Abstract
In vitro compartmentalization (IVC) has previously been used to evolve protein enzymes. Here, we demonstrate how IVC can be applied to select RNA enzymes (ribozymes) for a property that has previously been unselectable: true intermolecular catalysis. Libraries containing 1011 ribozyme genes are compartmentalized in the aqueous droplets of a water-in-oil emulsion, such that most droplets contain no more than one gene, and transcribed in situ. By coencapsulating the gene, RNA, and the substrates/products of the catalyzed reaction, ribozymes can be selected for all enzymatic properties: substrate recognition, product formation, rate acceleration, and turnover. Here we exploit the complementarity of IVC with systematic evolution of ligands by exponential enrichment (SELEX), which allows selection of larger libraries (≥1015) and for very small rate accelerations (kcat/kuncat) but only selects for intramolecular single-turnover reactions. We selected ≈1014 random RNAs for Diels–Alderase activity with five rounds of SELEX, then six to nine rounds with IVC. All selected ribozymes catalyzed the Diels–Alder reaction in a truly bimolecular fashion and with multiple turnover. Nearly all ribozymes selected by using eleven rounds of SELEX alone contain a common catalytic motif. Selecting with SELEX then IVC gave ribozymes with significant sequence variations in this catalytic motif and ribozymes with completely novel motifs. Interestingly, the catalytic properties of all of the selected ribozymes were quite similar. The ribozymes are strongly product inhibited, consistent with the Diels–Alder transition state closely resembling the product. More efficient Diels–Alderases may need to catalyze a second reaction that transforms the product and prevents product inhibition.
Keywords: emulsion, RNA, intermolecular catalysis
The Diels-Alder [4 + 2] cycloaddition reaction (1) between a 1,3 diene and an alkene dienophile is one of the most useful and important in synthetic chemistry, because it allows the formation of six-membered rings by making two simultaneous C-C bonds and at the same time generates up to four chiral centers (2). However, nature rarely seems to use this mechanism for C-C bond formation and instead prefers other mechanisms such as the aldol condensation reaction. Although >100 natural products have been proposed to be Diels-Alder cycloadducts (3), direct evidence from biosynthetic studies with purified or partially purified enzymes is available for only three naturally occurring Diels–Alder reactions (4).
However, several antibodies and RNAs that catalyze Diels–Alder reactions have been generated in the laboratory. The Diels–Alderase antibodies were raised against a hapten that mimicked either the Diels–Alder adduct or the transition state of the desired reactants (reviewed in ref. 5). Diels–Alderase ribozymes with both pyridyl-modified (6, 7) and unmodified RNA (8) were generated by using a variation on systematic evolution of ligands by exponential enrichment (SELEX) (9, 10). One substrate is physically tethered to the members of an RNA library, and active sequences are enriched by selection for the formation of a product, which remains tethered to the RNA and allows either selective amplification or affinity purification of the RNA. The crystal structures of a natural Diels–Alderase enzyme (11), a Diels–Alderase ribozyme (12), and several catalytic antibodies have now been elucidated (13–15), providing a structural framework for comparing the mechanisms of these different biopolymers.
Advantageously, SELEX allows selection of large libraries of >1015 variants, and, because selection is for an intramolecular single-turnover reaction (in cis reaction), it is possible to select for RNAs with very small rate accelerations (kcat/kuncat) (16). However, it is not a true selection for catalysis, and there is neither selection pressure for turnover of multiple substrate molecules, nor for the conversion of untethered substrates (in trans reaction).
We have developed a method, termed in vitro compartmentalization (IVC) (17), that has been used to select proteins for catalysis (18–23), binding (24–27), and regulation (28). IVC uses compartmentalization to link a genotype (a nucleic acid that can be replicated) and a phenotype (a functional trait such as a binding or catalytic activity). Instead of compartmentalizing genes in cells, as in nature, in IVC, the genes are compartmentalized in aqueous microdroplets dispersed in a water-in-oil emulsion (17). Compartmentalization allows the selection of enzymes acting on multiple substrate molecules free in solution (in trans reaction) and, therefore, allows direct selection for all aspects of catalysis simultaneously (substrate recognition, product formation, rate acceleration, and turnover).
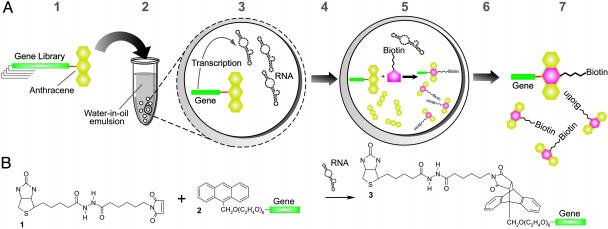
Here, we demonstrate the use of IVC for the selection of ribozymes. We have selected Diels–Alderase ribozymes that catalyze the reaction between dienes and dienophiles in solution in a truly bimolecular fashion and with multiple turnover (Fig. 1).
Fig. 1.
Selection of Diels–Alderase ribozymes by using IVC. (A) Schematic diagram of the selection procedure. A repertoire of genes (DNA) encoding ribozymes, each coupled to anthracene through a PEG linker, is created (1). Genes are compartmentalized within the aqueous droplets of a water-in-oil emulsion to give, on average, less than one gene per compartment (2). Genes are transcribed, giving ≈60 RNA molecules per gene (3). Mg2+ and biotin-maleimide are allowed to diffuse into the compartments (4). In compartments containing active Diels–Alderase ribozymes, the formation of the cycloadduct by reaction of biotin-maleimide is catalyzed, thereby biotinylating genes encoding active ribozymes (5). The emulsion is broken (6), and active genes are enriched by binding to streptavidin-coated magnetic beads (7) and are amplified by PCR to allow further rounds of selection. For multiple-turnover selections, free anthracene is emulsified with the gene repertoire. (B) The Diels–Alder cycloaddition of biotin-maleimide (1) and AHEG (2) covalently coupled to the gene to generate the adduct (3), thereby biotinylating the gene.
Methods
Ribozyme Genes. Two vectors were created: pIVEX-49mer containing the gene for a 49mer Diels–Alderase ribozyme (8) cloned downstream of a T7 promoter and the deleted variant, pIVEX-Δ49mer (Fig. 6, which is published as supporting information on the PNAS web site). Anthracene-labeled DNA for selection was prepared by PCR amplification of the above plasmids by using the primer LMB2-5 and the anthracene-labeled primer DA3-18 (for primer sequences and other information on the primers, see Table 2 and Fig. 7, which are published as supporting information on the PNAS web site). See Supporting Text, which is published as supporting information on the PNAS web site, for further details.
Synthesis of Substrates and Products. 9-anthracenylmethyl hexaethylene glycol (AHEG) 2 (Fig. 1B) was prepared as in ref. 29. Synthesis of 9-anthracenylmethyl hexaethylene glycol phosphoramidite is described in the Supporting Text.
The DA3-18 and J3-18 oligonucleotides were synthesized by using solid-phase phosphoramidite chemistry. After the incorporation of all of the deoxynucleotides, three units of spacer phosphoramidite-18 (Glen Research) were added, followed by 9-anthracenylmethyl hexaethylene glycol phosphoramidite, giving a total of 24 polyethylene oxide units between the anthracene and DNA moieties. The oligos were gel purified.
The Diels–Alder adduct rac4 (Fig. 8, which is published as supporting information on the PNAS web site) was prepared as in ref. 29 by using AHEG and 6-maleimidocaproic acid (MCA) (Fluka).
Library Construction. One microliter of a 1,000-fold dilution of DNA recovered after the fifth round of SELEX (8) was amplified by PCR with 10 μM each of the primers JA and anthracenylated J3-18 in a 50-μl reaction then gel purified, yielding a library of 176-bp DNA fragments encoding 157 base RNAs.
Ribozyme Selections. Compartmentalized transcription. A50 μl in vitro transcription reaction was prepared on ice containing 40 mM Hepes-KOH (pH 7.0), 9 mM MgCl2, 60 mM NaCl, 5 mM each NTP, 1 unit/μl RNAsin (Promega), 1 mg/ml BSA, 0.015 units/μl inorganic pyrophosphatase (Sigma), 30 μg/ml T7 RNA polymerase (prepared as in ref. 30), and 3.3 nM anthracene-labeled ribozyme genes (1 × 1011 genes total) in a 2-ml round-bottom microcentrifuge tube (Eppendorf), and 0.5 ml of ice-cold oil mix [0.5% wt/wt Triton X-100 (Fisher) and 4.5% wt/wt Span 80 (Fluka) in light mineral oil (Sigma)] were added. The mixture was emulsified by homogenization for 3 min at 25,300 rpm by using an Ultra-Turrax T8 Homogenizer (IKA) with a 5-mm diameter dispersing tool.
After transcription for 15 min at 37°C, 4 μl of 3.75 M MgCl2 dissolved in 50% ethanol was added directly to the emulsion and vortexed for 10 s. The emulsions were incubated another 15 min at ambient temperature, and 1 μl of 111 mM biotin-maleimide (Sigma) dissolved in DMSO was added directly to the emulsion and vortexed for 10 s. The reaction was incubated for 15 min at ambient temperature, and 75 μl of stop buffer (100 mM EDTA/5 mM 2-mercaptoethanol/25 ng/μl yeast tRNA) was added to the emulsion.
Breaking emulsions. Water-saturated diethyl ether (700 μl) was added and centrifuged at 25,000 × g for 1 min. The ether was removed, and the aqueous phase was extracted sequentially with 350 μl of neat 2-butanol, 700 μl of water-saturated 2-butanol, and 700 μl of water-saturated diethyl ether, then centrifuged under reduced pressure for 7 min. The aqueous phase was then purified on a G-25 spin column (General Electric Healthcare) to remove excess biotin maleimide.
Gene capture. M-280 streptavidin-coated dynabeads (50 μl) (Dynal) were washed twice in 2× binding and washing buffer plus Triton X-100 (BWT) buffer (10 mM Tris·HCl, pH 7.5/1 mM EDTA/2.0 M NaCl/0.1% vol/vol Triton X-100) and resuspended in 100 μl of 2× BWT buffer. The washed dynabeads were combined with 100 μl of DNA from above and incubated for 15 min at ambient temperature with gentle rotation. The beads were washed in turn with 200 μlof2× BWT/200 μl of GHCl Buffer (3 M guanidinium hydrochloride/25 mM sodium phosphate, pH 7.2/1 mM EDTA)/200 μlof2× BWT/200 μl of water and then resuspended in 25 μl of water.
PCR amplification of enriched genes. The selected DNA was PCR amplified from 10 μl of beads by using LMB2-A and J3-18 as primers. Reactions were heated to 94°C for 2 min, then cycled 20 times (94°C for 10 s; 50°C for 30 s; 72°C for 45 s), then 7 min at 72°C. After PCR, DNA was purified by using Wizard PCR preps (Promega) for use in further rounds of selection.
Assaying Diels–Alderase Activity of Selected Libraries. DNA amplified after selection was transcribed for1hina10-μl reaction in bulk solution (not emulsified) by using identical conditions to the emulsified transcriptions described above but with 5 nM DNA. The entire transcription reaction was assayed for activity in 100 μlof40 mM Hepes-KOH, pH 7.5/80 mM MgCl2/60 mM NaCl containing 100 μM AHEG and 500 μM biotin-maleimide in a 96-well plate by following the reduction in the absorbance at 365 nm over time.
Screening of the Selected Library. The selected library was amplified from beads by using the primers JBgl and JPst. The PCR products were ligated into pIVEX 2.2bNde at the BglII and PstI sites. The ligations were transformed into XL-10 Gold competent cells (Stratagene). Colonies were picked into 384-well plates (Genetix), containing 2×TY, 100 mg/ml ampicillin, and 8% glycerol (75 μl per well), and were incubated overnight at 37°C and stored at –70°C.
Bacteria from these plates were transferred by using pin replicators (Genetix) into 10-μl PCR reactions in 384-well plates and PCR amplified by using the primers LMB2-A and J3. Reactions were incubated for 10 min at 94°C, and then cycled 30 times (94°C for 30 s; 55°C for 30 s; 72°C for 45 s), then 7 min at 72°C. One microliter of crude PCR product was transcribed in a 6-μl reaction giving a final DNA concentration of ≈20 nM by using the same conditions as the selected pool screen above. Activity was assayed as above by using 5 μl of the transcription reaction in a 55-μl assay in 384-well plates, giving a final RNA concentration in the assay of ≈3 μM.
Large-Scale RNA Preparation. RNA for structural probing and kinetic assays was produced by in vitro transcription from a PCR product template as above, but in 50-ml volumes, and then gel-purified.
Structural Probing of Selected Clones. Structural probing by inline attack and partial nuclease digestion were performed essentially as described in refs. 31 and 32, respectively (see Supporting Text).
Kinetic Characterization of Selected Ribozymes. Determination of kinetic constants. Purified RNA was heated to 90°C for 2 min and cooled on ice. Activities were measured in 15 mM Tris·HCl, pH 7.4/80 mM MgCl2/75 mM NaCl with 30–500 μM AHEG and 0.5–8 mM 6-maleimidocaproic acid. Maximum substrate concentrations were limited by solubility. The reactions were performed in 384-well plates in 70 μl with a final RNA concentration of 3 μMbyfollowing the reduction in the absorbance at 365 nm over time and analyzed as described in the Supporting Text.
Determination of product inhibition constants. Purified RNA was heated and cooled and initial rates were measured in the same buffer as above by using a single concentration of each substrate: 100 μM AHEG and 500 μM 6-maleimidocaproic acid. Chemically synthesized racemic Diels–Alder product rac4 (Fig. 8) was added to the reactions from 0 to 1 mM giving a concentration of the more inhibitory R,R-enantiomer of between 0 and 500 μM. Initial rates were plotted against the inhibitor concentration and Ki values were determined by fitting the data to a competitive inhibition model as described in the Supporting Text.
Results
Model Selection. We used a gene encoding a 49mer ribozyme, rationally designed to incorporate the catalytic motifs identified in ribozymes selected by using SELEX (8), from which transcription is driven by a T7 promoter, to optimize the selection of ribozymes by IVC (Fig. 1). We also created a mutant, termed Δ49mer, in which there is a 40-bp deletion encompassing the T7 promoter and the first 6 bp of the ribozyme gene (Fig. 6).
We made anthracene-labeled PCR products of the 49mer and Δ49mer by amplifying the corresponding plasmids with an anthracene-labeled primer. The labeled PCR products were added to an in vitro transcription system in various ratios of 49mer to Δ49mer genes at concentrations of 3 pM-3 nM (108 to 1011 total genes), emulsified, and selected as described in Fig. 1.
Selections were performed under both single- and multiple-turnover conditions. Selective pressure for multiple turnover can be achieved by adding free anthracene-HEG substrate to the emulsion compartments (see Supporting Text).
We have previously shown that it is possible to deliver a substrate and change pH in the compartments of a preformed water-in-oil emulsion (20) and various solutes, including metal ions, have been delivered into preformed emulsion droplets via a nanodroplet delivery system (28). Here, the addition of MgCl2 to the oil phase after transcription was used to bring the free Mg2+ concentration within the compartments from a transcriptionally optimal 4 mM to a higher level necessary for catalytic activity (8). The biotin-maleimide substrate was added after transcription because transcription is inefficient in the presence of maleimide, probably because of the maleimide reacting with the active site cysteine in T7 RNA polymerase and also to prevent the uncatalyzed reaction generating a significant amount of product before an adequate amount of ribozyme has been transcribed, increasing the dynamic range of the selection. When either biotin-maleimide or Mg2+ was omitted, no enrichment was observed (data not shown).
DNA was amplified by PCR preselection and postselection (Fig. 2). A starting ratio of 1:5 49mer:Δ49mer genes was enriched to approximately a 1:1 ratio after selection (a 5-fold enrichment) under both single- and multiple-turnover conditions (data shown for single-turnover only). No enrichment was observed when the reactions were not emulsified, demonstrating that, as expected for in trans catalysis, compartmentalization of the genes in an emulsion is essential for enrichment. This relatively low enrichment is entirely due to the relatively high rate of the uncatalyzed reaction (kuncat = 3 M–1min–1; refs. 8 and 33) (see Supporting Text).
Fig. 2.
Model selection of the 49mer ribozyme by IVC. Genes from before and after selection were PCR amplified and analyzed by agarose gel electrophoresis. Selections were performed starting from 1:1 or 1:5 ratios of 49mer:Δ49mer genes either emulsified or unemulsified. The upper band is a heteroduplex between the 49mer and Δ49mer DNA.
We were able to measure consistent enrichment levels with up to 1011 genes in 50 ml of aqueous phase in a 550-μl emulsion (data not shown), implying that there are ≈3 × 1011 droplets, with an average (functional) volume of 170 al and an average diameter of 70 nm. A single molecule in a 170-al compartment is at a concentration of 9 nM. Under the selection conditions, we measured ≈60 RNA molecules transcribed per gene by gel quantitation, corresponding to an RNA concentration of ≈540 nM in a droplet.
Library Selection. cDNA recovered after the fifth round of SELEX (8), and therefore already enriched for active Diels–Alderase ribozymes, was amplified by PCR to generate DNA fragments with a single anthracene molecule attached to the distal end and from which transcription is driven off a proximal T7 promoter. A total of 1011 genes were selected as Fig. 1 (see Fig. 3A). Fifty-three individual clones with the highest activity from round six of the single-turnover selections and 12 clones with the highest activity from round nine of the multiple-turnover selections were sequenced.
Fig. 3.
Selection of 157mer libraries by IVC. (A) Selection progress under single- and multiple-turnover conditions. The pool of selected DNA from each round was transcribed and assayed for Diels–Alderase activity by following the reduction in absorbance at 365 nm. The dotted line indicates the rate of the wild-type ribozyme (0.4 μM/min). (B) Schematic showing the different classes of ribozymes isolated from the selections. The primer binding sites and, for the ribozymes possessing the same structural motif as the SELEX ribozymes (8), helices I, II, and III (hI, hII, and hIII) and the two halves of the internal loop (IL1 and IL2) of the ribozyme are labeled and shown as colored bars. Regions with no homology to the SELEX ribozymes are shown with a black line. SELEX groups corresponding to IVC classes are indicated. An additional 13 groups, comprising 24 clones, which do not correspond to any of the IVC classes, were identified by using SELEX (8). See also Fig. 9, which is published as supporting information on the PNAS web site. (C) Comparison of the internal loop sequence of ribozymes selected by SELEX and IVC displayed in format similar to a sequence logo (34). Residues are numbered as in ref. 8.
Most of the ribozymes isolated from the SELEX library after 11 rounds of selection shared a basic secondary structure motif (8, 35). That is, an internal loop (IL) flanked by two helical regions (hII and hIII), with a helical region (hI) extending from near the 5′ terminus up to helix hII (Fig. 4).
Fig. 4.
Secondary structure of ribozymes. Lowest energy secondary structures as predicted by mfold (36, 37) of the 49mer ribozyme (8), and two ribozymes from the IVC selections: F12 (class 1), which has the same structural motif and internal loop sequence as the 49mer ribozymes, and E03 (class 8), which has a novel secondary structure, not found in ribozymes from SELEX (8). The predicted secondary structures were confirmed by inline attack (31) and nuclease digestion (32) (see Figs. 10 and 11). Helices I, II, and III and the internal loop of the 49mer and F12 ribozymes are labeled.
Using IVC, nine distinct sequence classes were identified (Figs. 3B and 9). Clones from classes 1–7 have the same secondary structure motif found in nearly all of the ribozymes isolated by SELEX. However, only three of these classes (classes 1, 3, and 7) are the same as ribozymes from SELEX (groups 2, 7, and 5, respectively) (8).
There are also some interesting differences in the sequence of the IL between the ribozymes selected by IVC and SELEX. The data for all of the IL region sequences from the ribozymes selected by both SELEX (41 clones sequenced) and IVC (61 clones sequenced) are displayed in Fig. 3C in a format analogous to a sequence logo (34) with the height of a particular letter in a stack proportional to the frequency of its occurrence in the selected clones. The U12 variant, which is present in 25% of ribozymes selected by IVC (and in all of the class 2 ribozymes), is not seen at all in the ribozymes from SELEX. The G15 variant that is the most common in IVC is present in only one clone from the SELEX selections.
Furthermore, sequence alignments and mfold predictions (36, 37) of the class 8 and 9 sequences indicate that they do not share sequence or structural homologies with any of the ribozymes from SELEX (8, 35). The secondary structures of representative clones from class 1 (F12) and class 8 (E03) were confirmed by structural probing, indicating that class 8 ribozymes indeed have a completely novel fold (Fig. 4; also see Figs. 10 and 11, which are published as supporting information on the PNAS web site).
Kinetic Characterization of Selected Ribozymes. Representatives from three classes were characterized kinetically (Table 1): F12 and E12 (class 1) (49-mer IL sequence, and A15G mutation, respectively), G11 (class 2), and E03 (novel class 8 fold) (see also Figs. 12–15, which are published as supporting information on the PNAS web site). The kinetic parameters of ribozyme F12 were similar to the 49mer ribozyme (33), which has the same structural motif and internal loop sequence despite its greater length (157 bases). The other two ribozymes with the same basic structural motif, but with internal loop sequences that were seen rarely (clone E12) or never (G11) in SELEX, were slightly less efficient, both in terms of kcat and kcat/Km1(diene)Km2(dienophile). Clone E03, with the novel fold, had very similar kinetic parameters to clone F12 and the 49-mer.
Table 1. Kinetic parameters of Diels—Alderase ribozymes.
| Clone | Class | Km1 (diene), μM | Km2 (dienophile), μM | kcat, s-1 | kcat/Km1, M-1 s-1 | kcat/Km2, M-1 s-1 | kcat/Km1Km2, M-2 s-1 | Ki,* μM |
|---|---|---|---|---|---|---|---|---|
| 49mer† | 1 | 200 ± 35 | 5,200 ± 1,300 | 0.33 ± 0.06 | 1,667 | 64.1 | 3.2 × 105 | 11 (IC50)‡ |
| 49mer | 1 | 148 ± 32 | 3,960 ± 623 | 0.32 ± 0.11 | 2,175 | 81.4 | 5.5 × 105 | 31.4 ± 1.8 |
| F12 | 1 | 150 ± 32 | 1,650 ± 197 | 0.13 ± 0.04 | 878 | 79.9 | 5.3 × 105 | 29.3 ± 2.4 |
| E12 | 1§ | 86 ± 10 | 7,890 ± 646 | 0.05 ± 0.02 | 583 | 6.4 | 7.4 × 104 | 57.3 ± 2.9 |
| G11 | 2 | 396 ± 40 | 1,310 ± 65 | 0.01 ± 0.003 | 25 | 7.6 | 1.9 × 104 | 40.3 ± 9.4 |
| E03 | 8 | 213 ± 7 | 6,150 ± 618 | 0.11 ± 0.04 | 504 | 17.5 | 8.2 × 104 | 68.0 ± 2.7 |
All were strongly product inhibited by the Diels–Alder product rac4 (see Supporting Text), but the clones with sequences rarely or never seen in SELEX (clones E12, G11, and E03) had slightly higher Ki values.
Discussion
To date, most ribozymes produced by in vitro selection have relied on the same SELEX strategy based on selection for an intramolecular single-turnover reaction (in cis reaction) (for recent review, see ref. 38).
Here, we describe the selection of ribozymes for true intermolecular catalysis (in trans reaction) by using IVC as described in Fig. 1. We selected ribozymes that catalyze the Diels–Alder cycloaddition of anthracene (diene) and maleimide (dienophile).
This system was chosen because to set up, optimize, and characterize this ribozyme selection system, it was important to have a positive control ribozyme. The 49mer Diels–Alderase ribozyme, derived from a SELEX experiment, which catalyses the reaction between anthracene and maleimide, is the only known ribozyme that can catalyze a multiple-turnover reaction between two small nonnucleic acid molecules in trans (8). Using this 49mer ribozyme allowed us to establish an IVC selection system in which the enrichment factors were very close to those predicted based on the measured kinetic parameters of the 49mer and the uncatalyzed reaction.
We compared IVC and SELEX directly by selecting a library by using IVC that was previously selected with SELEX (8). The SELEX experiment started from a library of ≈2 × 1014 RNA molecules, each 157 bases long with the central 120 nucleotides randomized. To select a 1014-member library by using IVC would require a 1.1-liter emulsion. We chose an alternative strategy that exploits the complementary characteristics of both SELEX and IVC: SELEX allows selection of very large libraries and for very small rate accelerations (kcat/kuncat) but only selects for intramolecular single-turnover reactions; selection of very large libraries by IVC is more difficult and the threshold for selection (kcat/kuncat) is higher, but IVC selects for true intermolecular catalysis (16).
The IVC selections were started with 1011 molecules of cDNA from the 157-mer library purified after the fifth round of SELEX (8). This library should be enriched for RNAs that catalyze an intramolecular single-turnover reaction (in cis reaction). By then selecting this library with IVC, only RNAs capable of catalyzing true intermolecular catalysis (in trans reaction) are recovered. This strategy avoids the danger that the ribozymes isolated can only catalyze intramolecular single-turnover reactions, as seen in many SELEX experiments, including SELEX for Diels–Alderase ribozymes (6, 7). In our experiments, the library after the fifth round of SELEX already showed low, but detectable, Diels–Alderase activity in trans (Fig. 3A).
When selected by IVC, Diels–Alderase activity increased more quickly under single-turnover than under multiple-turnover conditions, consistent with the theoretical predictions of the enrichment rates under these two regimes (Fig. 3A; see Supporting Text). Individual clones were analyzed after round six and round nine of the single- and multiple-turnover selections, respectively. Hence, under the single-turnover regime, the library had undergone a total of 11 rounds of selection (5 by SELEX then 6 by IVC), the same number of rounds used to isolate ribozymes from the same library by using SELEX (8).
Most ribozymes isolated from the library after 11 rounds of SELEX shared the same small secondary structural motif consisting of three helices, an asymmetric internal loop, and the single-stranded 5′ end (Fig. 4; ref. 8). Nucleotides of the 5′ terminus were found to base pair with nucleotides on both sides of the internal loop, forming a lambda-shaped nested pseudoknot fold with a preformed binding pocket (12, 35). Using IVC, nine distinct sequence classes were identified (Figs. 3B and 9). Classes 1–7 contain the same secondary structure motif found in most of the SELEX ribozymes but differ from each other in the location of the internal loop and the lengths of the helices. However, only three classes from IVC are the same as ribozymes from SELEX (IVC classes 1, 3, and 7 are the same as SELEX groups 2, 7, and 5 respectively); the other IVC classes have not previously been observed. The most common class of ribozymes from both single- and multiple-turnover IVC selections (class 1) was identical to the most common class from SELEX (group 2).
The IVC-selected ribozymes also show some striking differences in the frequency of certain bases in the internal loop compared to SELEX ribozymes with the same structural motif (see Fig. 3C). U12 is found in 25% of all ribozymes selected by IVC (and is found in all of the class 2 ribozymes) but is not seen in any of the ribozymes from SELEX. Similarly, G15 is found in nearly half of the ribozymes selected by IVC (and in 75% of class 1 ribozymes), but this sequence was seen in only a single ribozyme from SELEX.
In two of the class 2 sequences, the length of transcribed RNA is only 106 nt instead of 156 nt for the rest of the class. Selective pressure, probably due to a PCR advantage for the shorter sequence, is beginning to reduce the RNA to a size that is more similar to the minimal 49mer motif that was engineered by using rational design (8).
In addition, primary sequence analysis and secondary structure predictions indicate that the class 8 (from the single-turnover selections) and class 9 (from the multiple-turnover selections) ribozymes have no structural or sequence homology with any of the ribozymes from SELEX (8), including the three orphan sequences that did not contain the minimum structure motif (clones 16, 50, and 53). The secondary structure of clone E03 (class 8) was confirmed by structural probing experiments and indicated that it indeed has a completely novel fold (Figs. 4, 10, and 11).
All of the Diels–Alderase ribozymes selected by IVC tested were capable of catalyzing the in trans reaction by using diene and dienophile free in solution under multiple-turnover conditions. They all exhibit quite similar kinetic parameters (Table 1). Notably, ribozyme E03, with the novel fold (class 8), has Km(diene), Km(dienophile), and kcat values that are very close to those of the other ribozymes.
The 49mer (derived from SELEX) and clone F12 (class 1), which both have the same internal loop sequence, were marginally the most efficient ribozymes, followed closely by ribozyme E03 (class 8), clone E12 (class 1 but with a different internal loop sequence), and clone G11 (class 2). From the kinetic parameters themselves, it is unclear why ribozymes such as E03 and G11 (not observed in SELEX) and E12 (seen in only a single ribozyme from SELEX) should have been selected by using IVC. It may be due to differences in relative rates when selecting for the in cis reaction (SELEX) and in trans reaction (IVC), or there may simply be less of a selective disadvantage for these ribozymes when selecting for the in trans reaction (IVC) compared with the in cis reaction (SELEX).
It is tempting to speculate that this similarity in rates is not purely a coincidence but is related fundamentally to the mechanism of the Diels–Alder reaction. There is kinetic evidence to suggest that neither ribozymes nor a range of other noncovalent catalysts of the Diels–Alder reaction (including several catalytic antibodies, b-cyclodextrin, and Rebek's tennis ball) achieve significant specific binding of transition states that is the hallmark of enzyme catalysis (5). Rate acceleration is thought to arise predominantly from binding of reactants, converting a second-order reaction of diene with dienophile into a pseudo-first-order reaction of the termolecular complex of catalyst, diene, and dienophile (5), i.e., catalysis by approximation (39).
The results from this and earlier studies of Diels–Alderases could go some way to explaining why nature seems to prefer other mechanisms of C-C bond formation, such as the aldol condensation reaction, to the Diels–Alder cycloaddition. All of the Diels–Alderase ribozymes in this study are strongly product inhibited with almost identical Ki(product) of ≈50 mM, which is lower than the Km for either substrate, consistent with the Diels–Alder transition state closely resembling the product. Stabilization of the transition state results, almost inevitably, in an increase in affinity for the product, reducing the rate of product release and causing product inhibition.
Indeed, in the most efficient artificial Diels–Alderase described to date, the catalytic antibody 1E9, uncatalyzed SO2 elimination from the Diels–Alder adduct is programmed to avoid product inhibition (40). Similarly, the natural Diels–Alderase, macrophomate synthase, catalyses the elimination of CO2 to prevent product inhibition (41). Macrophomate synthase and the two other examples of natural Diels–Alderases, solanapyrone synthase and lovastatin nonaketide synthase, also catalyze the conversion of the corresponding substrates into reactive Diels–Alder substrates, which are not released from the active sites (4). They are thus all producers of reactive substrates with an entropy trap for [4 + 2] cycloaddition.
A requirement for enzymes to evolve to catalyze, not only the Diels–Alder cycloaddition but also a second reaction in which the cycloadduct is the substrate, may present a significant evolutionary barrier. This barrier may be even higher if it is also necessary to catalyze a reaction to create a reactive substrate for the [4 + 2] cycloaddition.
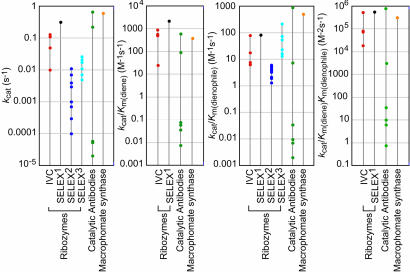
As can be seen from Fig. 5, none of the natural or artificial Diel–Alderases that have been characterized kinetically have kcat of ≥1 s–1, kcat/Km(diene) ≥ 104 M–1s–1, kcat/Km(dienophile) ≥ 103 M–1s–1, or kcat/Km(diene)Km(dienophile) ≥ 106 M–2s–1. It may be difficult for Diels–Alderases to pass this upper limit because of the nature of the Diels–Alder reaction itself. The Diels–Alderase ribozymes selected by IVC have kinetic constants that already approach this upper limit (Fig. 5).
Fig. 5.
Kinetic parameters of natural and artificial Diels–Alderases. The kcat, kcat/Km(diene), kcat/Km(dienophile), and kcat/Km(diene)Km(dienophile) for four ribozymes selected by IVC (red circles), the 49mer ribozyme derived by SELEX (SELEX1) (33) (black circle), nine ribozymes isolated from a different SELEX experiment (SELEX2) (6, 42) (blue circles), six ribozymes derived by mutation and SELEX of a ribozyme from the previous study (SELEX3) (7) (cyan circles), six catalytic antibodies, 1E9, 39A11, 13G5, 4D5, 22C8, and 7D4 (data summarized in ref. 5) (green circles), and macrophomate synthase (41) (orange circle). Data for the SELEX2 and SELEX3 ribozymes was determined under single-turnover conditions, and it was not possible to determine the Km(diene) because these ribozymes were only active when the diene was tethered to the RNA. Circles for enzymes with very similar kinetic parameters are overlayed.
However, with other systems that do not have such mechanistic limitations, it may be possible to use IVC to select ribozymes that are very fast and efficient. There is a view that ribozymes (and deoxyribozymes) may be inherently incapable of generating the same rate enhancements as those achieved by protein enzymes because of the lower chemical complexity of nucleic acids. In contrast, it has been argued that by using three or four catalytic strategies in parallel, a ribozyme or deoxyribozyme should theoretically be able to catalyze RNA cleavage by internal phosphoester transfer at rates at least equivalent to the protein enzyme RNase A (a nearly 1013-fold rate acceleration) (43).
However, attempts to improve ribozymes by in vitro selection with SELEX are likely to fail (see, for example, ref. 44) because there is neither selection for substrate recognition nor high kcat. Using IVC, however, it may prove possible to select for more efficient ribozymes, if they exist and, thereby, answer some important questions about the true catalytic potential of RNA.
Supplementary Material
Acknowledgments
We thank Catherine Isel for advice on RNA structural probing and for critically reading the manuscript, along with Eric Westhof. This work was funded by the Medical Research Council. A.D.G. is a member of the EC Framework 5 Marie Curie European Research Training Network on Directed Evolution of Functional Proteins.
Author contributions: J.J.A., B.T.K., A.J., and A.D.G. designed research; J.J.A. and B.T.K. performed research; A.J. contributed new reagents/analytic tools; J.J.A., A.J., and A.D.G. analyzed data; and J.J.A., B.T.K., A.J., and A.D.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AHEG, 9-anthracenylmethyl hexaethylene glycol; IL, internal loop; IVC, in vitro compartmentalization; SELEX, systematic evolution of ligands by exponential enrichment.
References
- 1.Diels, O. & Alder, K. (1928) Justus Liebigs Ann. Chem. 460, 95–122. [Google Scholar]
- 2.Nicolaou, K. C., Snyder, S. A., Montagnon, T. & Vassilikogiannakis, G. (2002) Angew. Chem. Int. Ed. 41, 1668–1698. [DOI] [PubMed] [Google Scholar]
- 3.Stocking, E. M. & Williams, R. M. (2003) Angew. Chem. Int. Ed. Engl. 42, 3078–3115. [DOI] [PubMed] [Google Scholar]
- 4.Pohnert, G. (2001) Chembiochem. 2, 873–875. [DOI] [PubMed] [Google Scholar]
- 5.Kim, S. P., Leach, A. G. & Houk, K. N. (2002) J. Org. Chem. 67, 4250–4260. [DOI] [PubMed] [Google Scholar]
- 6.Tarasow, T. M., Tarasow, S. L. & Eaton, B. E. (1997) Nature 389, 54–57. [DOI] [PubMed] [Google Scholar]
- 7.Tarasow, T. M., Kellogg, E., Holley, B. L., Nieuwlandt, D., Tarasow, S. L. & Eaton, B. E. (2004) J. Am. Chem. Soc. 126, 11843–11851. [DOI] [PubMed] [Google Scholar]
- 8.Seelig, B. & Jaschke, A. (1999) Chem. Biol. 6, 167–176. [DOI] [PubMed] [Google Scholar]
- 9.Tuerk, C. & Gold, L. (1990) Science 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 10.Bartel, D. P. & Szostak, J. W. (1993) Science 261, 1411–1418. [DOI] [PubMed] [Google Scholar]
- 11.Ose, T., Watanabe, K., Yao, M., Honma, M., Oikawa, H. & Tanaka, I. (2004) Acta Crystallogr. D. 60, 1187–1197. [DOI] [PubMed] [Google Scholar]
- 12.Serganov, A., Keiper, S., Malinina, L., Tereshko, V., Skripkin, E., Hobartner, C., Polonskaia, A., Phan, A. T., Wombacher, R., Micura, R., et al. (2005) Nat. Struct. Mol. Biol. 12, 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu, J., Deng, Q., Chen, J., Houk, K. N., Bartek, J., Hilvert, D. & Wilson, I. A. (1999) Science 286, 2345–2348. [DOI] [PubMed] [Google Scholar]
- 14.Heine, A., Stura, E. A., Yli-Kauhaluoma, J. T., Gao, C., Deng, Q., Beno, B. R., Houk, K. N., Janda, K. D. & Wilson, I. A. (1998) Science 279, 1934–1940. [DOI] [PubMed] [Google Scholar]
- 15.Haynes, M. R., Stura, E. A., Hilvert, D. & Wilson, I. A. (1994) Science 263, 646–652. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths, A. D. & Tawfik, D. S. (2000) Curr. Opin. Biotechnol. 11, 338–353. [DOI] [PubMed] [Google Scholar]
- 17.Tawfik, D. S. & Griffiths, A. D. (1998) Nat. Biotechnol. 16, 652–656. [DOI] [PubMed] [Google Scholar]
- 18.Ghadessy, F. J., Ong, J. L. & Holliger, P. (2001) Proc. Natl. Acad. Sci. USA 98, 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, Y. F., Tawfik, D. S. & Griffiths, A. D. (2002) Nucleic Acids Res. 30, 4937–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths, A. D. & Tawfik, D. S. (2003) EMBO J. 22, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen, H. M., Tawfik, D. S. & Griffiths, A. D. (2004) Protein Eng. Des. Sel. 17, 3–11. [DOI] [PubMed] [Google Scholar]
- 22.Ghadessy, F. J., Ramsay, N., Boudsocq, F., Loakes, D., Brown, A., Iwai, S., Vaisman, A., Woodgate, R. & Holliger, P. (2004) Nat. Biotechnol. 22, 755–759. [DOI] [PubMed] [Google Scholar]
- 23.Doi, N., Kumadaki, S., Oishi, Y., Matsumura, N. & Yanagawa, H. (2004) Nucleic Acids Res. 32, e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doi, N. & Yanagawa, H. (1999) FEBS Lett. 457, 227–230. [DOI] [PubMed] [Google Scholar]
- 25.Sepp, A., Tawfik, D. S. & Griffiths, A. D. (2002) FEBS Lett. 532, 455–458. [DOI] [PubMed] [Google Scholar]
- 26.Yonezawa, M., Doi, N., Kawahashi, Y., Higashinakagawa, T. & Yanagawa, H. (2003) Nucleic Acids Res. 31, e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertschinger, J. & Neri, D. (2004) Protein Eng. Des. Sel. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- 28.Bernath, K., Magdassi, S. & Tawfik, D. S. (2005) J. Mol. Biol. 345, 1015–1026. [DOI] [PubMed] [Google Scholar]
- 29.Stuhlmann, F. & Jaschke, A. (2002) J. Am. Chem. Soc. 124, 3238–3244. [DOI] [PubMed] [Google Scholar]
- 30.He, B., Rong, M., Lyakhov, D., Gartenstein, H., Diaz, G., Castagna, R., McAllister, W. T. & Durbin, R. K. (1997) Protein Expression Purif. 9, 142–151. [DOI] [PubMed] [Google Scholar]
- 31.Soukup, G. A. & Breaker, R. R. (1999) RNA 5, 1308–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isel, C., Marquet, R., Keith, G., Ehresmann, C. & Ehresmann, B. (1993) J. Biol. Chem. 268, 25269–25272. [PubMed] [Google Scholar]
- 33.Seelig, B., Keiper, S., Stuhlmann, F. & Jaschke, A. (2000) Angew. Chem. Int. Ed. Engl. 39, 4576–4579. [PubMed] [Google Scholar]
- 34.Schneider, T. D. & Stephens, R. M. (1990) Nucleic Acids Res. 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keiper, S., Bebenroth, D., Seelig, B., Westhof, E. & Jaschke, A. (2004) Chem. Biol. 11, 1217–1227. [DOI] [PubMed] [Google Scholar]
- 36.Zuker, M. (2003) Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathews, D. H., Sabina, J., Zuker, M. & Turner, D. H. (1999) J. Mol. Biol. 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 38.Joyce, G. F. (2004) Annu. Rev. Biochem. 73, 791–836. [DOI] [PubMed] [Google Scholar]
- 39.Westheimer, F. H. (1985) Adv. Phys. Org. Chem. 21, 1–35. [Google Scholar]
- 40.Hilvert, D. (1991) Ciba Found. Symp. 159, 174–183, discussion 183–187. [PubMed] [Google Scholar]
- 41.Ose, T., Watanabe, K., Mie, T., Honma, M., Watanabe, H., Yao, M., Oikawa, H. & Tanaka, I. (2003) Nature 422, 185–189. [DOI] [PubMed] [Google Scholar]
- 42.Tarasow, T. M., Tarasow, S. L. & Eaton, B. E. (2000) J. Am. Chem. Soc. 122, 1015–1021. [Google Scholar]
- 43.Emilsson, G. M., Nakamura, S., Roth, A. & Breaker, R. R. (2003) RNA 9, 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santoro, S. W. & Joyce, G. F. (1997) Proc. Natl. Acad. Sci. USA 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.