. (G. B.) Spec. Rep. Ser. 259:1-42. [Google Scholar]
- 49.Wheeler, P. R., and C. Ratledge. 1992. Control and location of acyl-hydrolysing phospholipase activity in pathogenic mycobacteria. J. Gen. Microbiol. 138:825-830. [DOI] [PubMed] [Google Scholar]