Abstract
Inhibitor of apoptosis (IAP) proteins, which bind to caspases via their baculoviral IAP repeat domains, also bear RING domains that enable them to promote ubiquitylation of themselves and other interacting proteins. Here we show that the RING domain of cIAP1 allows it to bind directly to the RING of X-linked IAP, causing its ubiquitylation and degradation by the proteasome, thus revealing a mechanism by which IAPs can regulate their abundance. Expression of a construct containing the RING of cellular IAP1 was able to deplete melanoma cells of endogenous X-linked IAP, promoted apoptosis, and also markedly reduced their clonogenicity when treated with cisplatin. Cross control of protein levels by RING domains may therefore enable their levels to be manipulated therapeutically.
Keywords: apoptosis, ubiquitin, homeostasis, E3 ligase
Inhibitor of apoptosis (IAP) proteins were initially identified in baculoviruses, where they prevent defensive apoptosis of the host cell (1), thereby increasing the time available for viral replication. Cellular IAP (cIAP) homologues, which all bear one to three baculoviral IAP repeat (BIR) domains, have been identified in yeasts and metazoans. Those that bear a RING domain in addition to BIR domains [X-linked IAP (XIAP), cIAP1, cIAP2, and ML-IAP/Livin] appear to function as cell death inhibitors (reviewed in ref. 2).
The RING domains of IAPs can act as E3 ubiquitin ligases to promote the ubiquitylation of associated proteins such as TNF receptor-associated factors (TRAFs), Smac/Diablo, and caspases (3–6). However, the importance of the RING domain for the antiapoptotic activity of the IAPs is unclear; on the one hand, a RING-less DIAP1 protein overexpressed in Drosophila had increased antiapoptotic activity (7); on the other hand, alleles of DIAP1 with mutations in the RING finger are null for Reaper-induced cell death, although more potent at blocking Hid-induced cell death (5, 8, 9).
Our initial experiments showed that cIAP1 and XIAP can heterodimerize via a RING–RING interaction, but we also observed that expression of a stably integrated cIAP1 gene caused a specific reduction in the abundance of endogenous XIAP. Deletion studies revealed that the RING finger of cIAP1 was necessary and sufficient to cause loss of XIAP in a proteasome-dependent manner. cIAP1 RING-stimulated depletion of XIAP was seen in several cell types and associated with greatly increased sensitivity of several melanoma cells to cisplatin. Because several other E3 ligases such as BRCA1, BARD1, and RAG1 can interact via their RING fingers, it is possible that other RING-containing E3 ligases act to regulate the abundance of each other following heterodimerization. In this regard, it is striking that the E3 ligase activity of BRCA1 is greatly enhanced by heterodimerization with BARD1 (10, 11), suggesting a possible mechanism for homeostatic control of protein levels by RING domains.
Materials and Methods
Transfections and Constructs. The complete sequence of all constructs used can be obtained upon request. Full length IAPs, RING fingers, or SOCS boxes were cloned into pcDNA5 FRT TO (Invitrogen), pGEX 6P3, pRev TRE, and pProEX HTb vectors. The pEF Flag XIAP domain and pEF Flag CrmA constructs are described in refs. 14 and 15, respectively. Cells were transfected with Effectene (Qiagen, Valencia, CA) by using the manufacturer's protocols.
Stable Cell Lines. Stable cell lines were established with the Flp-In T-REx 293 cell line (Invitrogen). Stable melanoma cell lines were generated with pRev TRE EGFP cIAP1 RING/RING ΔC and pRev Tet Off (Clontech).
Immunoprecipitations, Western Blot Analysis, and Apoptosis Assays. Cell lysates were prepared throughout by using death-induced signaling complex (DISC) lysis buffer except where stated otherwise, and immunoprecipitations were performed as described in ref. 17.
Apoptosis Assays. Apoptosis assays were performed as described in ref. 17.
Results
cIAP1 Can Bind to and Reduce the Abundance of XIAP. To test whether endogenous XIAP could bind to cIAP1, we generated cells that expressed N-terminally Flag epitope-tagged cIAP1 by using a FLP recombinase system that yields isogenic stable cell lines that can be regulated by doxycyclin. After a 30-h induction with doxycyclin, Flag-tagged cIAP1 was present at readily detectable levels (Fig. 1a, lane 3, third and lowest blots). Flag bead immunoprecipitates probed with anti-XIAP showed that stably expressed cIAP1 can bind to endogenous XIAP (Fig. 1a, top two blots) as well as to endogenous TRAF2 (18). This experiment revealed that IAPs can heterodimerize as well as homodimerize but also suggested that induction of cIAP1 caused a reduction in the levels of endogenous XIAP (Fig. 1a, fourth blot). See Supporting Text, which is published as supporting information on the PNAS web site.
Fig. 1.
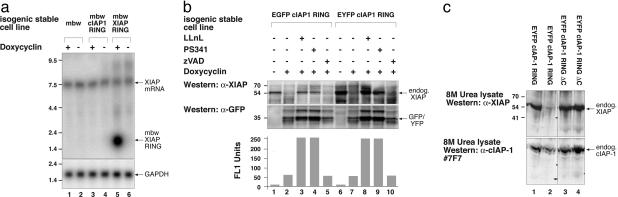
cIAP1 binds to and induces loss of endogenous XIAP. (a) Stable isogenic cell lines expressing cIAP1 or EGFP were established using the Flp-In T-REx system, and were induced for 30 h with doxycyclin. Lysates were immunoprecipitated with anti-FLAG beads and blots probed with anti-XIAP, anti-Flag, and anti-TRAF2. The experiment was repeated with three independently generated cIAP1 stable lines. (b) Degradation of endogenous XIAP induced by the minimal RING domain. Stable isogenic cell lines were cultured for 16 h with or without doxycyclin to induce expression of the relevant construct (second blot). Lysates alone were run on 4–20% gradient SDS/PAGE gels and blotted with anti-XIAP, anti-HSP70, anti-Bcl-w (mbw), anti-c-cbl, anti-TRAF2, and anti-TRAF4. (c) Stable cell lines expressing mbw or mbw cIAP1 RING were induced for the indicated times, and lysates were probed as before. (d) The more highly expressed cIAP1 RING fusion is more effective at promoting degradation of XIAP than full length cIAP1. Note that the lysates were probed with an antibody raised to ML-IAP RING that crossreacts with cIAP1 RING. *, Nonspecific cross-reactive.
The RING Domain Is Sufficient for Reduction of XIAP Abundance by cIAP1. To determine which part of cIAP1 bound to XIAP, we constructed fusion proteins in which the RING domain of cIAP1 was attached to the C terminus of a nonfunctional carrier protein, the well characterized cytoplasmic, monomeric, and nonfunctional Bcl-w ΔC10 (19, 20), which we chose due to the availibility of in-house antibodies and expression systems for this protein (hereafter referred to as mbw). Remarkably, expression of the mbw cIAP1 RING protein caused the complete loss of all endogenous XIAP (Fig. 1b top blot, lanes 3, 5, and 17). Six independent constructs containing the cIAP1 RING were able to promote complete degradation of endogenous XIAP (Fig. 1b, lanes 3, 5, and 17; and Figs. 2 and 4), even though they were expressed to different levels. The highly similar cIAP2 RING was also able to promote degradation of endogenous XIAP.
Fig. 2.
XIAP regulation by cIAP1 is proteasome-dependent and does not occur at the level of transcription. (a) Inducible isogenic stable cell lines mbw, mbw cIAP1 short RING, and mbw XIAP RING constructs were treated for 16 h with or without doxycyclin to induce expression of the relevant construct. RNA was isolated and Northern blot performed with the XIAP RING probe. (b) Proteasomal inhibitors impair cIAP1-induced degradation of endogenous XIAP. Stable cell lines expressing EGFP cIAP1 RING or EYFP cIAP1 RING were induced with or without doxycyclin for 8 h. LLnL (50 μM), PS341 (1 μM), or zVAD-fmk (50 μM) were added at the same time as the doxycyclin for the period of the induction. (c) Stable cell lines expressing EYFP cIAP1 RING or EYFP cIAP1 RING ΔC were either induced or not and then lysed directly in 8-M urea lysis buffer. The lysates were run on an SDS/PAGE gel and probed with antibodies to XIAP or cIAP1.
Fig. 4.
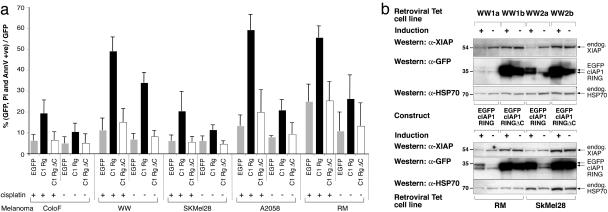
cIAP1 RING sensitizes melanoma cell lines to cisplatin. (a) Melanoma cell lines were transiently transfected with pcDNA5 FRT TO EGFP, EGFP cIAP1 RING, or EGFP cIAP1 RING ΔC mutants together with pcDNA6 Tet repressor at a ratio of 2:3. After ≈16 h, doxycyclin was used to induce expression of the relevant construct. Where indicated, 24 h after transfection, the induced cells were treated for a further 16 h with 8 μg/ml (ColoF, WW, SKMel28, and RM) or 4 μg/ml (A2058) cisplatin. Cells were then harvested, stained for annexin V/propidium iodide uptake, and run on a FACS scanner. The means of the results of three (ColoF, A2058), four (WW, SKMel28), and five (RM) independent experiments are represented as mean ± SD. (b) Stable inducible WW, RM, and Sk Mel28 cells were induced to express EGFP cIAP1 RING or EGFP cIAP1 RING ΔC for 24 h. Western blots indicate depletion of XIAP by the expressed constructs.
A construct in which the mbw carrier was fused to the RING of XIAP was also able to reduce endogenous XIAP (Fig. 1b, lane 15) but not quite as dramatically as the mbw cIAP1 RING protein, particularly when expression levels are taken into account. Neither the mbw carrier itself (Fig. 1b, lanes 11–14), nor an unrelated E3 ligase domain, the SOCS1 SOCS box alone (Fig. 1b, lanes 9 and 10), nor the SOCS box, fused to mbw in either the N- (Fig. 1b, lanes 1 and 2) or C-terminal position (Fig. 1b, lanes 7 and 8), promoted degradation of endogenous XIAP, suggesting interactions were restricted to RING E3 ligases. Furthermore, the effect was specific, because the cIAP1 RING domain caused loss of XIAP but did not alter the abundance of any of the RING domain proteins c-cbl, TRAF2, or TRAF4 (Fig. 1b, lower three blots) or of an unrelated protein, Hsp70.
In a kinetic experiment, increased levels of mbw cIAP1 RING, following addition of doxycyclin, correlated with a decrease in the abundance of endogenous XIAP (Fig. 1c, upper blot), such that by 6 h, XIAP could no longer be detected (Fig. 1c, lane 8). This effect was due to the cIAP1 RING, because expression of the mbw-only construct had no effect on XIAP, even though it was present at higher levels than the mbw RING (Fig. 1c, α-mbw Western analysis, second blot).
We were surprised that the mbw cIAP1 RING construct, which bears only the RING of cIAP1, seemed to be more efficient than full length cIAP1 in causing depletion of XIAP. To determine whether this could be due to different levels of mbw RING and full length cIAP1 protein, we compared them directly by Western analysis by using an anti-ML-IAP antibody that crossreacts with the RING finger of cIAP1. Fig. 1d shows that, even when expressed as isogenes, the abundance of the mbw cIAP1 RING construct was much greater than that of full length cIAP1 (Fig. 1d, compare lanes 2 and 6). Therefore, full length cIAP1 is probably less able to reduce endogenous XIAP, because it cannot attain the same levels as the mbw cIAP1 RING construct, possibly because full length cIAP1 contains additional domains that render it more susceptible to degradation than the carrier-RING fusion protein.
The Reduction of XIAP Is Mediated by the Proteasome in a Caspase-Independent Manner. To determine whether cIAP1 reduced the abundance of XIAP protein by decreasing XIAP mRNA, we carried out Northern analysis (Fig. 2a) of inducible stable cell lines. As a control for the induction of the constructs, we included the mbw XIAP RING construct and probed with an XIAP RING probe that can detect both endogenous XIAP message and the fusion construct message. As expected, the mbw XIAP RING RNA was readily induced by doxycyclin (Fig. 2a, lane 5). Induction of the mbw-cIAP RING construct reduced the XIAP protein to undetectable levels (Fig. 1b, top blot) but did not affect levels of XIAP messenger RNA (Fig. 2a, lane 3).
As with mbw cIAP1 RING (data not shown), both EGFP cIAP1 RING and EYFP cIAP1 RING induced degradation of endogenous XIAP, and this degradation was blocked by proteasome inhibitors PS341 or LLnL but not by the caspase inhibitor zVAD-fmk (Fig. 2b Bottom), indicating the proteasome was required for XIAP depletion caused by the cIAP1 RING. It remained posible that cIAP1 RING induced a ubiquitin-dependent translocation of XIAP, or that the loss of XIAP is a postlysis event and does not occur within a cell. To disprove these possibilities, we lysed the cells directly in an 8-M urea lysis buffer and analyzed them by SDS/PAGE gel. As before, cIAP1 RING induced degradation of XIAP (Fig. 2c Upper, lane 3). By using a cIAP1 monoclonal antibody that we have developed, we also show that endogenous cIAP1 levels are relatively unaffected by cIAP1 RING expression. This is reminiscent of the XIAP RING being less efficient at inducing loss of XIAP than cIAP1 RING (Fig. 1b, lane 16).
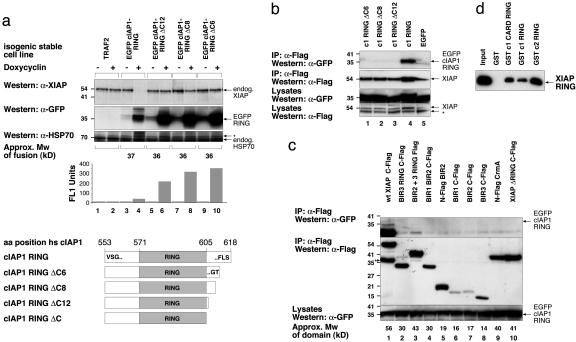
The C-Terminal Residues of cIAP1 RING Are Required for Binding to XIAP and for Promoting XIAP Degradation. To further delineate the residues of cIAP1 RING required to promote degradation of endogenous XIAP, we made point and deletion mutants. Inducible stable cell lines expressing constructs with the cIAP1 RING fused to EGFP or mbw (data not shown) were generated and tested for their ability to degrade endogenous XIAP. As with the mbw cIAP1 RING fusions, inducible stable cell lines expressing EGFP cIAP1 RING promoted complete degradation of endogenous XIAP. However, when the C-terminal 13 amino acids were removed from any of the cIAP1 RING finger constructs (ΔC), their ability to promote degradation of endogenous XIAP was abrogated. Western analysis of lysates from all of the stable lines confirmed that cIAP1 RING finger did not affect the levels of caspases 3 or 9 or TRAFs 2 or 4, confirming the specificity of the action of cIAP1's RING (data not shown). Remarkably, even deletion of just the last six amino acids of the cIAP1 RING completely destroyed the ability of a stably expressed EGFP cIAP1 RING fusion construct to promote degradation of endogenous XIAP (Fig. 3a), even when induced for 30 h or more (data not shown). Deletion of six amino acids does not interfere with any of the Zn-coordinating residues of the cIAP1 RING finger and does not affect the structure of the bacterially expressed recombinant protein as judged by far UV-CD spectra analysis (data not shown). Tellingly, there appears to be an inverse correlation between the stability of the EGFP RING protein and its ability to degrade XIAP (determined by Western analysis), which suggests destruction of XIAP and self destruction are related events.
Fig. 3.
The C-terminal six residues of cIAP1 are essential for binding to and degradation of XIAP. (a) Inducible isogenic stables stable cell lines were generated with TRAF2, EGFP cIAP1 RING, or EGFP cIAP1 RING ΔC mutant constructs and treated for 16 h with or without doxycyclin. *, Nonspecific cross-reactive band. (b) EGFP cIAP1 RING ΔC mutants are unable to interact with full length XIAP. Cells were transiently transfected with equal amounts of Flag-tagged XIAP and EGFP cIAP1 RING constructs, lysates used for Flag bead immunoprecipitation (IP) were run on SDS/PAGE gels, and membranes were blotted with anti-GFP and then subsequently with anti-Flag. *, anti-Flag nonspecific cross-reactive band. (c) XIAP proteins lacking either the RING or XIAP BIR1 are unable to interact with cIAP1 RING. 293T cells were transiently transfected with 0.6 μg of XIAP domain constructs and 0.4 μg of EGFP cIAP1 RING constructs and lysates and IPs were run on SDS/PAGE gels and transferred to poly(vinylidene difluoride) membranes, and blotted with anti-GFP, and then subsequently with anti-Flag. *, The residual EGFP cIAP1 RING signal from previous anti-GFP blot. (d) Recombinant RING proteins fused to GST were purified and incubated with recombinant XIAP RING in PBS buffer and 0.2% Tween. XIAP ring was detected with anti-His.
To see whether binding was required for degradation, we analyzed the cIAP1 deletion mutants for their ability to bind XIAP in coimmunoprecipitation assays. Significantly, deletion mutants that were unable to promote degradation of endogenous XIAP were unable to bind XIAP (Fig. 3b, compare lanes 4 and 1, 2, 3, and 5), confirming that RING–RING interactions are required for XIAP degradation.
To determine which regions of XIAP were required for cIAP1 interaction, Flag-tagged domains of XIAP were cotransfected with EGFP cIAP1 RING and the Flag-tagged constructs immunoprecipitated with anti-Flag beads. As expected, cIAP1 RING bound tightly to full length XIAP but did not bind an XIAP ΔRING construct, even though both XIAP constructs were expressed to equivalent levels (Fig. 3c, compare lanes 1 and 10). This shows that the RING of XIAP is required for it to bind to the RING of cIAP1. Surprisingly, deletion of BIR1 alone from XIAP almost completely abolished binding, although the BIR2 BIR3 RING protein was expressed to the same levels as wild-type XIAP (Fig. 3c, lane 3), indicating that, in addition to the RING of XIAP, BIR1 of XIAP is also important for maximal binding to the RING of cIAP1. To prove that the interaction between XIAP RING and cIAP1 RING was direct, we purified recombinant GST RING proteins and tested them for their ability to bind recombinant XIAP RING, in the presence of 0.2% Tween. As expected, cIAP1 RING, cIAP1 CARD RING (see cartoon Fig. 1b), and cIAP2 RING all specifically bound XIAP RING, but the GST control did not (Fig. 3d).
Reduction of IAPs by EGFP cIAP RING Sensitizes Melanoma Cells to Cisplatin. To test whether induced reduction of XIAP could affect the survival of melanoma cells, we transiently transfected five different melanoma lines with constructs that could subsequently be induced with doxycyclin. Cells were transfected with inducible EGFP RING constructs together with the Tet repressor construct at a ratio of 2:3 and induced with doxycyclin 16 h later. Expression of EGFP or an EGFP cIAP1 RING ΔC mutant lacking the 13 C-terminal amino acids caused no increase in cell death above the ≈7% background, whereas EGFP cIAP1 RING fusion was sufficient to cause 34% of WW melanoma cells to die, as determined by binding of annexin V and uptake of propidium iodide (Fig. 4a). Expression of EGFP cIAP1 RING fusion in A2058 and RM cells also caused a significant increase in amount of cell death, but to a lesser extent.
Significantly, when cisplatin was added to the cells, the amount of cell death was greater in all five lines expressing the EGFP cIAP1 RING fusion construct than in the cisplatin-treated controls. For example, after 16-h exposure to 16 μg/ml cisplatin, 60% of the EGFP cIAP1 RING expressing A2058 cells had died, whereas the same levels of cisplatin caused <20% of the EGFP or EGFP deleted-RING-expressing A2058 cells to die.
Probably because expression of the EGFP RING construct caused cell death, attempts to generate stable lines expressing EGFP cIAP1 RING failed. Therefore, we established stable cell lines by using a retroviral doxycyclin regulatable system. Induction with the retroviral system could not be regulated as tightly as in the isogenic cell lines, evidenced by the significant levels of the nontoxic EGFP cIAP1 RING ΔC constructs seen even when uninduced. Importantly, no loss of endogenous XIAP was observed in these ΔC lines.
Two independent EGFP cIAP1 RING (WW1a and WW2a) and RING mutant (WW1b and WW2b) polyclonal lines were generated from the melanoma cell line WW, and polyclonal lines were also generated from melanoma lines SKMel28 and RM. As before, induced expression of the wild-type cIAP1 RING first, reduced levels of endogenous XIAP (Fig. 4b); second, enhanced killing by cisplatin (data not shown); and last, appeared to be less stable than the ΔC mutant.
To see whether EGFP cIAP1 RING increased the number of cells that ultimately died in response to cisplatin and whether it was not just accelerating the rate of cell death, we performed a clonogenic survival assay. Cells (5 × 104) of the WW stable cell lines were plated, and doxycyclin was removed to induce the construct, after which cells were treated with 1 μg/ml cisplatin for 24 h. At this time, cell death measurable by annexin/propidium iodide uptake was negligible. Thirty-six hours after removal of the cisplatin, the cells were replated to determine whether they could form colonies. Two independent WW EGFP cIAP1 RING ΔC mutant cell lines (WW1b and WW2b) formed 77 and 55 colonies, whereas the WW lines expressing WT EGFP cIAP1 RING (WW1a and WW2a) formed only 17 and 8 colonies, respectively, indicating that depletion of IAPs by EGFP cIAP1 RING can significantly reduce clonogenic survival of drug-treated melanoma cells.
Discussion
These experiments reveal that, in an interaction mediated by their RING domains, one IAP, cIAP1, can directly bind to another IAP, XIAP, and reduce its levels by targeting it for degradation by the proteasome. This mechanism of IAP regulation might explain why cIAP1 protein levels are elevated in XIAP-deleted mice (21), and why XIAP-deleted mice have such a minor phenotype. Reciprocal regulation of cIAP1 by XIAP might also explain why cIAP1 RING constructs that are able to bind to and promote degradation of XIAP are considerably less stable than mutants that cannot bind. Similar interactions between cIAP1 and cIAP2 would also explain how cIAP1 is able to posttranscriptionally down-regulate cIAP2 in vivo (22).
This mechanism could therefore achieve homeostatic regulation of IAP levels and explain why it has been difficult to achieve stable high-level expression of IAPs in cells. In physiological circumstances, this mechanism might allow levels of IAPs to be matched to other proteins in a complex, such as TRAFs (18, 22, 23), so that excess IAPs are automatically destroyed.
IAP RING regulation is specific, because levels of other RING domain proteins, such as c-cbl, TRAF2, and TRAF4, were not affected by expression of cIAP1 RING. The RING domains in these proteins are positioned in the N-terminal half of the proteins rather than at the C terminus as in the IAPs and have different flanking sequences (Fig. 5a). Based on the structure of the BRCA1 BARD1 complex (24) and data presented in this paper, it is likely that the sequences flanking the RING domain determine whether RINGs heterodimerize. It is likely, therefore, that cIAP1 regulates the levels of other IAPs that have very similar flanking sequences, such as cIAP2 and ML-IAP, in addition to XIAP. Indeed, the cIAP1 RING can also heterodimerize with cIAP2 RING (data not shown), and deletion of cIAP1 results in a dramatic increase in cIAP2 levels in vivo (22). We show, however, that cIAP1 RING does not significantly regulate endogenous cIAP1 levels, nor is XIAP RING very efficient at depleting endogenous XIAP, suggesting that RING heterodimerization induces degradation and leaves open the role of RING homodimerization. Because constructs bearing the cIAP1 RING finger alone are so much more potent than full length cIAP1 at promoting degradation of endogenous XIAP, it seems likely that that a cIAP1 fragment generated, for example, by proteolytic cleavage (25, 26) is the functional domain of cIAP1 that promotes XIAP degradation under physiological conditions. Such a cIAP1 fragment can readily be detected in transient transfection assays (data not shown) and has also been observed in apoptotic cells (27), and indeed generation of such a fragment could even account for the loss of XIAP observed in apoptotic thymocytes (28).
Fig. 5.
Lineup of RING fingers and a model for XIAP or cIAP1 RING–RING interaction. A subset of 13 (from >50 RING finger proteins) was selected to emphasize the conservation across large phylogenetic differences of the C-terminal extension of certain RING fingers, exactly 13 amino acids after the final zinc-coordinating cysteine. For comparison, three RING finger proteins whose levels were not decreased with cIAP1 RING (c-cbl, TRAF2, and TRAF4), and which have different flanking sequences, are shown. Lowercase, RING consensus residues; uppercase, flanking residues of RING; yellow, similar charged residues; pink, similar polar residues; gray, similar hydrophobic; white on black, conserved zinc-coordinating residues. Structures from 1JM7 (24) showing the BARD1 BRCA1 RING–RING interaction, produced with rasmol (16). A schematic showing a speculative model for the C-terminal 13 amino acids of cIAP1 RING interacting with the N-terminal helix preceding the XIAP RING domain.
Earlier models for IAP function suggested that IAPs ubiquitinate and degrade proteins that bind to their BIR domains, such as caspases and IAP antagonists. Although mammalian IAPs have been shown to promote ubiquitylation of caspase-3, caspase-7, and Diablo/smac in vitro (3, 4, 6, 29, 30), XIAP does not act as an E3 ligase for Diablo/smac in intact cells, even though it binds to it with extremely high affinity (6, 31). Contrary to these models, Diablo/smac actually inhibits the E3 ligase activity of XIAP (31, 32). The results presented here suggest that an important functional target of RING-mediated ubiquitylation by IAPs are other IAP molecules, rather than other IAPs or IAP antagonists. In this regard, it has been shown that the RING domain of DIAP1 is required to inhibit cell death induced by HID (8, 9) and is also required for HID to induce degradation of DIAP1 (33), so it will be interesting to see whether HID might cause degradation of DIAP1 by promoting its dimerization and RING–RING interaction.
RING domains recruit E2 ubiquitin-conjugating enzymes (UBCs) to substrates, and two reports have described different UBCs binding to, or facilitating autoubiquitylation by, IAPs (6, 34), but how the heterodimer of cIAP1 and XIAP interacts with its cognate UBC is not yet known. It is noteworthy, however, that the ubiquitin ligase activity of the BRCA1-BARD1 RING heterodimer is significantly greater than the ubiquitin ligase activity of either RING domain alone (35). Thus it is plausible that the formation of an XIAP-cIAP1 RING heterodimer activates the E3 ligase activity of these RINGs.
Structural studies of the RINGs of RAG1 homodimers and BRCA1-BARD1 heterodimers have shown that α-helical regions N- and C-terminal to the core RING domains are critical for RING–RING interaction (refs. 24 and 36 and Fig. 5). Because the amino acids on both the N- and C-terminal flanks of the core RING of cIAP1 are predicted to form α helices, these regions may mediate the interactions between cIAP1 and XIAP, and MDM2 and MDMX, and possibly many other RING-bearing proteins, in a similar way (Fig. 5a). Consistent with this model, deletion of six residues in the predicted C-terminal α helix of cIAP1 prevents interaction of cIAP1 RING with XIAP.
RING-containing fragments of several IAPs, generated either artificially or by endogenous proteases, are highly apoptotic (7, 27, 37). It has been shown that cIAP1 RING was toxic when expressed in baby hamster kidney and Chinese hamster ovary cells and consistent with these observations (27), the cIAP1 RING finger was extremely cytotoxic in transient transfection assays in 293T, NT2 (J.S., unpublished data) and several melanoma cell lines. Our findings that the RING of an IAP can specifically bind to and target other intact IAPs for destruction but leave many other RING finger proteins unaffected strongly suggests that the proapoptotic activity of the RING fragments is due to depletion of endogenous IAPs, although it does not exclude other possibilities. Although removal or inactivation of DIAP1 is sufficient to cause apoptosis of insect cells, deletion of XIAP does not cause the death of mammalian cells but can increase their sensitivity to chemotherapeutic agents. However, recent reports with specific smac mimetics (38–40) suggest that tumor lines may require IAPs for cell survival, because smac mimetics can induce cell death as well as sensitizing cells to chemotherapy at lower doses. The similarities with our own observations suggest that complete inhibition of IAPs is an effective way of killing certain cancer cells, and that incomplete inhibition sensitizes them to chemotherapy. Either way, these observations may offer a therapeutic approach for diseases featuring overexpression of IAPs, such as small-cell carcinoma of the lung and melanoma (41–43).
The similarity of the regions flanking the RINGs of IAPs to those of other RING-bearing proteins raises the possibility that the levels of these proteins are also under homeostatic control. If so, it may be possible to increase or decrease the levels of other RING-bearing proteins by expressing constructs with or without the RING domains or by designing drugs that enhance or interfere with the functioning of the RING domains.
Supplementary Material
Acknowledgments
We thank A. Strasser, B. Callus, J. Zhang, K. Satterley, and D. Krebs for stimulating discussions and technical help; and Y. Lazebnik (The Walter and Eliza Hall Institute), P. Meier (Institute of Cancer Research, London), W. Langdon (University of Western Australia, Perth), C. House (PeterMac, Melbourne), and L. O'Reilly (The Walter and Eliza Hall Institute) for antibodies. This project was supported by the National Health and Medical Research Council 257502 and by a Leukemia and Lymphoma Society Center grant. J.S. and P.G.E. are funded by a National Health and Medical Research Council career development grant.
Author contributions: J.S. and D.L.V. designed research; J.S., T.K., D.C., C.L.D., and M.P. performed research; J.S., P.G.E., and D.C.S.H. contributed new reagents/analytic tools; J.S. and D.L.V. analyzed data; and J.S. and D.L.V. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IAP, inhibitor of apoptosis; BIR, baculoviral IAP repeat; XIAP, X-linked IAP; cIAP, cellular IAP; TRAF, TNF receptor-associated factor.
References
- 1.Crook, N. E., Clem, R. J. & Miller, L. K. (1993) J. Virol. 67, 2168–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silke, J. & Vaux, D. L. (2001) J. Cell Sci. 114, 1821–1827. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki, Y., Nakabayashi, Y. & Takahashi, R. (2001) Proc. Natl. Acad. Sci. USA 98, 8662–8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li, X., Yang, Y. & Ashwell, J. D. (2002) Nature 416, 345–347. [DOI] [PubMed] [Google Scholar]
- 5.Wilson, R., Goyal, L., Ditzel, M., Zachariou, A., Baker, D. A., Agapite, J., Steller, H. & Meier, P. (2002) Nat. Cell Biol. 4, 445–450. [DOI] [PubMed] [Google Scholar]
- 6.Hu, S. & Yang, X. (2003) J. Biol. Chem. 278, 10055–10060. [DOI] [PubMed] [Google Scholar]
- 7.Hay, B. A., Wassarman, D. A. & Rubin, G. M. (1995) Cell 83, 1253–1262. [DOI] [PubMed] [Google Scholar]
- 8.Goyal, L., McCall, K., Agapite, J., Hartwieg, E. & Steller, H. (2000) EMBO J. 19, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lisi, S., Mazzon, I. & White, K. (2000) Genetics 154, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallery, D. L., Vandenberg, C. J. & Hiom, K. (2002) EMBO J. 21, 6755–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia, Y., Pao, G. M., Chen, H. W., Verma, I. M. & Hunter, T. (2003) J. Biol. Chem. 278, 5255–5263. [DOI] [PubMed] [Google Scholar]
- 12.Zhang, J. G., Farley, A., Nicholson, S. E., Willson, T. A., Zugaro, L. M., Simpson, R. J., Moritz, R. L., Cary, D., Richardson, R., Hausmann, G., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 2071–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilton, D. J., Richardson, R. T., Alexander, W. S., Viney, E. M., Willson, T. A., Sprigg, N. S., Starr, R., Nicholson, S. E., Metcalf, D. & Nicola, N. A. (1998) Proc. Natl. Acad. Sci. USA 95, 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verhagen, A. M., Silke, J., Ekert, P. G., Pakusch, M., Kaufmann, H., Connolly, L. M., Day, C. L., Tikoo, A., Burke, R., Wrobel, C., et al. (2001) J. Biol. Chem. 277, 445–454. [DOI] [PubMed] [Google Scholar]
- 15.Ekert, P. G., Silke, J. & Vaux, D. L. (1999) EMBO J. 18, 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayle, R. & Milner-White, E. J. (1995) Trends Biochem. Sci. 20, 374–376. [DOI] [PubMed] [Google Scholar]
- 17.Silke, J., Ekert, P. G., Day, C. L., Hawkins, C. J., Baca, M., Chew, J., Pakusch, M., Verhagen, A. M. & Vaux, D. L. (2001) EMBO J. 20, 3114–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothe, M., Pan, M. G., Henzel, W. J., Ayres, T. M. & Goeddel, D. V. (1995) Cell 83, 1243–1252. [DOI] [PubMed] [Google Scholar]
- 19.Hinds, M. G., Lackmann, M., Skea, G. L., Harrison, P. J., Huang, D. C. S. & Day, C. L. (2003) EMBO J. 22, 1497–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson-Annan, J., O'Reilly, L. A., Crawford, S. A., Hausmann, G., Beaumont, J. G., Parma, L. P., Chen, L., Lackmann, M., Lithgow, T., Hinds, M. G., et al. (2003) J. Cell Biol. 162, 877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harlin, H., Reffey, S. B., Duckett, C. S., Lindsten, T. & Thompson, C. B. (2001) Mol. Cell. Biol. 21, 3604–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conze, D. B., Albert, L., Ferrick, D. A., Goeddel, D. V., Yeh, W. C., Mak, T. & Ashwell, J. D. (2005) Mol. Cell. Biol. 25, 3348–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uren, A. G., Pakusch, M., Hawkins, C. J., Puls, K. L. & Vaux, D. L. (1996) Proc. Natl. Acad. Sci. USA 93, 4974–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brzovic, P. S., Rajagopal, P., Hoyt, D. W., King, M. C. & Klevit, R. E. (2001) Nat. Struct. Biol. 8, 833–837. [DOI] [PubMed] [Google Scholar]
- 25.Yang, Q. H., Church-Hajduk, R., Ren, J., Newton, M. L. & Du, C. (2003) Genes Dev. 17, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, S., Kalkum, M., Overholtzer, M., Stoffel, A., Chait, B. T. & Levine, A. J. (2003) Genes Dev. 17, 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clem, R. J., Sheu, T. T., Richter, B. W., He, W. W., Thornberry, N. A., Duckett, C. S. & Hardwick, J. M. (2001) J. Biol. Chem. 276, 7602–7608. [DOI] [PubMed] [Google Scholar]
- 28.Yang, Y., Fang, S., Jensen, J. P., Weissman, A. M. & Ashwell, J. D. (2000) Science 288, 874–877. [DOI] [PubMed] [Google Scholar]
- 29.Huang, H., Joazeiro, C. A., Bonfoco, E., Kamada, S., Leverson, J. D. & Hunter, T. (2000) J. Biol. Chem. 275, 26661–26664. [DOI] [PubMed] [Google Scholar]
- 30.MacFarlane, M., Merrison, W., Bratton, S. B. & Cohen, G. M. (2002) J. Biol. Chem. 277, 36611–36616. [DOI] [PubMed] [Google Scholar]
- 31.Silke, J. H., Kratina, T., Ekert, P. G., Pakusch, M. & Vaux, D. L. (2004) J. Biol. Chem. 279, 4313–4321. [DOI] [PubMed] [Google Scholar]
- 32.Creagh, E. M., Murphy, B. M., Duriez, P. J., Duckett, C. S. & Martin, S. J. (2004) J. Biol. Chem. 279, 26906–26914. [DOI] [PubMed] [Google Scholar]
- 33.Yoo, S. J., Huh, J. R., Muro, I., Yu, H., Wang, L., Wang, S. L., Feldman, R. M., Clem, R. J., Muller, H. A. & Hay, B. A. (2002) Nat. Cell Biol. 4, 416–424. [DOI] [PubMed] [Google Scholar]
- 34.Yang, Q. H. & Du, C. (2004) J. Biol. Chem. 279, 16963–16970. [DOI] [PubMed] [Google Scholar]
- 35.Hashizume, R., Fukuda, M., Maeda, I., Nishikawa, H., Oyake, D., Yabuki, Y., Ogata, H. & Ohta, T. (2001) J. Biol. Chem. 276, 14537–14540. [DOI] [PubMed] [Google Scholar]
- 36.Bonetta, L. (2001) Nat. Med. 7, 1106. [DOI] [PubMed] [Google Scholar]
- 37.Nachmias, B., Ashhab, Y., Bucholtz, V., Drize, O., Kadouri, L., Lotem, M., Peretz, T., Mandelboim, O. & Ben-Yehuda, D. (2003) Cancer Res. 63, 6340–6349. [PubMed] [Google Scholar]
- 38.Schimmer, A. D., Welsh, K., Pinilla, C., Wang, Z., Krajewska, M., Bonneau, M. J., Pedersen, I. M., Kitada, S., Scott, F. L., Bailly-Maitre, B. C., et al. (2004) Cancer Cell 5, 25–35. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Z., Cuddy, M., Samuel, T., Welsh, K., Schimmer, A., Hanaii, F., Houghten, R., Pinilla, C. & Reed, J. C. (2004) J. Biol. Chem. 279, 48168–48176. [DOI] [PubMed] [Google Scholar]
- 40.Carter, B. Z., Gronda, M., Wang, Z., Welsh, K., Pinilla, C., Andreeff, M., Schober, W. D., Nefzi, A., Pond, G. R., Mawji, I. A., et al. (2005) Blood 105, 4043–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vucic, D., Stennicke, H. R., Pisabarro, M. T., Salvesen, G. S. & Dixit, V. M. (2000) Curr. Biol. 10, 1359–1366. [DOI] [PubMed] [Google Scholar]
- 42.Kasof, G. M. & Gomes, B. C. (2001) J. Biol. Chem. 276, 3238–3246. [DOI] [PubMed] [Google Scholar]
- 43.Hofmann, H. S., Simm, A., Hammer, A., Silber, R. E. & Bartling, B. (2002) J. Cancer Res. Clin. Oncol. 128, 554–560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.