Abstract
In plants, protein synthesis occurs in the cytosol, mitochondria, and plastids. Each compartment requires a full set of tRNAs and aminoacyl-tRNA synthetases. We have undertaken a systematic analysis of the targeting of organellar aminoacyl-tRNA synthetases in the model plant Arabidopsis thaliana. Dual targeting appeared to be a general rule. Among the 24 identified organellar aminoacyl-tRNA synthetases (aaRSs), 15 (and probably 17) are shared between mitochondria and plastids, and 5 are shared between cytosol and mitochondria (one of these aaRSs being present also in chloroplasts). Only two were shown to be uniquely chloroplastic and none to be uniquely mitochondrial. Moreover, there are no examples where the three aaRS genes originating from the three ancestral genomes still coexist. These results indicate that extensive exchange of aaRSs has occurred during evolution and that many are now shared between two or even three compartments. The findings have important implications for studies of the translation machinery in plants and on protein targeting and gene transfer in general.
Keywords: GFP, mitochondria, plastids, protein targeting, translation
In plants, protein synthesis occurs in three cellular compartments, the cytosol, the mitochondria, and the chloroplasts. Thus, complete sets of transfer RNAs (tRNAs) and of aminoacyl-tRNA synthetases (aaRSs), which catalyze the addition of amino acid to their cognate tRNAs, have to be present in these three cellular compartments. In plants, all aaRSs are nuclear encoded and are posttranslationally targeted to their respective compartments. For mitochondrial and chloroplastic enzymes, targeting usually depends on a characteristic transit peptide sequence localized at the N terminus of the protein.
All cytosolic tRNAs are nuclear encoded. All plastid tRNAs are assumed to be encoded by the plastid genome in photosynthetic plants but not in nonphotosynthetic plants such as Epifagus virginiana (1, 2). Land plant mitochondrial genomes also lack a number of tRNA genes, and the corresponding tRNAs, which are nuclear encoded, are imported from the cytosol. These mitochondrial-imported tRNAs are typical eukaryotic cytosolic tRNAs and show minimal sequence similarity with the prokaryotic-type mitochondrial tRNAs they have replaced. Moreover, two types of mitochondrial-encoded tRNAs are found: the native tRNAs that are authentic mitochondrial tRNAs, and the chloroplast-like (cp-like) tRNAs that are highly similar to their plastid counterparts (97-100% sequence identity). It is presumed that these cp-like genes were acquired by the mitochondrial genome by intracellular transfer from the plastid genome (1). Because fidelity of translation depends on the accuracy of the aminoacylation reaction, it is generally admitted that a strong coevolution must exist between aaRSs and their cognate tRNAs. In plant mitochondria, this question is particularly pertinent, because different tRNA isoacceptors with very different origins coexist. This diversity raises important questions about the set of aaRSs present in each cellular compartment.
To determine which aaRS is found in which cellular compartment, we have used the well studied plant Arabidopsis thaliana. A. thaliana is the only higher plant, together with rice, whose nuclear genome has been completely sequenced, thus permitting the identification of all aaRS genes. Moreover, the mitochondrial tRNA population is now well known. The tRNAs corresponding to 11 amino acids are encoded by the mitochondrial genome, of which 4 are cp-like tRNAs. The tRNAs corresponding to 7 amino acids are encoded by nuclear genes (which all are of ancestrally eukaryotic origin) and are imported into mitochondria. Last, both native and imported isoacceptors are found for two amino acids, isoleucine and glycine (3). Previous results for a few mitochondrial aaRSs (4-9) indicated that, in some cases, enzymes were “shared” between different compartments, prompting us to carry out a systematic study to determine the probable localization for all Arabidopsis aaRSs. The first step was an in silico analysis to identify all aaRS genes and those encoding potential targeting sequences. Subsequently, these N-terminal sequences were expressed, fused to GFP or red fluorescent protein (RFP), and the localization of the fusion protein was determined by in vivo and in vitro experiments. The results show an unprecedented potential for dual targeting of proteins to two compartments, reaching the ultimate situation of an enzyme, an AlaRS, localized in all three compartments.
Materials and Methods
Bioinformatics. The Arabidopsis aaRS sequences were downloaded through The Arabidopsis Information Resource database (www.arabidopsis.org). For the other species, the aaRS sequences were downloaded through ENZYME (www.expasy.org/enzyme) and SwissProt databases (www.expasy.org/sprot). The list of species and sequences used is shown in Tables 3 and 4, which are published as supporting information on the PNAS web site. The entire proteins sequences were aligned by using clustalw at the European Bioinformatics Institute server (www.ebi.ac.uk/clustalw). The phylogeny was performed by using the phylip programs (http://evolution.genetics.washington.edu/phylip.html) The trees were built by using the neighbor joining method. The number of boot-strap replicates used was 100. Trees were drawn with treedyn (www.treedyn.org). Prediction of targeting sequences was carried out with predotar (10), targetp (11), and mitoprot (12).
PCR Amplification and Cloning. Sequences encoding putative aaRS targeting sequences were amplified from genomic DNA of Arabidopsis thaliana Col-0 if existing cDNA sequences showed there were no introns in the relevant region of the gene. If cDNA sequences did reveal the presence of introns, or if no cDNA sequences were available, RT-PCR was carried out on Col-0 leaf or suspension cell RNA as described in ref. 8. The amplified sequences were cloned in-frame with GFP or RFP reading frames in pOL vectors as described for previous fusions in refs. 8 and 13. For in vitro import experiments, the targeting sequence-GFP reading frame was subcloned into a pBS or pCRII vector in such a direction that it could be transcribed from T7 promoter.
In Vivo GFP or RFP Targeting Analyses. Transient expression in tobacco protoplasts and visualization of GFP and RFP fluorescence by confocal microscopy was performed as described in refs. 8 and 13.
Isolation of Mitochondria and Chloroplasts. Mitochondria were extracted from potato tubers (5) or 5-day-old Arabidopsis cell cultures (14). Chloroplasts were extracted from leaves of 10-day-old pea plants (15) or from 5-day-old Arabidopsis cell cultures (14).
In Vitro Import into Isolated Mitochondria and Chloroplasts. The constructs were used as templates for in vitro transcription/translation carried out with a TNT Coupled Reticulocyte Lysate System (Promega) in the presence of [35S]methionine. Import of [35S]labeled fusion proteins into purified potato mitochondria was performed according to Duchene et al. (5). Import into purified pea chloroplasts was performed according to Bruce et al. (15). All experiments were analyzed by SDS/PAGE.
Western Blot Analysis. Protein extracts were prepared in 10 mM Tris·HCl, pH 7.5/5 mM EDTA/0.3% (wt/vol) SDS/5% (vol/vol) 2-mercaptoethanol, then separated by SDS/PAGE, electrotransferred onto Immobilon-P membranes (Millipore, Bedford, MA), and submitted to immunological detection. Antisera were used at a1/10,000 dilution. Binding of the primary antibodies was revealed by chemoluminescence by using peroxidase-conjugated secondary antibodies and ECL reagents (Amersham Pharmacia Biotech).
Results
Forty-Five Expressed aaRS Genes Were Found in the A. thaliana Nuclear Genome. The analysis of the A. thaliana genome revealed 53 genes presenting similarities with aaRSs (Table 1). The expression of 51 of 53 could be proven by expressed sequence tags (ESTs) or by whole-genome array (16), and 6 sequences of the 51 probably have no aaRS function (Table 1). The remaining 45 aaRS genes must provide 19 or 20 aaRS activities to the cytosol, mitochondria, and plastids, which suggests that many of these enzymes have to be targeted to at least two of these three cellular compartments. Sequence homology with well characterized aaRSs from other eukaryotes and bacteria enabled unambiguous attribution of the probable amino acid specificity of each of these enzymes. For 13 amino acids of the 20, only two aaRS genes have been found in A. thaliana (Table 1). For glutamine (Gln), only one expressed cytosolic GlnRS was found. This observation is in accordance with the fact that Gln-tRNAGln formation was shown to occur by transamidation in chloroplasts, and led to the suggestion that the same route of Gln-tRNA synthesis is also prevalent in mitochondria, although it has been shown in yeast that the cytosolic GlnRS is imported into mitochondria and is involved in mitochondrial Gln-tRNAGln formation (17).
Table 1. Fifty-three predicted genes with similarities to aminoacyl-tRNA synthetases were identified in A. thaliana.
| Amino acid | AGI no. | EST, or whole-genome array* | Full-length mRNA | Potential targeting sequence | Closest similarity to aaRSs from |
|---|---|---|---|---|---|
| Ala | At1g50200 | + | − | + | Cytosol |
| At5g22800 | + | − | + | Cyanobacteria | |
| Cys | At5g38830† | + | − | Eubacteria‡ | |
| At3g56300† | + | − | Eubacteria‡ | ||
| At2g31170† | + | + | + | Eubacteria‡ | |
| Asp | At4g33760 | + | + | + | Cyanobacteria |
| At4g31180† | + | + | Cytosol | ||
| At4g26870† | + | + | Cytosol | ||
| Glu | At5g26710 | + | + | Cytosol | |
| At5g64050 | + | + | + | Cyanobacteria | |
| Phe | At4g39280 | + | + | Cytosol | |
| At1g72550 | + | + | Cytosol | ||
| At3g58140 | + | + | + | Mitochondria | |
| Gly | At3g48110 | + | + | + | Eubacteria |
| At1g29880 | + | + | + | Cytosol | |
| At3g44740 | +*§ | − | |||
| At1g29870 | − | − | |||
| His | At3g02760 | + | − | Cytosol | |
| At3g46100 | + | + | + | ArChaea | |
| Ile | At5g49030 | + | + | + | Cyanobacteria |
| At4g10320 | + | − | Cytosol | ||
| Lys | At3g11710 | + | + | Cytosol | |
| At3g13490 | + | + | + | Cyanobacteria | |
| Leu | At4g04350 | + | + | + | Eubacteria |
| At1g09620 | + | + | Cytosol | ||
| Met | At4g13780 | + | + | Cytosol | |
| At3g55400 | + | + | + | Cyanobacteria | |
| At2g40660 | +§ | + | |||
| Asn | At5g56680† | + | + | Cyanobacteria | |
| At4g17300† | + | + | + | Cyanobacteria | |
| At1g70980† | + | − | Cyanobacteria | ||
| At3g07420 | +§ | + | |||
| At1g68420 | +*§ | − | |||
| At5g38750 | − | − | |||
| Pro | At5g52520 | + | − | + | Cytosol |
| At3g62120 | + | + | Cytosol | ||
| Gln | At1g25350 | + | + | Cytosol | |
| At5g19720 | +*§ | − | |||
| Arg | At4g26300 | + | − | + | Cytosol |
| At1g66530 | + | + | Cytosol | ||
| Ser | At5g27470 | + | + | Cytosol | |
| At1g11870 | + | + | + | Mitochondria | |
| Thr | At5g26830† | + | + | + | Cytosol |
| At2g04842 | +* | − | + | Cyanobacteria | |
| At1g17960† | + | + | Cytosol | ||
| Val | At5g16715 | + | − | + | Eubacteria |
| At1g14610 | + | + | + | Cytosol | |
| At1g27160 | +*§ | − | |||
| Trp | At3g04600 | + | + | Cytosol | |
| At2g25840 | + | + | + | Cyanobacteria | |
| Tyr | At3g02660 | + | − | + | Eubacteria |
| At2g33840† | + | + | Archaea | ||
| At1g28350† | + | − | Archaea |
Identification codes are from the Arabidopsis Genome Initiative annotation at The Arabidopsis Information Resource (http://arabidopsis.org).
Whole-genome array. Indications about the expression of these genes [full-length mRNA, EST, whole genome array] and the aaRS domains were obtained from http://mips.gsf.de, http://arabidopsis.org, www.tigr.org, www.ncbi.nlm.nih.gov, and www.rarf.riken.go.jp. Similarities to proteins from other organisms were estimated from distance matrices by following clustalw alignments to all other annotated aminoacyl-tRNA synthetases in SwissProt (29).
Close paralogues resulting from recent duplications.
Weak similarities (see also ref. 8).
In italics are the two genes for which no expression could be found and the six others that we predict to be pseudogenes, or at least to encode proteins with no aaRS activity because they lack essential aaRS domains.
Using different computer prediction programs [predotar (10), mitoprot (12), and targetp (11)] and alignment of protein sequences to homologues, a potential organelle targeting sequence was identified for half of the sequences (23 of 45) (Tables 1 and 2). It should be stressed that the extremely high sequence conservation of these essential enzymes makes the identification of N-terminal targeting information relatively straightforward. For only four amino acids were two aaRs with targeting sequences identified (Table 2). For 16 of these 23 sequences, putative full-length mRNAs were found. For the other genes, 5′ RACE or RT-PCR on their transcripts were performed to confirm the likely N-terminal sequence of the protein.
Table 2. Twenty-three aaRS sequences have a potential mitochondrial and/or chloroplastic targeting sequence.
| Amino acid
|
Mitochondrial tRNA
|
predotar
|
targetp
|
mitoprot
|
Experimental localization
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| AGI no. | Mit | Chloro | Mit | Chloro | Mit | Mit | Chloro | Ref. and remarks | ||
| Cys | Native | At2g31170 | 0.21 | 0.70 | 0.40 | 0.80 | 0.97 | + | + | 8 |
| Met | Native | At3g55400 | 0.35 | 0.93 | 0.19 | 0.83 | 0.91 | + | + | 6 |
| Asn | cp-like | At4g17300 | 0.29 | 0.97 | 0.35 | 0.83 | 1.00 | + | + | 8 |
| His | cp-like | At3g46100 | 0.50 | 0.01 | 0.59 | 0.22 | 0.99 | + | + | 4 |
| Glu | Native | At5g64050 | 0.37 | 0.07 | 0.77 | 0.29 | 1.00 | + | + | — |
| Lys | Native | At3g13490 | 0.26 | 0.56 | 0.51 | 0.49 | 0.89 | + | + | — |
| Pro | Native | At5g52520 | 0.36 | 0.19 | 0.63 | 0.72 | 0.94 | + | + | — |
| Tyr | Native | At3g02660 | 0.13 | 0.84 | 0.19 | 0.86 | 0.98 | + | + | — |
| Ser | Native + cp-like | At1g11870 | 0.48 | 0.00 | 0.52 | 0.09 | 0.99 | + | + | — |
| Asp | cp-like | At4g33760 | 0.50 | 0.01 | 0.32 | 0.36 | 0.99 | + | + | — |
| Ala | Imported | At1g50200 | 0.69 | 0.02 | 0.70 | 0.74 | 0.99 | + | + | AlaRS-1:also cyto (7) |
| At5g22800 | 0.04 | 0.00 | 0.19 | 0.63 | 0.87 | + | + | AlaRS-2 | ||
| Phe | Imported | At3g58140 | 0.39 | 0.48 | 0.16 | 0.76 | 0.94 | + | + | — |
| Leu | Imported | At4g04350 | 0.01 | 0.03 | 0.25 | 0.92 | 0.83 | − | + | LeuRS-2 |
| Arg | Imported | At4g26300 | 0.04 | 0.61 | 0.10 | 0.91 | 0.74 | − | + | — |
| Thr | Imported | At5g26830 | 0.91 | 0.00 | 0.72 | 0.31 | 1,00 | + | − | ThrRS-1:also cyto (9) |
| At2g04842 | 0.18 | 0.96 | 0.14 | 0.89 | 1.00 | + | + | ThrRS-2 | ||
| Val | Imported | At1g14610 | 0.88 | 0.04 | 0.84 | 0.24 | 0.99 | + | − | ValRS-1:also cyto (9) |
| At5g16715 | 0.79 | 0.60 | 0.11 | 0.94 | 1.00 | ? | + | ValRS-2 | ||
| Trp | Imported | At2g25840 | 0.18 | 0.39 | 0.27 | 0.85 | 0.96 | + | + | — |
| Gly | Imported + native | At1g29880 | 0.41 | 0.01 | 0.91 | 0.04 | 0.92 | + | − | GlyRS-1:also cyto (5) |
| At3g48110 | 0.09 | 0.15 | 0.22 | 0.82 | 0.99 | + | + | GlyRS-2 (5) | ||
| Ile | Imported + native | At5g49030 | 0.07 | 0.52 | 0.13 | 0.77 | 0.55 | ? | + | |
The origin of their cognate mitochondrial tRNAs is indicated. The targeting predictions obtained with different programs are indicated, as well as the experimental results. Mit, mitochondrial; Chloro, chloroplastic.
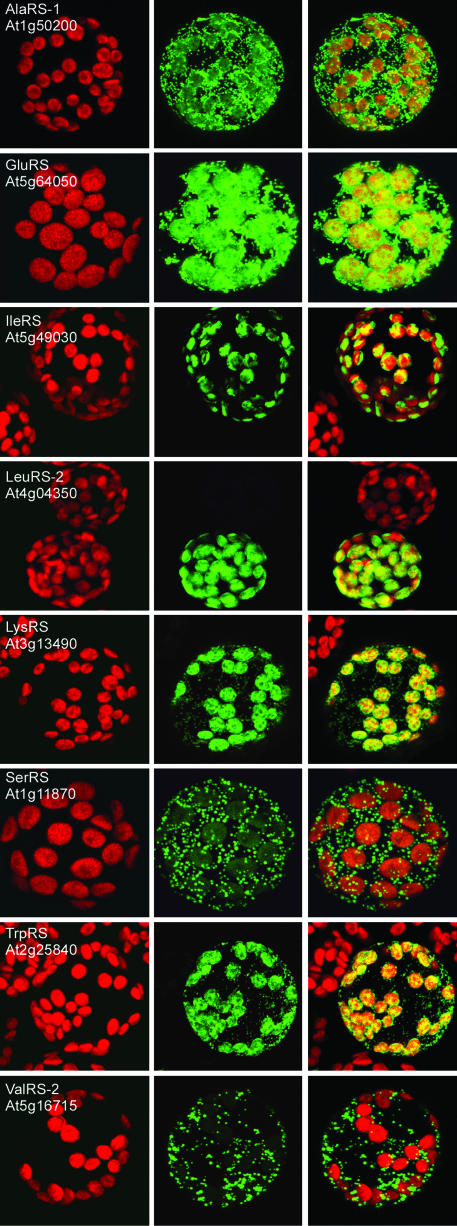
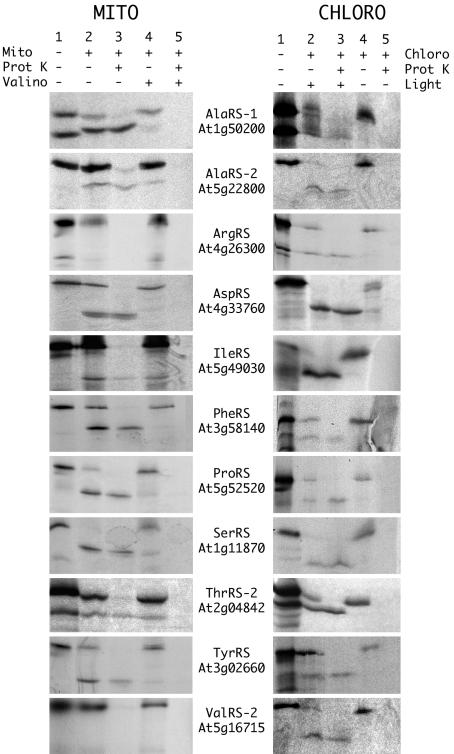
Targeting of aaRSs to Mitochondria and Chloroplasts. To analyze the intracellular localization of the putative organellar aaRSs, their N-terminal sequences were fused to GFP and RFP reporter genes. The localization of the fusion proteins was determined by transient expression in tobacco protoplasts for all constructs (Fig. 1; see also Fig. 5, which is published as supporting information on the PNAS web site) and by in vitro import into isolated organelles for one of each pair of constructs (Fig. 2; see also Fig. 6, which is published as supporting information on the PNAS web site). GFP and RFP fusions always gave the same localization in vivo. In vitro import was almost always in agreement with the localization by fluorescence, with a few exceptions discussed below.
Fig. 1.
Mitochondrial and/or chloroplast import of targeting sequence GFP or RFP fusions in tobacco protoplasts. The images are false-color maximum projections from a confocal microscope. The red channel (Left) shows chlorophyll autofluorescence, the green channel (Middle) shows GFP or RFP fluorescence (RFP for GluRS and ThrRS, GFP for all others); in Right, the two channels are superimposed. Several of the images include untransformed protoplasts in the field of view to demonstrate the lack of background fluorescence under the conditions used. The scale of the images and cell size varies slightly, but the chloroplasts are a uniform 5 μm in these protoplasts.
Fig. 2.
In vitro import into organelles of targeting sequence-GFP fusions. In vitro transcriptions/translations products (lane 1) were incubated in the presence of mitochondria or of chloroplasts and were partially processed into smaller polypeptides (lane 2), which corresponded to the fusion proteins upon cleavage of the predicted targeting sequences. The addition of proteinase K to the import medium reduced the signals corresponding to the preproteins but did not affect the signals corresponding to the processed proteins that were protected by mitochondrial or chloroplastic membranes (lane 3). The addition of valinomycin, which is known to inhibit mitochondrial protein import, or darkness, which is known to inhibit chloroplastic protein import, prevented the formation of the processed proteins (lanes 4 and 5). In these conditions, all radioactive signals were digested by proteinase K (lanes 5), showing that the signal observed in lane 3 represented genuine imported proteins.
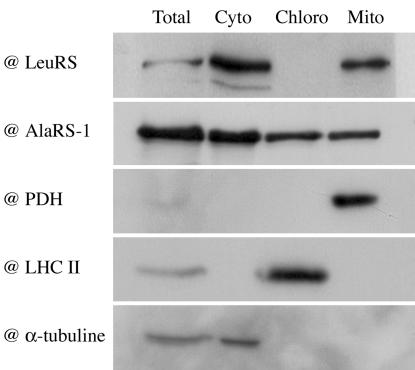
We have already shown that the targeting sequences from AsnRS (At4g17300), CysRS (At2g31170) (8), GlyRS-2 (At3g48110) (5), HisRS (At3g46100) (4), and MetRS (At3g55400) (6) allow targeting to both mitochondria and chloroplasts. Our systematic study demonstrates that such dual targeting is the general rule for plant organellar aaRSs, and we have identified at least 11 previously uncharacterized dual-targeted aaRSs: two AlaRSs (At1g50200 and At5g22800), AspRS (At4g33760), PheRS (At3g58140), GluRS (At5g64050), LysRS (At3g13490), ProRS (At5g52520), SerRS (At1g11870), ThrRS (At2g04842), TrpRS (At2g25840), and TyrRS (At3g02660). The AlaRS gene At1g50200 had already been shown to encode both cytosolic and mitochondrial forms of the enzyme, but the fluorescence data (Fig. 1), the in vitro import (Fig. 2), and Western blots (Fig. 3) all indicate that some of the “mitochondrial” form is also in plastids. For all of the dual-targeted proteins listed above, the in vitro import gave clear unequivocal results, whereas the fluorescence localization was often less easy to interpret. All of these GFP and RFP constructs gave strong mitochondrial signals (verified by colocalization with formate dehydrogenase-GFP or CoxIV-RFP fusions (13, 18) (data not shown), but many of them (AspRS, PheRS, ProRS, SerRS, and TyrRS) gave plastid fluorescence signals that were weak (see SerRS result in Fig. 1) and that varied between experiments and between cells within the same experiment. This weak fluorescence was not due to passenger protein effects as results with GFP and RFP fusions were identical. Equally, transient expression by Agrobacterium infiltration of leaves in situ gave the same variable, weak plastid fluorescence (data not shown).
Fig. 3.
Western blot analysis. Total (Total), cytosolic (Cyto), chloroplastic (Chloro), or mitochondrial (Mito) proteins extracts were prepared from A. thaliana suspension cells. Immunodetection was performed with bean chloroplastic leucyl-tRNA synthetase (@LeuRS) (19), A. thaliana alanyl-tRNA synthetase (@AlaRS-1) (7), maize mitochondrial pyruvate dehydrogenase (@PDH E1-α) (GT Monoclonal Antibodies), Chlamydomonas chloroplastic light harvesting complex II (@LHC II) and α-tubuline (@α-tubuline) (Amersham Pharmacia Biotech).
Two other aaRSs are presumably dual targeted, but our results are not fully conclusive. Targeting of the IleRS (At5g49030) construct to chloroplasts was observed both in vivo and in vitro (Figs. 1 and 2). In contrast, targeting to mitochondria was observed only in vitro. The ValRS-2 (At5g16715) targeting sequence localized GFP and RFP to mitochondria, with weak fluorescence sometimes observed in chloroplasts. In vitro, the same GFP fusion was clearly imported into chloroplasts but not into mitochondria. These two cases are the only ones where we observed conflicting results in vivo and in vitro.
Contrary to all other N-terminal sequences tested here, in vivo and in vitro experiments showed only plastid targeting for LeuRS-2 (At4g04350) and ArgRS (At4g26300) N-terminal sequences (Figs. 1 and 2). No targeting sequence could be detected for the other LeuRS gene, At1g09620, and its full-length mRNA (AY056261) corresponds to a protein without a targeting sequence. Antibodies were available for this enzyme (19) and tested with A. thaliana protein extracts. Western blots showed that the corresponding enzyme is localized both in the cytosol and the mitochondria, even if no targeting sequence could be identified (Fig. 3).
Phylogenetic Analysis and Evolutionary Origins. Multiple alignments and distance trees were performed to see relationships between each A. thaliana aaRS and aaRSs from other organisms (Table 1; see also Fig. 7, which is published as supporting information on the PNAS web site). Cytosolic enzymes are usually most similar to other cytosolic enzymes and very different from their organellar counterparts. Two exceptions are the CysRS and AsnRS enzymes, likely to be derived from organellar origins (8). The organellar enzymes are generally closest to eubacterial enzymes. Nine of them show clear, specific affinities with cyanobacterial sequences and are therefore strongly predicted to originate from the plastid ancestor. Five others also appear to be most related to eubacterial sequences and are therefore likely to originate from organelle ancestors. Surprisingly, only two enzymes (PheRS and SerRS) show specific similarities to mitochondrial enzymes from yeast or animals. Finally, the organellar ProRS and ArgRS enzymes are most similar to their homologues from the eukaryotic cytosol and presumably arose by duplication of the gene for the cytosolic enzyme and subsequent acquisition of an organelle targeting sequence.
Discussion
In this paper, we have identified 45 potential aminoacyl-tRNA synthetases from A. thaliana and predicted or analyzed their intracellular localization. Half of these enzymes have an organellar targeting sequence, and we have determined, both by in vivo and in vitro approaches, their mitochondrial and/or chloroplastic localization. Astonishingly, rather than being an isolated exception, dual targeting appeared to be a general rule. Among the 24 identified organellar aaRSs, 15 (and probably 17) are shared between mitochondria and plastids, and 5 are shared between cytosol and mitochondria (one of these aaRSs being present in chloroplasts as well). Only two were shown to be uniquely chloroplastic, and none to be uniquely mitochondrial. A few other proteins have been shown to be dual targeted to mitochondria and chloroplasts (20, 21) but, in general, targeting is highly specific; for example, a similarly sized study of targeting of pentatricopeptide repeat proteins to plant organelles did not reveal a single example of dual targeting (22).
Dual targeting was verified by localization of GFP and/or RFP fluorescence in living cells and import of the fusion protein into isolated mitochondria or chloroplasts. Problems with the use of either technique can be found in the literature: absence of GFP targeting in vivo or missorting into mitochondria in vitro (23-25). To help avoid misinterpretations, we used complementary in vivo and in vitro approaches for each construct. In almost all cases, the same results were obtained, but low levels of fluorescence were often observed in chloroplasts for constructs shown to be dual targeted in vitro. Mistargeting has been reported into isolated mitochondria but has never been observed with isolated chloroplasts (24). It should be noted that, in addition to the in vitro support for chloroplast targeting in these cases, the enzyme activities are absolutely essential in plastids for translation to occur.
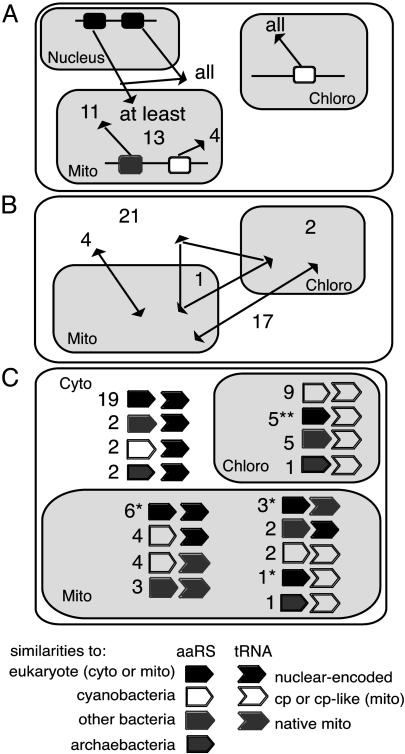
Plant cells have received three sets of aaRSs, one for each compartment where translation occurs. During evolution, the number of aaRSs has been reduced to only 45 aaRSs in A. thaliana, half of them being targeted to two translational compartments. The reduction of the number of aaRSs in plant cells is the result of a loss of all aaRS genes in the organellar genomes and transfer of only some of them to the nucleus. Indeed, nuclear-encoded aaRSs can be of eukaryotic or prokaryotic origin, in contrast to nuclearencoded tRNAs that are all of eukaryotic type. There are no remaining cases of coexistence of the three aaRS genes originating from the three ancestral genomes. This loss of genes was associated with extensive sharing and exchange of aaRS genes during plant evolution, resulting in numerous “mismatches” between aaRSs and substrate tRNAs of different genetic origin (Fig. 4). Given the extreme importance of tRNA recognition to translation fidelity, these mismatches are surprising and can only have occurred if they have little or no negative consequences. Of the 17 aaRSs likely to be shared by both organelles, 14 are clearly of eubacterial origin. These aaRSs, for the most part, aminoacylate organelle-encoded tRNAs that are themselves derived from the endosymbiont organelle ancestors and are therefore of eubacterial origin. In the same vein, all five of the aaRSs shared between cytosol and mitochondria, which are typical eukaryotic enzymes, correspond to mitochondrially imported cytosolic tRNAs. It is clear that sharing of aaRSs between mitochondria and chloroplasts has been facilitated by their similar eubacterial tRNAs, whereas sharing between the cytosol and organelles is relatively less frequent, except where import of the cytosolic tRNA makes it possible. However, some “cross-kingdom” exchanges have occurred. For example, the dual-targeted organellar TrpRS is close to Synechocystis TrpRS and likely to be of plastid origin. In most plants, the mitochondrial tRNATrp is a cp-like tRNA, but in Arabidopsis, tRNATrp is imported from the cytosol (3). Thus, in Arabidopsis mitochondria a eubacterial TrpRS is aminoacylating an imported cytosolic tRNA. In the other direction, the organellar ProRS shows clear affinities with eukaryotic cytosolic enzymes and, yet, plant organelles have retained eubacterial tRNAPro.
Fig. 4.
Putative origins of Arabidopsis aaRSs and their cognate tRNAs. Extensive sharing between the different cellular compartments are observed in A. thaliana at the level of tRNAs (A) and aaRSs (B). In most cases, the tRNA/aaRS couples are in accordance with the generally admitted coevolution between aaRSs and tRNAs, but numerous mismatches are also found (C). Some aaRSs have cognate tRNAs of different origins, and all of the cases are represented in C. For example, mitochondrial tRNAsSer are native or cp-like, so the SerRS is represented twice, once associated with native tRNAs and once with cp-like tRNAs. *, one is similar to mitochondrial aaRSs. **, two are similar to mitochondrial aaRSs.
One final surprising finding is the targeting of two AlaRSs, two GlyRSs, two ThrRSs and probably two ValRSs to plant mitochondria. In each case, one enzyme is of eukaryotic cytosolic type and the other of eubacterial type. It is tempting to speculate that one of the enzymes in each pair plays a role other than aminoacylation. Because all four pairs of aaRSs correspond to tRNAs imported into mitochondria from the cytosol, an attractive hypothesis is that some of these enzymes are needed for import of cytosolic tRNAs into mitochondria. Coimport of tRNA with the corresponding aminoacyl-tRNA synthetase has been described in yeast (26), and in plants, point mutations in tRNAAla and tRNAVal blocked both the aminoacylation of these tRNAs and their import into mitochondria (27, 28). In this context, it is interesting to note that although GlyRS-1 is efficiently and incontrovertibly imported into mitochondria, it has no apparent aminoacylation activity within the organelle, this activity being assured by GlyRS-2 (5).
In conclusion, this study has revealed an unprecedented degree of cross-compartment sharing of aminoacyl-tRNA synthetases. It will be interesting to see whether other components of the cytosolic and organellar translation machineries are shared to a similar extent. The results have important implications for the control of translation in each compartment because aaRS activity and tRNA-charging levels are a major control point in bacterial and eukaryotic translation systems. Because of their high conservation and ubiquitous distribution, aminoacyl-tRNA synthetases have been popular choices for deriving phylogenetic relationships between distantly related organisms. The obvious ease with which these enzymes can be incorporated into different translation systems implies that transfer of aaRS genes may be even more common than has been presumed hitherto.
Supplementary Material
Acknowledgments
We thank A. Eschbach for help in bioinformatics and C. de Vitry (Institut de Biologie Physico-Chimique-CNRS, Paris) for the generous gift of LHCII antibodies.
Author contributions: A.-M.D., L.M.-D., and I.D.S. designed research; A.-M.D., A.G., B.H., D.L., N.M.P., and M.Z. performed research; A.-M.D., V.C., L.M.-D., and I.D.S. analyzed data; and A.-M.D. and I.D.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: aaRS, aminoacyl-tRNA synthetases; cp-like, chloroplast-like; RFP, red fluorescent protein.
References
- 1.Maréchal-Drouard, L., Weil, J. H. & Dietrich, A. (1993) Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 13-32. [Google Scholar]
- 2.Wolfe, K. H., Morden, C. W. & Palmer, J. D. (1992) Proc. Natl. Acad. Sci. USA 89, 10648-10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duchêne, A. M. & Maréchal-Drouard, L. (2001) Biochem. Biophys. Res. Comm. 285, 1213-1216. [DOI] [PubMed] [Google Scholar]
- 4.Akashi, K., Grandjean, O. & Small, I. (1998) FEBS Lett. 431, 39-44. [DOI] [PubMed] [Google Scholar]
- 5.Duchêne, A. M., Peeters, N., Dietrich, A., Cosset, A., Small, I. D. & Wintz, H. (2001) J. Biol. Chem. 276, 15275-15283. [DOI] [PubMed] [Google Scholar]
- 6.Menand, B., Maréchal-Drouard, L., Sakamoto, W., Dietrich, A. & Wintz, H. (1998) Proc. Natl. Acad. Sci. USA 95, 11014-11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mireau, H., Lancelin, D. & Small, I. (1996) Plant Cell 8, 1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peeters, N. M., Chapron, A., Giritch, A., Grandjean, O., Lancelin, D., Lhomme, T., Vivrel, A. & Small, I. (2000) J. Mol. Evol. 50, 413-423. [DOI] [PubMed] [Google Scholar]
- 9.Souciet, G., Menand, B., Ovesna, J., Cosset, A., Dietrich, A. & Wintz, H. (1999) Eur. J. Biochem. 266, 848-854. [DOI] [PubMed] [Google Scholar]
- 10.Small, I., Peeters, N., Legeai, F. & Lurin, C. (2004) Proteomics 4, 1581-1590. [DOI] [PubMed] [Google Scholar]
- 11.Emanuelsson, O., Nielsen, H., Brunak, S. & Von Heijne, G. (2000) J. Mol. Biol. 300, 1005-1016. [DOI] [PubMed] [Google Scholar]
- 12.Claros, M. G. & Vincens, P. (1996) Eur. J. Biochem. 241, 779-786. [DOI] [PubMed] [Google Scholar]
- 13.Mollier, P., Hoffmann, B., Debast, C. & Small, I. (2002) Curr. Genet. 40, 405-409. [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto, W., Spielewoy, N., Bonnard, G., Murata, M. & Wintz, H. (2000) Plant Cell Physiol. 41, 1157-1163. [DOI] [PubMed] [Google Scholar]
- 15.Bruce, B. D., Perry, S., Froelich, J. & Keegstra, K. (1994) Plant Molecular Biology Manual (Kluwer, Dordrecht, The Netherlands), pp. 1-15.
- 16.Yamada, K., Lim, J., Dale, J. M., Chen, H., Shinn, P., Palm, C. J., Southwick, A. M., Wu, H. C., Kim, C., Nguyen, M., et al. (2003) Science 302, 842-846. [DOI] [PubMed] [Google Scholar]
- 17.Rinehart, J., Krett, B., Rubio, M. A., Alfonzo, J. D. & Soll, D. (2005) Genes Dev. 19, 583-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambard-Bretteville, F., Small, I., Grandjean, O. & Colas des Francs-Small, C. (2003) Biochem. Biophys. Res. Commun. 311, 966-971. [DOI] [PubMed] [Google Scholar]
- 19.Dietrich, A., Souciet, G., Colas, B. & Weil, J. H. (1983) J. Biol. Chem. 258, 12386-12393. [PubMed] [Google Scholar]
- 20.Peeters, N. & Small, I. (2001) Biochim. Biophys. Acta. 1541, 54-63. [DOI] [PubMed] [Google Scholar]
- 21.Silva-Filho, M. C. (2003) Curr. Opin. Plant Biol. 6, 589-595. [DOI] [PubMed] [Google Scholar]
- 22.Lurin, C., Andres, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyere, C., Caboche, M., Debast, C., Gualberto, J., Hoffmann, B., et al. (2004) Plant Cell. 16, 2089-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, M. N. & Whelan, J. (2004) Plant Mol. Biol. 54, 193-203. [DOI] [PubMed] [Google Scholar]
- 24.Rudhe, C., Chew, O., Whelan, J. & Glaser, E. (2002) Plant J. 30, 213-220. [DOI] [PubMed] [Google Scholar]
- 25.Chew, O. & Whelan, J. (2004) Trends Plant Sci. 9, 318-319. [DOI] [PubMed] [Google Scholar]
- 26.Tarassov, I. & Martin, R. (1996) Biochimie 78, 502-510. [DOI] [PubMed] [Google Scholar]
- 27.Delage, L., Duchêne, A. M., Zaepfel, M. & Maréchal-Drouard, L. (2003) Plant J. 34, 623-633. [DOI] [PubMed] [Google Scholar]
- 28.Dietrich, A., Maréchal-Drouard, L., Carneiro, V., Cosset, A. & Small, I. (1996) Plant J. 10, 913-918. [DOI] [PubMed] [Google Scholar]
- 29.Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., Martin, M. J., Michoud, K., O'Donovan, C., Phan, I., et al. (2003) Nucleic Acids Res. 31, 365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.