Abstract
Adhesive interactions of cells are critical to tissue integrity. We show that infection with Porphyromonas gingivalis, a major pathogen in the periodontal disease periodontitis, interferes with both cell-matrix and cell-cell adhesion in the oral keratinocyte cell line HOK-16. Thus, infected cells showed reduced adhesion to extracellular matrix, changes in morphology from spread to rounded, and impaired motility on purified matrices in Transwell migration assays and scratch assays. Western blot analysis of P. gingivalis-challenged HOK-16 cells revealed proteolysis of focal contact components (e.g., focal adhesion kinase), adherens junction proteins (e.g., catenins), and adhesion signaling molecules (e.g., the tyrosine kinase SRC). Proteolysis was selective, since important components of adherens junctions (E-cadherin) or signaling molecules (extracellular signal-regulated kinases ERK1/2) were not degraded. The virulence factors gingipains, cysteine proteinases expressed by P. gingivalis, are likely responsible for this proteolytic attack, since they directly digested specific proteins in pull-down experiments, and their proteolytic activity was blocked by the cysteine proteinase inhibitor N-α-p-tosyl-l-lysine chloromethyl ketone and also by a caspase inhibitor. Proteolysis was strain dependent, such that ATCC 33277 and 381 had high proteolytic potential, whereas W50 showed almost no proteolytic activity. These findings may help explain the formation of gingival pockets between cementum and periodontal epithelium, a hallmark of periodontitis. Furthermore, they illustrate a new pathogenetic paradigm of infection whereby bacteria may disrupt the integrity of epithelia.
The junctional epithelium, a specialized tissue consisting of a basal and a suprabasal layer of nonkeratinizing, nondifferentiating oral keratinocytes, mediates the attachment of the gingival epithelium to the enamel tooth surface. By forming a basal lamina, the junctional epithelium provides a substrate that enables oral keratinocytes to adhere both to subepithelial connective tissue and to the tooth via hemidesmosomes (39). The basal lamina is a network of extracellular matrix (ECM) polymers like laminins and type IV collagen and further contains proteoglycans, calcium-binding proteins, and structural and adhesive proteins (45). We have recently detected laminin-5 as a new ECM component of the tooth basal lamina (29).
Keratinocytes bind to laminin-5 via two integrin receptors, α3β1 and α6β4. Integrin α3β1 is involved in the regulation of cell spreading and migration, whereas α6β4, a hemidesmosomal component, anchors keratinocytes tightly to the ECM (6, 7, 15). Integrins, together with other cell surface proteins, mediate the adhesion of epithelial cells to the ECM (18). Upon ligand binding, integrins stimulate a diverse set of signaling molecules to efficiently regulate adhesion, survival, proliferation, differentiation, and migration (19, 36).
Intercellular contacts between epithelial cells are mediated by morphologically distinct adhesion complexes such as tight junctions, gap junctions, desmosomes, and adherens junctions. The last complex contains E-cadherins, transmembrane cell-cell adhesion receptors, which link adjacent cells via calcium-dependent homophilic interactions between extracellular domains. Intracellularly, E-cadherins are linked to the actin cytoskeleton and to signaling cascades through a multiprotein complex consisting mainly of catenins (4, 46, 49).
By maintaining a continuous epithelial attachment around the tooth, the junctional epithelium forms a structural barrier against noxious agents passing from the oral cavity into the tooth-supporting tissue (3, 43). Thus, oral keratinocytes play an important role as the first barrier against offending bacterial agents, which are the cause of periodontal diseases (52). Periodontal diseases, especially periodontitis, represent a group of chronic infectious diseases that lead to inflammation of the gingiva, destruction of periodontal tissue, formation of periodontal pockets, and loss of alveolar bone, with eventual exfoliation of teeth (52). Initiation of periodontal disease is associated with the formation of bacterial plaque on the tooth surface, bringing oral pathogens in direct contact with oral keratinocytes. Tissue destruction is caused both by direct effects of the microorganism and by the inflammatory host response.
In vitro studies have shown that some of these pathogens can bind to and invade oral keratinocytes, where they undergo several rounds of cell division before they exit the host and infect neighboring epithelial cells (28). Whether oral pathogens use the same mechanism in the clinical disease remains to be conclusively established. A major etiological agent of adult periodontitis is Porphyromonas gingivalis, a gram-negative, in vitro-invasive, anaerobic bacterium (5, 28). It is equipped with a repertoire of virulence factors, such as proteinases, fimbriae, lectin-type adhesins, and hemagglutinating factors (33, 42, 50). Based on in vitro studies (33, 42, 50), it can be speculated that these virulence factors may enable P. gingivalis to colonize periodontal pockets in vivo.
Bacterial pathogens use a multitude of strategies, on both the cellular and molecular levels, to interact with mammalian cells and thus influence their hosts (13). In this study, we investigated whether oral bacteria may affect the adhesive contacts of oral keratinocytes with ECM and with each other. Oral keratinocyte-ECM interactions are an important determinant of periodontal tissue organization, self-renewal, remodeling, and repair. Thus, interference with these interactions would have devastating effects on gingival health and may be a strategic target for oral pathogens. Analyzing Actinobacillus actinomycetemcomitans and P. gingivalis, we found that the latter species interferes with several fundamental adhesive and migratory properties of oral keratinocytes in contact with laminin-5. Most probably responsible for these effects are gingipains, bacterially associated proteinases known as potent virulence factors of P. gingivalis (33).
MATERIALS AND METHODS
Bacterial strains and keratinocyte culture.
Bacterial strains A. actinomycetemcomitans Y4, Escherichia coli HB101, and P. gingivalis ATCC 33277, W50, and 381 were cultured as described previously (23, 50). HOK-16 B-BaP-T1, obtained from N.-H. Park (University of California-Los Angeles), is an immortalized oral keratinocyte cell line derived from primary normal human oral keratinocytes (32). Cells were grown in Dulbecco's modified Eagle's medium (4.5 g of glucose/liter) containing 10% fetal calf serum (FCS) and 0.4 μg of hydrocortisone per ml at 37°C with 5% CO2.
Antibodies, extracellular matrix molecules, and reagents.
Rabbit anti-focal adhesion kinase (FAK) immunoglobulin G (IgG) (Pharmingen) was used for immunoprecipitations, and monoclonal anti-FAK antibody (Transduction Laboratories, Lexington, Ky.) was used for Western blotting. Monoclonal antibodies to p130 Crk-associated substrate (CAS), paxillin, and p85 were purchased from Transduction Laboratories. Polyclonal antisera to E-cadherin, ERK1/2, β- and γ-catenin, p120, SRC, and integrin subunits α3 and β4 were from Santa Cruz Biotechnology. Mouse monoclonal anti-α-tubulin antibody was from Sigma, and fluorescein isothiocyanate-conjugated anti-mouse IgG was from Molecular Probes. Laminin-5 deposited by the rat bladder carcinoma cell line 804G was purified as described previously (27). Nα-p-Tosyl-l-lysine chloromethyl ketone (TLCK), camptothecin, and mouse epidermal growth factor (EGF) were purchased from Sigma. Calpeptin and caspase inhibitors I and III were from Calbiochem.
Migration and adhesion assays.
Scratch assays were performed in 24-well plates which were coated overnight at 4°C with 0.4 μg of laminin-5 per ml in phosphate-buffered saline (PBS). HOK-16 cells (4 × 105/well) in migration medium (MM; culture medium without FCS) were seeded and incubated at 37°C for 2 h. Then, cell layers were wounded with a plastic pipette tip and washed three times with MM. The denuded surfaces were recoated with 0.4 μg of laminin-5 per ml in MM for 1 h at 37°C. Cell layers were washed once with MM and were then infected with bacteria at a multiplicity of infection (MOI) of 1,000. Where indicated, EGF at 1 ng/ml was added. Culture plates were incubated for 15 h at 37°C. Photographs of identical locations within each scratch were taken before and 15 h after infection.
In Transwell migration assays, the underside of the filters (8.0-μm pore size; Costar, Cambridge, Mass.) was coated with 0.4 μg of laminin-5 per ml in PBS overnight at 4°C. Filters were washed twice with PBS containing 0.2% Tween 20 (PBST) and then blocked with 5% dry milk in PBST at room temperature for 2 h. HOK-16 cells (1.2 × 105 cells/filter) in MM were incubated in suspension with bacteria at the indicated MOI for 30 min at room temperature before plating on filters which were washed twice with PBS after blocking. EGF (1 ng/ml) was added to the lower chamber only. Cells were maintained at 37°C for 15 h and were then fixed and stained with the LeukoStat kit (Fisher, Pittsburgh, Pa.). The uncoated side of each filter was wiped with a cotton swab to remove cells which had not migrated through the filter. Filters were viewed under bright-field optics to count stained cells in eight fields (with a 20× objective) from each of two filters for each condition. The mean number of cells per field was determined, and results from at least three experiments were expressed as the mean relative cell migration ± standard deviation (SD), with that of nonstimulated cells set at 1.
Adhesion assays were performed as described previously by Goodwin and Pauli (20) with minor modifications. Microtiter 96-well plates were coated with 1 μg of laminin-5 per ml overnight at 4°C, washed twice with PBS, and blocked with 5% dry milk in PBS for 2 h. HOK-16 cells (6 × 104 cells/100 μl/well) in MM were incubated in suspension with bacteria for 1 h at 37°C before seeding in triplicate in wells, which were washed twice with PBS after blocking. Cells were maintained at 37°C for 30 min, and then 2 × 100 μl of Percoll flotation medium (73 ml of Percoll [Pharmacia; density, 1.13 g/ml]), 27 ml of distilled water, and 900 mg of NaCl) were added to each well for 15 min. Adherent cells were fixed for 15 min with 50 μl/well of 25% glutaraldehyde (Sigma), washed with PBS, and stained with crystal violet (0.5% in 20% methanol) for 10 min. Excess dye was washed off with water, and absorbance was measured at 595 nm. Bars represent mean absorbance ± SD of each condition tested in triplicate after subtracting background (cell adhesion to wells blocked with milk). Experiments were done three times.
Immunofluorescence microscopy.
HOK-16 cells in MM were seeded 4 h prior to infection or camptothecin treatment or infected and immediately seeded on laminin-5-coated (0.4 μg/ml) glass coverslips. Infected or camptothecin-treated HOK-16 cells were incubated for various times at 37°C before they were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. The following steps were all performed at room temperature. Cells were permeabilized with 0.1% Triton X-100 in PBS for 20 min, and the fixing step was repeated, followed by blocking in 5% FCS in PBS for 20 min. Cells were then incubated with mouse monoclonal anti-α-tubulin antibody (Sigma) in 2.5% FCS in PBS (FCS-PBS) for 2 h. Cells were washed three times with FCS-PBS and incubated with fluorescein isothiocyanate-conjugated anti-mouse IgG (Molecular Probes) in FCS-PBS. After 1 h, cells were washed two times with FCS-PBS and once with PBS and mounted on glass slides. Samples were analyzed with an Axiovert S100 Zeiss immunofluorescence microscope fitted with an automated XY stage and an Axiocam color digital camera.
Survival and apoptosis assay.
HOK-16 cells (1.2 × 105 cells/well) in MM were incubated in suspension with bacteria at the indicated MOI for 30 min at room temperature before plating in 24-well plates coated with 0.4 μg of laminin-5 per ml. Cells were incubated at 37°C for 15 h and then washed with PBS. Adherent cells were detached by trypsinization (150 μl of trypsin per well). Culture medium (400 μl) containing trypan blue (20 μl) was added to each well, and the cells were evenly distributed. Settled cells that excluded trypan blue were counted in eight fields per well, and the mean number of cells was determined. Survival experiments were done three times, and the results are expressed as mean relative cell survival ± SD, with the value for noninfected surviving cells set as 1.
HOK-16 cells in MM were seeded on laminin-5-coated (0.4 μg/ml) glass coverslips 4 h prior to camptothecin treatment. Then, cells were incubated with 4 μM camptothecin or vehicle (dimethyl sulfoxide). After various times, cells were fixed, and either permeabilized and apoptotic cells were stained with the Apoptag peroxidase in situ apoptosis detection kit (Intergen Company, Purchase, N.Y.), or cells were subjected to tubulin immunofluorescence staining.
Proteolysis studies and Western blotting.
Proteolysis due to P. gingivalis infection was analyzed in HOK-16 cells seeded in serum-free medium 3 h prior to infection. Where indicated, 2 h before infection, calpeptin (50 μg/ml), caspase inhibitor I or III (both at 20 μM), or dimethyl sulfoxide was added. Infected HOK-16 cells (an MOI of 1,000) were incubated at 37°C for the indicated times before lysis in 40 mM Tris (pH 7.5)-150 mM NaCl-1% Triton X-100-1% sodium deoxycholate-0.1% sodium dodecyl sulfate (SDS)-10% glycerol-6 mM EDTA-100 mM NaF-1 mM sodium orthovanadate-10 mM sodium pyrophosphate-1 mM phenylmethylsulfonylfluoride-10 μg of leupeptin per ml-1 tablet/50 ml of complete protease inhibitor cocktail (Roche Diagnostics Corporation). Protein concentrations were determined with the DC protein assay (Bio-Rad), and samples were prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Equal amounts of total protein were separated by SDS-PAGE and blotted, and blots were incubated with the primary antibodies diluted 1:2,000 in 5% milk in PBS containing 0.1% Tween 20. Blots were washed extensively in 5% milk in PBS containing 0.1% Tween 20, and the secondary antibody was diluted 1:2,000. After several washes, proteins were visualized with the ECLplus system (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom).
To study proteolysis of immunoprecipitated proteins, HOK-16 cells were lysed in the buffer described above, lysates were centrifuged at maximum speed for 10 min, and antibodies were added to the supernatant for 3 h at 4°C on a rotary shaker. Immunocomplexes were collected with protein-G Sepharose for 1 h at 4°C and then washed four times with ice-cold PBS. Where indicated, equal aliquots of the captured immunocomplexes were incubated with vehicle (dimethyl sulfoxide), TLCK (150 μg/ml), or caspase inhibitor I (100 μM) for 30 min on ice. Then, immunocomplexes were exposed to 107 bacterial cells (P. gingivalis ATCC 33277) at 37°C for the indicated times. Reactions were terminated by boiling in 2× SDS-PAGE sample buffer for 5 min. SDS-PAGE and Western blotting were performed as described above.
RESULTS
Oral keratinocyte adhesion and spreading are inhibited by P. gingivalis.
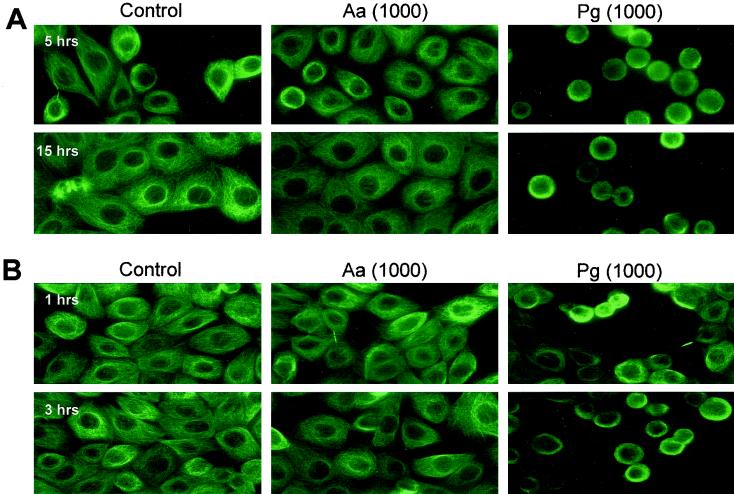
To study whether oral pathogens affect the adhesion and migration properties of oral keratinocytes, we analyzed the human oral keratinocyte cell line HOK-16 and focused on the invasive organisms A. actinomycetemcomitans Y4 and P. gingivalis ATCC 33277. E. coli HB101 acted as a nonoral, noninvasive control. HOK-16 cells plated on laminin-5-coated glass coverslips in the presence of pathogens at an MOI of 1,000 were fixed at distinct times after plating, and the microtubule cytoskeleton was stained (Fig. 1). Both bacteria-free and A. actinomycetemcomitans-infected HOK-16 cells were completely adhered and spread between 2 and 5 h from seeding (Fig. 1A). In contrast, P. gingivalis-infected HOK-16 cells, although tethered to laminin-5, were unable to spread even after 15 h of incubation (Fig. 1A).
FIG. 1.
P. gingivalis blocks spreading on laminin-5 and loosens keratinocyte-ECM and keratinocyte intercellular interactions. (A) HOK-16 cells infected with A. actinomycetemcomitans (Aa) or P. gingivalis (Pg) ATCC 33277 at an MOI of 1,000 were seeded on laminin-5-coated glass coverslips for the indicated times before immunostaining with an anti-α-tubulin antibody. Controls were bacteria free. (B) Four hours after seeding on laminin-5, HOK-16 cells were infected at an MOI of 1,000 and further incubated for the indicated times. Microtubules were visualized by immunofluorescence staining. Controls were bacteria free.
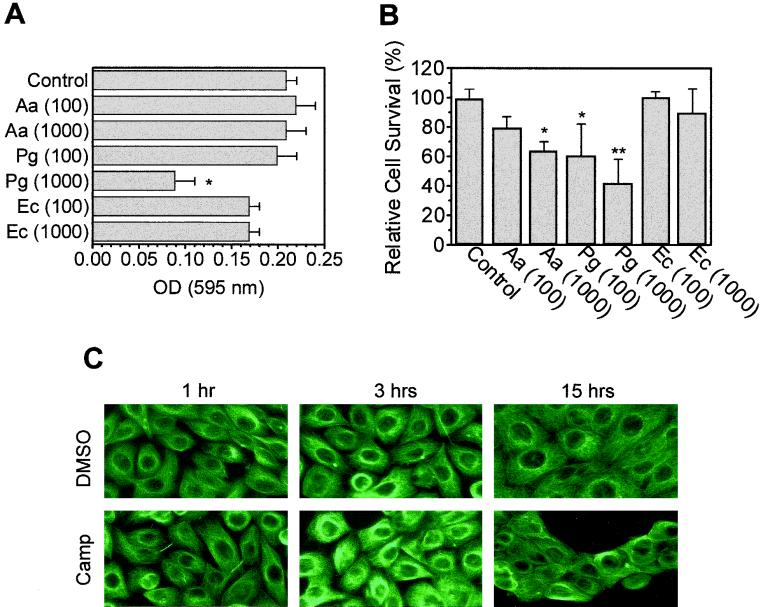
If HOK-16 cells were allowed to adhere and spread on laminin-5 prior to bacterial infection, addition of P. gingivalis but not A. actinomycetemcomitans caused cell rounding within 1 h, and almost all oral keratinocytes were affected after 3 h (Fig. 1B). In adhesion assays, HOK-16 cells preincubated with P. gingivalis at an MOI of 1,000 reduced HOK-16 cell adhesion to laminin-5 more than 50%, whereas other bacteria tested (E. coli and A. actinomycetemcomitans) had either a minor or no effect on HOK-16 cell adhesion (Fig. 2A).
FIG. 2.
P. gingivalis blocks keratinocyte adhesion to laminin-5. (A) Adhesion assays with HOK-16 cells infected with A. actinomycetemcomitans (Aa), E. coli (EC), or P. gingivalis (Pg) ATCC 33277 at an MOI of 100 or 1,000. Infected cells were incubated for 1 h at 37°C before aliquots were seeded in triplicate in laminin-5-coated (1 μg/ml) 96-well plates. Cells were allowed to adhere for 30 min, then nonadherent cells were removed, and adherent cells were fixed and stained. Results are mean absorbance at 595 nm ± SD for one of four experiments. Controls were bacteria free. *, significantly different from control value (P < 0.05, Student's t test). (B) Survival assay with HOK-16 cells. Cells were infected at the indicated MOI and seeded in laminin-5-coated 24-well plates. After 15 h, cells were washed and trypsinized, and cells excluding trypan blue were counted in each well. Results are expressed as mean relative cell survival ± SD (n = 3), with survival of bacteria-free HOK-16 cells (control) set at 100%. *, significantly different from control value at P < 0.1; **, significantly different from control value at P < 0.05 (Student's t test). (C) Apoptosis in HOK-16 cells was induced by 4 μM camptothecin (Camp) 4 h after seeding on laminin-5-coated coverslips. Controls were treated with vehicle (dimethyl sulfoxide [DMSO]). At the indicated times, cells were subjected to immunostaining with an anti-α-tubulin antibody. Induction of apoptosis was confirmed in parallel samples and was detected as early as 3 h after camptothecin treatment (data not shown). Note the detachment in sheets observed at 15 h and the lack of morphological changes such as those induced by P. gingivalis (Fig. 1).
Reduced oral keratinocyte adhesion to laminin-5 triggered by P. gingivalis could be indirect and caused by a loss in cell viability. Therefore, we investigated whether the two oral pathogens could affect the survival of oral keratinocytes in contact with laminin-5. At an MOI of 1,000, both A. actinomycetemcomitans and P. gingivalis decreased the number of surviving HOK-16 cells on laminin-5 to about 60% and 40% of the noninfected control value, respectively (Fig. 2B). These results are consistent with previous reports that these bacteria can induce apoptosis in lymphocytes and macrophages (17, 24, 31), but they do not explain the selective inhibition of cell spreading by P. gingivalis, since both strains induced similar losses in cell viability. Furthermore, HOK-16 cells treated with 4 μM camptothecin, a selective inducer of apoptosis (26), either spread on laminin-5 or completely lost contact with ECM, but they were not tethered to laminin-5 like P. gingivalis-infected cells (Fig. 2C). These findings suggest that P. gingivalis-induced loss of HOK-16 cell adhesion to laminin-5 was triggered by a mechanism other than cell death.
Altogether, these results indicated that P. gingivalis inhibits initiation of cell spreading on ECM and that it reverses and loosens ECM contacts and possibly cell-cell adhesion in adhered and spread oral keratinocytes.
Oral keratinocyte migration is inhibited by P. gingivalis.
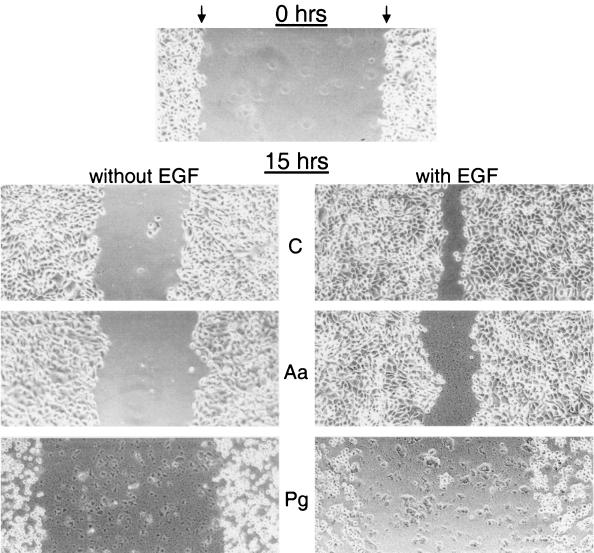
Since the dramatic negative effect of P. gingivalis on HOK-16 cell adhesion on laminin-5 is expected to also influence cell migration, we analyzed the motility of infected HOK-16 cells with two different assay systems. In the scratch migration assay, considered an in vitro model for keratinocyte migration during wound healing or reepithelialization (51), P. gingivalis-infected HOK-16 cells showed remarkably impaired migration on laminin-5. Furthermore, dramatic changes in cell morphology were observed (Fig. 3, left panels). Scratch closure on laminin-5 was also partially inhibited by A. actinomycetemcomitans (Fig. 3, left panels). In the presence of EGF, a well-documented chemoattractant involved in wound healing (51), at 1 ng/ml, migration of control cells on laminin-5 was markedly stimulated (Fig. 3, right panels). This EGF-stimulated motility was likewise blocked by P. gingivalis and A. actinomycetemcomitans infection, but again, P. gingivalis alone caused reduced contact with the ECM and a change in morphology from well spread to rounded (Fig. 3, right panels).
FIG. 3.
A. actinomycetemcomitans and P. gingivalis inhibit oral keratinocyte scratch closure on laminin-5. Scrape wounds were made in serum-free HOK-16 cell cultures infected with A. actinomycetemcomitans (Aa) or P. gingivalis (Pg) plated on laminin-5. Bacteria were added at an MOI of 1,000. Controls (C) were bacteria free. Wounded cultures were allowed to reepithelialize for 15 h in the absence (left panels) or the presence (right panels) of EGF (1 ng/ml). Micrographs show wounded, nonfixed cell layers at 0 h and 15 h after treatment. Arrows indicate the edges of denuded surface (space between arrows).
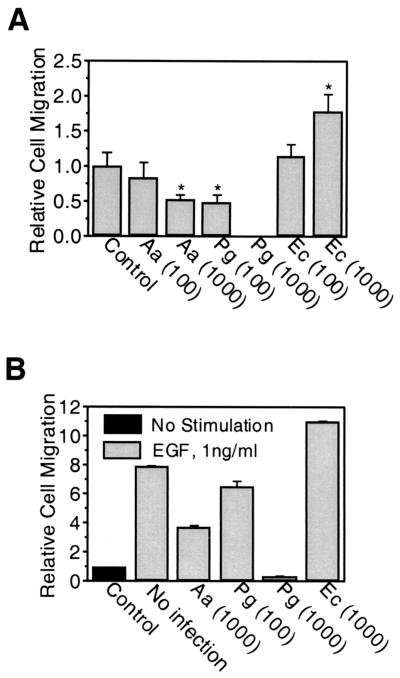
Transwell chamber assays allow a more quantitative analysis of effects on cell motility. Thus, HOK-16 cell migration on laminin-5 was studied in the presence of P. gingivalis, A. actinomycetemcomitans, or E. coli. The two oral strains both showed dose-dependent inhibition of HOK-16 cell spontaneous migration (i.e., in the absence of serum or any other stimulus). However, A. actinomycetemcomitans had only mild effects (up to 50% inhibition), whereas P. gingivalis blocked migration completely at an MOI of 1,000 (Fig. 4A). In contrast, the nonoral species E. coli had a stimulatory effect on HOK-16 cell migration (Fig. 4A). Medium conditioned by P. gingivalis- or A. actinomycetemcomitans-infected HOK-16 cells had no inhibitory effect on HOK-16 cell migration (data not shown), indicating that the decrease in migration activity was not triggered by a soluble factor released either by the pathogen or by the host. Treatment with EGF induced an eightfold increase in HOK-16 cell migration on laminin-5 (Fig. 4B). Similar to spontaneous migration, chemotaxis (migration induced by soluble molecules) was inhibited in a dose-dependent manner by both P. gingivalis and A. actinomycetemcomitans, whereas E. coli further enhanced chemotactic migration (Fig. 4B).
FIG. 4.
A. actinomycetemcomitans and P. gingivalis inhibit oral keratinocyte Transwell migration on laminin-5. (A) Migration assays performed in Transwell chambers with laminin-5-coated filters. HOK-16 cells were infected with A. actinomycetemcomitans (Aa), E. coli (Ec), or P. gingivalis (Pg) at an MOI of 100 or 1,000 and seeded in the upper chamber. After 15 h, cells which had migrated from the upper chamber through the filter to the lower chamber were counted. Results are expressed as mean relative cell migration ± SD (n = 3), with the value for noninfected cells (control) set at 1. *, significantly different from control value (P < 0.05, Student's t test). (B) Transwell migration assays performed as in panel A except that EGF was present in the lower chamber at 1 ng/ml. Results are expressed as mean relative cell migration ± SD (n = 3), with the value for noninfected, nonstimulated cells (control) set at 1. All values were significantly different from the EGF no-infection value (P < 0.05, Student's t test).
Discrete proteolysis of focal contact and adherens junction proteins is induced by P. gingivalis.
The observed loss of cell-ECM and cell-cell interactions in HOK-16 cells could be due to several mechanisms, e.g., stimulation of host signaling pathways upon adhesion to the host and invasion of the host by P. gingivalis, direct interaction of invaded bacteria with the host cytoskeleton, and bacterial proteolysis of host proteins involved in cell-ECM and cell-cell interactions. Since P. gingivalis is well known for its high protease activity (33), we started to investigate whether oral keratinocyte proteins are proteolytically processed upon bacterial challenge.
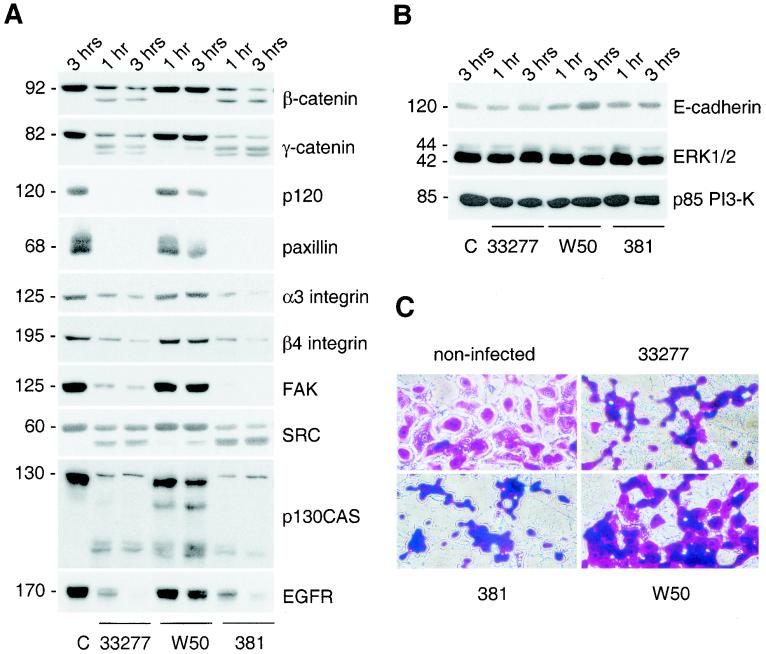
Total cell lysates of HOK-16 cells infected for various times with P. gingivalis were analyzed by immunoblotting of equal amounts of total protein. As shown in Fig. 5A and quantified in Table 1, several structural components of cell-cell contacts (β-catenin, γ-catenin, and p120), focal contacts (paxillin, α3 integrin subunit), and hemidesmosomes (β4 integrin subunit) underwent discrete proteolysis in oral keratinocytes infected with P. gingivalis strain ATCC 33277. Furthermore, several signaling molecules (FAK, SRC, and p130CAS) involved in the regulation of cell-ECM and cell-cell interactions (19, 46) were proteolytically processed upon bacterial challenge (Fig. 5A). The receptor for EGF was also proteolytically attacked. Proteolysis was apparent as early as 30 min after infection (earliest time point analyzed; data not shown). In contrast, no proteolysis was detectable in HOK-16 cells infected with A. actinomycetemcomitans for up to 3 h (data not shown).
FIG. 5.
P. gingivalis invasion induces discrete proteolysis of select focal contact and adherence junction components. (A and B) HOK-16 cells were seeded 3 h prior to infection with the indicated P. gingivalis strain (ATCC 33277, W50, or 381) at an MOI of 1,000. Infected cells were further incubated for the indicated times before lysis. Equal amounts of total protein were analyzed by Western blotting. Control cells (lane C) were bacteria-free HOK-16 cells. (C) HOK-16 cells were seeded 3 h prior to infection with the indicated P. gingivalis strain (ATCC 33277, W50, or 381) at an MOI of 1,000. Three hours after infection, cells were fixed and stained with crystal violet.
TABLE 1.
P. gingivalis invasion induces discrete proteolysis of select focal contact and adherence junction componentsa
| Strain | Time of addition (h) | % Reduction vs control
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Catenin | γ-Catenin | p120 | paxillin | α3 integrin | β4 integrin | FAK | SRC | p130CAS | EGF receptor | ||
| ATCC 33277 | 1 | 52.6 | 67.0 | 99.8 | 96.9 | 56.4 | 75.6 | 88.3 | 39.0 | 95.5 | 67.8 |
| 3 | 69.3 | 74.1 | 99.7 | 98.3 | 73.9 | 93.3 | 92.8 | 31.9 | 97.2 | 99.1 | |
| W50 | 1 | 9.1 | 0 | 0 | 18.7 | 6.8 | 0 | 16.9 | 0 | 40.9 | 15.6 |
| 3 | 15.8 | 0 | 37.63 | 43.3 | 3.1 | 6.1 | 2.7 | 97.3 | 57.4 | 36.4 | |
| 381 | 1 | 67.7 | 78.6 | 97.9 | 98.8 | 71.2 | 66.8 | 98.0 | 57.2 | 94.1 | 64.5 |
| 3 | 85.6 | 82.7 | 98.9 | 99.2 | 88.0 | 87.7 | 99.2 | 64.0 | 90.3 | 90.1 | |
HOK-16 cells were treated and infected with P. gingivalis strain ATCC 33277, W50, or 381 as described for Fig. 5A. At different time points, cells were lysed, and equal amounts of total protein were analyzed by Western blotting (see Fig. 5A). The Western blot signals from Fig. 5A were quantified by densitometry. Values represent reduction from control values, with control signals set at 100%.
P. gingivalis-induced proteolysis was selective, since proteins such as E-cadherin, the regulatory subunit p85 of phosphoinositide 3-kinase (PI3-K), and extracellular signal-regulated kinases 1 and 2 (ERK1 and -2) were not affected (Fig. 5B). Furthermore, proteolytic capacity varied in different P. gingivalis strains tested, as W50 caused almost no proteolysis, whereas 381 was as effective as ATCC 33277 (Fig. 5A and Table 1). These variations might be due to strain-dependent differences in the capacity to attach and possibly invade oral keratinocytes (23) (see Discussion).
In accordance with the proteolysis data, only a small number of HOK-16 cells rounded up upon W50 challenge compared to cells infected by ATCC 33277 or strain 381 (Fig. 5C). These data further suggest that attachment and invasion of oral keratinocytes by P. gingivalis is associated with selective protein degradation, resulting in reduced cell-ECM and cell-cell adhesion and thus migration defects. Medium conditioned by ATCC 33277-infected HOK-16 cells and bacterial spent medium had no inhibitory effect on HOK-16 cell migration (data not shown), indicating either that soluble bacterial proteinases were not involved in the observed process or that their proteolytic activity was not high enough to cause protein degradation within the tested time frame. Therefore, either proteolytic activity originates from P. gingivalis membranes or the bacterial attachment and invasion processes as such trigger activation of a host proteinase(s).
Since P. gingivalis uptake into host cells might be integrin dependent and triggers Ca2+ release (50), oral keratinocyte infection might lead to integrin-mediated stimulation of calpains, a family of calcium-dependent cysteine proteinases which trigger limited proteolysis of various focal contact structural proteins and signaling enzymes, regulating platelet morphology and aggregation (38, 55) or T-cell adhesion and spreading (35). Therefore, we next investigated whether the observed proteolysis was triggered by calpains. To this end, HOK-16 cells were incubated with the calpain-specific inhibitor calpeptin (50 μg/ml) before infection with ATCC 33277. Cells were lysed at different times, and equal amounts of total lysate protein were analyzed by Western blotting, testing for the same proteins as in Fig. 5A. However, calpeptin did not block proteolysis in HOK-16 cells infected with ATCC 33277 (data not shown), indicating that other proteinases, of host or bacterial origin, are responsible for proteolysis in P. gingivalis-infected HOK-16 cells.
Protein degradation in oral keratinocytes is triggered by P. gingivalis-associated proteinase activity.
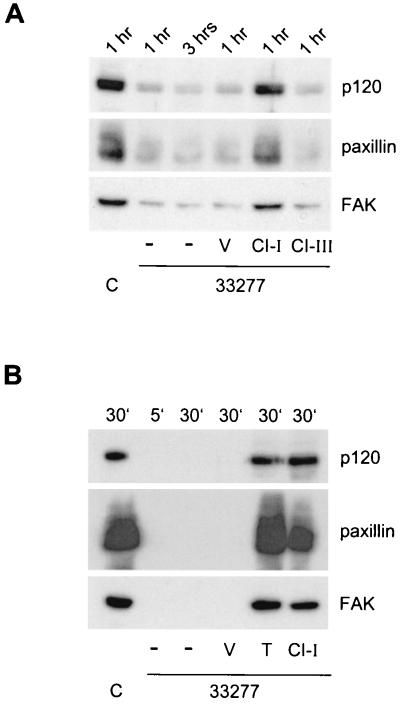
P. gingivalis-expressed cysteine proteinases have either Lys-Xaa or Arg-Xaa specificity (34). Interestingly, the Arg-specific gingipain catalytic domain shows topological similarities to caspase-1 (12). To test whether the observed proteolysis in HOK-16 cells was triggered by a caspase-like proteinase activity, we incubated seeded HOK-16 cells with caspase inhibitor I or III (20 μM) before infection with ATCC 33277. After different times, cells were lysed, and total cell lysates were subjected to Western blot analysis, testing three of the formerly proteolysed proteins, p120, paxillin, and FAK (Fig. 6A). Compared with dimethyl sulfoxide-treated control cells, proteolysis of all three proteins was reduced by caspase inhibitor I but not by caspase inhibitor III (Fig. 6A and Table 2). Caspase inhibitor I is highly specific for caspase-1, caspase-3, caspase-4, and caspase-7, whereas caspase inhibitor III is a broad-spectrum inhibitor with no specificity for a particular caspase. Thus, ATCC 33277-induced proteolysis is partially triggered by caspase-like proteinase activity of host or bacterial origin.
FIG. 6.
Proteolysis is triggered by P. gingivalis-associated proteinase activity. (A) HOK-16 cells were seeded 3 h prior to infection with P. gingivalis ATCC 33277 (MOI of 1,000). One hour after seeding, cells were treated with caspase inhibitor I (CI-I), caspase inhibitor III (CI-III), or dimethyl sulfoxide (vehicle, V). Controls (C) were bacteria free. Infected cells were further incubated for the indicated times before lysis. Equal amounts of total protein were analyzed by Western blotting with antibodies to p120, paxillin, or FAK. (B) HOK-16 cell proteins were immunoprecipitated, washed, and incubated at 37°C with 107 P. gingivalis ATCC 33277 cells for 5 min or 30 min. Reactions were stopped and prepared for SDS-PAGE, and samples were analyzed by Western blotting. Controls (C) were bacteria free. Where indicated, reaction mixtures were incubated with dimethyl sulfoxide (vehicle, V), TLCK (T, 150 μg/ml), or caspase inhibitor I (CI-I, 20 μM) before incubation with ATCC 33277.
TABLE 2.
Proteolysis is triggered by P. gingivalis-associated proteinase activitya
| Treatment | Duration | % Reduction vs control
|
||
|---|---|---|---|---|
| p120 | paxillin | FAK | ||
| None | 1 h | 83.2 | 74.7 | 85.2 |
| DMSO | 1 h | 87.2 | 83.9 | 88.4 |
| CI-I | 1 h | 19.9 | 22.2 | 31.8 |
| CI-III | 1 h | 84.6 | 84.1 | 82.1 |
| None | 5 min | 100 | 100 | 100 |
| None | 30 min | 100 | 100 | 100 |
| TLCK | 30 min | 0 | 0 | 0 |
| CI-I | 30 min | 0 | 24.9 | 0 |
HOK-16 cells were treated with vehicle, caspase inhibitor I (CI-I), or caspase inhibitor III (CI-III) and infected with P. gingivalis strain ATCC 33277 as described for Fig. 6A. At different time points, cells were lysed, and equal amounts of total protein were analyzed by Western blotting (see Fig. 6A), or HOK-16 proteins were immunoprecipitated, incubated with caspase inhibitor I or TLCK, and then challenged with ATCC 33277, as described for Fig. 6B. Samples were analyzed by Western blotting. Western blot signals from Fig. 6A and 6B were quantified by densitometry. Values represent reduction from control (bacteria free) values, with the control signal set at 100%.
To directly assess the potential of bacterially associated proteinases to cleave oral keratinocyte proteins, we used a keratinocyte-free system. To this end, immunoprecipitated HOK-16 cell proteins were incubated with P. gingivalis ATCC 33277 for different times, followed by Western blot analysis. As shown in Fig. 6A, all proteins tested (p120, paxillin, and FAK) were extensively degraded within 5 min. The cysteine proteinase inhibitor TLCK efficiently blocked proteolysis induced by P. gingivalis, as expected. Importantly, caspase inhibitor I was also able to block proteolysis, to almost the same extent as TLCK (Table 2). Thus, the caspase-like proteinase activity which led to proteolysis in HOK-16 cells (Fig. 6A) seems to be of bacterial origin and not host related. The fact that inhibition by caspase inhibitor I was not complete may be due to the fact that an additional noncaspase-like proteinase activity may also be involved, or the inhibitor may be less active with caspase-like proteinases.
These findings demonstrate that a P. gingivalis cysteine proteinase(s) can degrade intracellular oral keratinocyte proteins independent of host proteolytic activity and further support the possibility that such activity may be responsible for P. gingivalis-triggered reductions in the adhesiveness and motility of HOK-16 cells on laminin-5.
DISCUSSION
We studied the effects of periodontal pathogens on oral keratinocyte adhesive interactions with laminin-5, an ECM component that acts as a potent substrate for both adhesion and migration of epithelial cells (21). Laminin-5 is located at the interface between gingival and tooth surfaces, at the site of the internal basement membrane. The striking aspect of our data is that adhesion of the oral keratinocyte cell line HOK-16 to laminin-5 was decreased by more than 50% when cells were infected by P. gingivalis, whereas invasion by A. actinomycetemcomitans had no inhibitory effect on cell adhesion. The P. gingivalis-triggered decrease in adhesiveness was accompanied by a marked change in cell morphology from well spread to loosely tethered to the substrate. P. gingivalis not only inhibited initiation of cell spreading on ECM, but also reversed and loosened cell-ECM and cell-cell interactions in HOK-16 cells firmly adhered to laminin-5. None of these effects was induced by A. actinomycetemcomitans.
Our data suggest the possibility that P. gingivalis may play a role not only in inducing junctional epithelium detachment in wounded, regenerating, or healing tissue, but also in initiating detachment of healthy tissue. These possibilities remain to be tested experimentally, especially because HOK-16 cells are derived from gingival tissue and as such may represent a mixture of keratinocytes originating from junctional epithelium, gingival epithelium, and sulcular epithelium.
Loss of adhesion to ECM affects cell survival and motility. Thus, we determined the number of surviving HOK-16 cells and analyzed their migratory behavior after bacterial challenge. Infection of HOK-16 cells with A. actinomycetemcomitans caused about a 40% decrease in surviving cells compared to a 50% inhibition of Transwell migration. The smaller number of migrating cells in the presence of A. actinomycetemcomitans might well be due to increased HOK-16 cell death upon bacterial infection. However, the reduced number of surviving cells is unlikely to be the sole reason for the P. gingivalis-triggered reduction in motility, since the proportion of dead cells (about 60%) was greatly exceeded by the percentage of inhibition (100%) in Transwell migration assays.
Both spontaneous and chemotactic migration of HOK-16 cells was impaired or completely inhibited in the presence of A. actinomycetemcomitans or P. gingivalis, respectively. In periodontitis, it was proposed that epithelial cells take on a migratory phenotype, penetrating periodontal connective tissue in an apical direction (48). Our results suggest that, in the presence of P. gingivalis, this mechanism may not occur, since motility is inhibited by this bacterium. However, it is possible that the P. gingivalis-induced detachment may simulate a cell migratory effect as a wave of detaching cells progresses in the apical direction. Inhibition of oral keratinocyte motility by oral pathogens would result in decreased wound healing, leaving the wound surface unprotected from further infections and thus preventing optimal periodontal tissue regeneration. It still remains to be seen how other bacteria may contribute to modifying the adhesion and migration of oral keratinocytes, since a diverse microbial population is present in periodontal pockets.
Which molecular events induced by P. gingivalis make HOK-16 cells lose their adhesion to neighboring cells and to the ECM? We focused on the most important virulence factor of P. gingivalis, its proteolytic activity, mediated by gingipains (33). Both Arg-gingipain and Lys-gingipain enzymes occur in soluble and membrane-associated forms and are responsible for 85% of the overall proteolytic activity (30, 33, 42). In our system, soluble proteinases appeared to have no role, since neither P. gingivalis spent medium nor conditioned medium of P. gingivalis-infected HOK-16 cells had any effect on cell migration. Membrane-bound forms of gingipains have the potential to proteolyze both extracellular and intracelluar oral keratinocyte proteins, since P. gingivalis was shown to invade the cytoplasm of its host cells in vitro (5).
Indeed, our results show that several structural and regulatory adhesive molecules are proteolytically cleaved in P. gingivalis-infected HOK-16 cells. Cleavage of these molecules seems to be responsible for the adhesion defects, because they are part of complexes that establish and regulate adhesion. For instance, major targets of gingipains are catenins, which are found at cell-cell adherens junctions, where they anchor the cadherin complex to the actin cytoskeleton (4). Some members of the catenin family (β-catenin and p120) have additional signaling roles in the cytoplasm and the nucleus during development and cancer (1, 56). Cadherins act as transmembrane homophilic receptors in adherens junctions. Even though E-cadherin was not proteolysed in P. gingivalis-infected cells, adherens junctions could not be supported any longer because the catenins were missing (4, 46). Therefore, our data are consistent with the possibility that P. gingivalis may actively disrupt cell-cell adhesion.
P. gingivalis induced proteolysis of several components of focal contacts, which are essential for adhesion and migration over ECM. Perhaps a critical component proteolysed is paxillin, a protein acting as a molecular adapter and providing multiple docking sites at the plasma membrane for an array of signaling and structural proteins of focal contacts (47). Though many other focal contact molecules were proteolytically attacked, damaging paxillin may by itself explain the ECM adhesion defect we observed. Transmembrane proteins like integrins also underwent proteolysis upon P. gingivalis challenge. Integrin subunits α3 and β4, components of focal contacts and hemidesmosomes, respectively, regulate the adhesion and migration of epithelial cells on laminin-5 (21). The receptor for EGF, a chemotactic factor which plays an important role in the regulation of proliferation and motility of epithelial cells, was also proteolysed.
In addition to the loss of structural components, it is likely that dysregulation of adhesion and migration plays an important role in these defects, since several signaling molecules (CAS, FAK, and SRC) which are involved in the regulation of cell-ECM and cell-cell interactions (37) were proteolytically attacked. Baron and Schwartz reported ubiquitination and degradation of the platelet-derived growth factor receptor as a result of lost cell-ECM interactions (2). However, when in parallel experiments the level of FAK in suspended versus attached cells was analyzed, no difference in FAK protein levels was detected (2). Thus, loss of HOK-16 cell adhesion by itself could trigger proteolysis, but this would probably be less extensive than the proteolysis observed in our study. In addition, suspended cells show low activities of the Rho-like GTPases, ERK1/2, FAK, and SRC (22, 40), indicating that loss of cell-ECM interactions triggers termination of cell signals and protein activities rather than stimulation of degradation.
Furthermore, our results correlate with the findings that P. gingivalis proteinases caused loss of fibronectin and its main integrin receptor, α5β1, and of CD14 in human gingival fibroblasts (41, 44) and degraded cadherins, β1 integrin subunit, and occludin in canine kidney cells (25). Also, P. gingivalis biofilms have been shown to degrade cytokines (14). Furthermore, protease-active P. gingivalis protein preparations induced N-cadherin proteolysis and loss of cell adhesion and activated components of the complement system, matrix metalloproteinases, and platelets (8-11, 53). Thus, the importance of P. gingivalis proteolytic activity was recognized some time ago. Our studies address this proteolytic activity in the context of cell-cell and cell-ECM interactions to clarify the effects of P. gingivalis-induced proteolysis on important cellular mechanisms of cell migration and cell adhesion.
Importantly, P. gingivalis-induced proteolysis was selective. Proteolytic selectivity may depend upon the availability of target proteins in the cytoplasm, proteinase specificity (Arg-gingipain versus Lys-gingipain), or the cytoplasmic location of gingipains. Among the molecules spared, p85 PI3-K and ERK1/2 are particularly significant. PI3-K, consisting of a regulatory p85 domain and a catalytic p110 domain, plays an important role in mitogenic signaling and cell survival, cytoskeletal remodeling, metabolic control, and vesicular trafficking (54). ERK1/2, mitogen-activated protein kinases 1/2, are involved in the regulation of cell growth, differentiation, and motility (16). The fact that p85 PI3-K and ERK1/2 were not proteolytically attacked suggests that P. gingivalis infection selectively disables some cellular functions but not others. It is tempting to speculate that if these mechanisms of intracellular infection and resulting adhesion disabling occur in vivo, they may result in a strategy whereby P. gingivalis alters rather than kills epithelial cells in ways that make them vulnerable to further infection and bacterial propagation.
Severe host protein processing and morphological changes occurred only in the presence of strains 381 and ATCC 33277, whereas strain W50 was relatively inactive in comparison, within the experimental time frame. As published by Huang et al. (23), bacterial strain W50 attached much less effectively (0.12% of 381 and 0.47% of ATCC 33277 attachment) and showed less invasion (0.58% of 381 and 1.35% of ATCC 33277 invasion) than 381 and ATCC 33277. Thus, W50 is far less adhesive and invasive than 381 and ATCC 33277, indicating a possible correlation between the ability of an individual strain to adhere and invade a host and its capacity to proteolyse host proteins. Furthermore, studies with caspase inhibitor I indicate that the proteolytic activity responsible for oral keratinocyte protein processing is associated with the oral pathogen rather than with the host. Taken together, our results strongly indicate that P. gingivalis, via a bacterial proteinase(s), can trigger loss of oral keratinocyte adhesion by degrading host proteins which are involved in the regulation of cell-cell and cell-ECM interactions.
A hallmark of bacterially induced periodontal disease is the progressive detachment of the junctional epithelium from the tooth surface, leading to periodontal pockets. Our results provide a possible molecular explanation for how oral keratinocyte adhesion and migration can be manipulated by bacterial pathogens. These results suggest a novel pathogenic mechanism whereby P. gingivalis may cause disorganization of infected epithelia. However, these findings have not been tested in an in vivo model system. Thus, the significance of these proposed pathogenetic mechanisms in oral health remains to be determined by further studies.
Acknowledgments
We thank Lana Mossberger for secretarial assistance and Peter Polverini for useful comments.
This study was supported by NIH grant GM46902 to V.Q.
Editor: B. B. Finlay
REFERENCES
- 1.Anastasiadis, P. Z., S. Y. Moon, M. A. Thoreson, D. J. Mariner, H. C. Crawford, Y. Zheng, and A. B. Reynolds. 2000. Inhibition of RhoA by p120 catenin. Nat. Cell Biol. 2:637-644. [DOI] [PubMed] [Google Scholar]
- 2.Baron, V., and M. A. Schwartz. 2000. Cell adhesion regulates ubiquitin-mediated degradation of the platelet-derived growth factor receptor beta. J. Biol. Chem. 275:39318-39323. [DOI] [PubMed] [Google Scholar]
- 3.Bartold, P. M., L. J. Walsh, and A. S. Narayanan. 2000. Molecular and cell biology of the gingiva. Periodontology 24:28-55. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, J. 1999. Cadherins and catenins: role in signal transduction and tumor progression. Cancer Metast. Rev. 18:15-30. [DOI] [PubMed] [Google Scholar]
- 5.Belton, C. M., K. T. Izutsu, P. C. Goodwin, Y. Park, and R. J. Lamont. 1999. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell. Microbiol. 1:215-223. [DOI] [PubMed] [Google Scholar]
- 6.Borradori, L., and A. Sonnenberg. 1999. Structure and function of hemidesmosomes: more than simple adhesion complexes. J. Investig. Dermatol. 112:411-418. [DOI] [PubMed] [Google Scholar]
- 7.Carter, W. G., P. Kaur, S. G. Gil, P. J. Gahr, and E. A. Wayner. 1990. Distinct functions for integrins α3β1 in focal adhesions and α6β4/bullous pemphigoid antigen in a new stable anchoring contact (SAC) of keratinocytes: relation to hemidesmosomes. J. Cell Biol. 111:3141-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Z., C. A. Casiano, and H. M. Fletcher. 2001. Protease-active extracellular protein preparations from Porphyromonas gingivalis W83 induced N-cadherin proteolysis, loss of cell adhesion, and apoptosis in human epithelial cells. J. Periodontol. 72:641-650. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, M. A., M. Maceys, J. M. Slaney, and G. L. Howells. 1993. Platelet activation by Protease I of Porphyromonas gingivalis W83. FEMS Microbiol. Lett. 110:167-173. [DOI] [PubMed] [Google Scholar]
- 10.DeCarlo, A. A., Jr., L. J. Windsor, M. K. Bodden, G. J. Harber, B. Birkedal-Hansen, and H. Birkedal-Hansen. 1997. Activation and novel processing of matrix metalloproteinases by a thiol-proteinase from the oral anaerobe Porphyromonas gingivalis. J. Dent. Res. 76:1260-1270. [DOI] [PubMed] [Google Scholar]
- 11.Discipio, R. G., P. J. Daffner, M. Kawahara, R. Pike, J. Travis, T. E. Hugli, and J. Potempa. 1996. Cleavage of human complement component C5 by cysteine proteinases from Porphyromonas (Bacteroides) gingivalis. Prior oxidation of C5 augments proteinase digestion of C5. Immunology 87:660-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichinger, A., H. G. Beisel, U. Jacob, R. Huber, F. J. Medrano, A. Babula, J. Potempa, J. Travis, and W. Bode. 1999. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 18:5453-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finlay, B. B., and P. Cossart. 1997. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276:718-725. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher, J., S. Nair, S. Poole, B. Henderson, and M. Wilson. 1998. Cytokine degradation by biofilms of Porphyromonas gingivalis. Curr. Microbiol. 36:216-219. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs, E., J. Dowling, J. Segre, S. H. Lo, and Q.-C. Yu. 1997. Integrators of epidermal growth and differentiation: distinct functions of β1 and β4 integrins. Curr. Opin. Gen. Dev. 7:672-682. [DOI] [PubMed] [Google Scholar]
- 16.Garrington, T. P., and G. L. Johnson. 1999. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell Biol. 11:211-218. [DOI] [PubMed] [Google Scholar]
- 17.Geatch, D. R., J. I. Harris, P. A. Heasman, and J. J. Taylor. 1999. In vitro studies of lymphocyte apoptosis induced by the periodontal pathogen Porphyromonas gingivalis. J. Periodont. Res. 34:70-78. [DOI] [PubMed] [Google Scholar]
- 18.Geiger, B., A. Bershadsky, R. Pankov, and K. M. Yamada. 2001. Transmembrane crosstalk between the extracellular matrix—cytoskeleton crosstalk. Nat. Rev. Mol. Cell. Biol. 2:793-805. [DOI] [PubMed] [Google Scholar]
- 19.Giancotti, F. G., and E. Ruoslahti. 1999. Integrin signaling. Science 285:1028-1032. [DOI] [PubMed] [Google Scholar]
- 20.Goodwin, A. E., and B. U. Pauli. 1995. A new adhesion assay with buoyancy to remove nonadherent cells. J. Immunol. Methods 187:213-219. [DOI] [PubMed] [Google Scholar]
- 21.Hintermann, E., M. Bilban, A. Sharabi, and V. Quaranta. 2001. Inhibitory role of α6β4-associated erbB2 and phosphoinositide 3-kinase in keratinocyte haptotactic migration dependent on α3β1 integrin. J. Cell Biol. 153:465-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howe, A., A. E. Aplin, S. K. Alahari, and R. L. Juliano. 1998. Integrin signaling and cell growth control. Curr. Opin. Cell Biol. 10:220-231. [DOI] [PubMed] [Google Scholar]
- 23.Huang, G. T., D. Kim, J. K. Lee, H. K. Kuramitsu, and S. K. Haake. 2001. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via agonistic mechanisms. Infect. Immun. 69:1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato, S., K. Nakashima, M. Inoue, J. Tomioka, K. Nonaka, T. Nishihara, and Y. Kowashi. 2000. Human epithelial cell death caused by Actinobacillus actinomycetemcomitans infection. J. Med. Microbiol. 49:739-745. [DOI] [PubMed] [Google Scholar]
- 25.Katz, J., V. Sambandam, J. H. Wu, S. M. Michalek, and D. F. Balkovetz. 2000. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect. Immun. 68:1441-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann, S. H. 1998. Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim. Biophys. Acta 1400:195-211. [DOI] [PubMed] [Google Scholar]
- 27.Koshikawa, N., G. Giannelli, V. Cirulli, K. Miyazaki, and V. Quaranta. 2000. Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. J. Cell Biol. 148:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer, D. H., K. P. Mintz, and P. M. Fives-Taylor. 1997. Models of invasion of enteric and periodontal pathogens into epithelial cells: a comparative analysis. Crit. Rev. Oral Biol. Med. 8:389-409. [DOI] [PubMed] [Google Scholar]
- 29.Mullen, L. M., D. W. Richards, and V. Quaranta. 1999. Evidence that laminin-5 is a component of the tooth surface internal basal lamina, supporting epithelial cell adhesion. J. Periodont. Res. 34:16-24. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama, K., T. Kadowaki, K. Okamoto, and K. Yamamoto. 1995. Construction and characterization of arginine-specific cysteine proteinase (arg-gingipain)-deficient mutants of Porphyromonas gingivalis. J. Biol. Chem. 270:23619-23626. [DOI] [PubMed] [Google Scholar]
- 31.Nonaka, K. A., N. Ishisaki, T. Okahashi, S. Koseki, M. Kato, K. Muro, T. Nakashima, T. Nishihara, and Y. Kowashi. 2001. Involvement of caspases in apoptotic cell death of murine macrophages infected with Actinobacillus actinomycetemcomitans. J. Periodont. Res. 36:40-47. [DOI] [PubMed] [Google Scholar]
- 32.Park, N. H., B. M. Min, S. L. Li, M. Z. Huang, H. M. Cherrick, and J. Doniger. 1991. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis 12:1627-1631. [DOI] [PubMed] [Google Scholar]
- 33.Potempa, J., and J. Travis. 1996. Porphyromonas gingivalis proteinases in periodontitis, a review. Acta Biochim. Pol. 43:455-466. [PubMed] [Google Scholar]
- 34.Potempa, J., A. Banbula, and J. Travis. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontal 24:153-192. [DOI] [PubMed] [Google Scholar]
- 35.Rock, M. T., A. R. Dix, W. H. Brooks, and T. L. Roszman. 2000. Beta1 integrin-mediated T cell adhesion and cell spreading are regulated by calpain. Exp. Cell Res. 261:260-270. [DOI] [PubMed] [Google Scholar]
- 36.Roskelley, C. D., A. Srebrow, and M. J. Bissell. 1995. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr. Opin. Cell Biol. 7:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlaepfer, D. D., C. R. Hauck, and D. J. Sieg. 1999. Signaling through focal adhesion kinase. Prog. Biophys. Mol. Biol. 71:435-478. [DOI] [PubMed] [Google Scholar]
- 38.Schoenwaelder, S., Y. Yuan, P. Cooray, H. H. Salem, and S. P. Jackson. 1997. Calpain cleavage of focal adhesion proteins regulates the cytoskeletal attachment of integrin αIIbβ3 (platelet glycoprotein Iib/IIIa) and the cellular retraction of fibrin clots. J. Biol. Chem. 272:1694-1702. [DOI] [PubMed] [Google Scholar]
- 39.Schroeder, H. E. 1991. Oral structural biology. Thieme Medical Publishers, New York, N.Y.
- 40.Schwartz, M. A., Schaller, M. D., and M. H. Ginsberg. 1995. Integrins: emerging paradigms of signal transduction. Annu. Rev. Cell Dev. Biol. 11:549-599. [DOI] [PubMed] [Google Scholar]
- 41.Scragg, M. A., S. J. Cannon, M. Rangarajan, D. M. Williams, and M. A. Curtis. 1999. Target disruption of fibronectin-integrin interactions in human gingival fibroblasts by the RI protease of Porphyromonas gingivalis W50. Infect. Immun. 76:1837-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 43.Squier, C. A. 1991. The permeability of oral mucosa. Crit. Rev. Oral Biol. Med. 2:13-32. [DOI] [PubMed] [Google Scholar]
- 44.Tada, H., S. Sugawara, E. Nemoto, N. Takahashi, T. Imamura, J. Potempa, J. Travis, H. Shimauchi, and H. Takada. 2002. Proteolysis of CD14 on human gingival fibroblasts by arginine-specific cysteine proteinases from Porphyromonas gingivalis leading to down-regulation of lipopolysaccharide-induced interleukin-8 production. Infect. Immun. 70:3304-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timpl, R. 1996. Macromolecular organization of basement membranes. Curr. Opin. Cell Biol. 8:618-624. [DOI] [PubMed] [Google Scholar]
- 46.Troyanovsky, S. M. 1999. Mechanism of cell-cell adhesion complex assembly. Curr. Opin. Cell Biol. 11:561-566. [DOI] [PubMed] [Google Scholar]
- 47.Turner, C. E. 1998. Paxillin. Int. J. Biochem. Cell Biol. 30:955-959. [DOI] [PubMed] [Google Scholar]
- 48.Uitto, V. J., K. Airola, M. Vaalamo, N. Johansson, E. E. Putins, J. D. Firth, J. Salonen, C. Lopez-Otin, U. Saarialho-Kere, and V. M. Kahari. 1998. Collagenase-3 (matrix metalloproteinase-13) expression is induced in oral mucosal epithelium during chronic inflammation. Am. J. Pathol. 152:1489-1499. [PMC free article] [PubMed] [Google Scholar]
- 49.Vasioukhin, V., C. Bauer, M. Yin, and E. Fuchs. 2000. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell 100:209-219. [DOI] [PubMed] [Google Scholar]
- 50.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wells, A. 2000.Tumor invasion: role of growth factor-induced cell motility. Adv. Cancer Res. 78:31-101. [DOI] [PubMed] [Google Scholar]
- 52.Williams, R. C. 1990. Periodontal disease. N. Engl. J. Med. 322:373-376. [DOI] [PubMed] [Google Scholar]
- 53.Wingrove, J. A., R. G. DiScipio, Z. Chen, J. Potempa, J. Travis, and T. E. Hugli. 1992. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J. Biol. Chem. 267:18902-18907. [PubMed] [Google Scholar]
- 54.Wymann, M. P., and L. Pirola. 1998. Structure and function of phosphoinositide 3-kinases. Biochim. Biophys. Acta 1436:127-150. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, Y., S. M. Dopheide, C. Ivanidis, H. H. Salem, and S. P. Jackson. 1997. Calpain regulation of cytoskeletal signaling complexes in Von Willebrand factor-stimulated platelets. J. Biol. Chem. 272:21847-21854. [DOI] [PubMed] [Google Scholar]
- 56.Zhurinsky, J., M. Shtutman, and A. Ben-Ze'ev. 2000. Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. J. Cell Sci. 113:3127-3139. [DOI] [PubMed] [Google Scholar]