Abstract
Class II major histocompatibility (MHC-II) genes are prototype targets of IFN-γ. IFN-γ activates the expression of the non-DNA-binding master regulator of MHC-II, class II transactivator (CIITA), which is crucial for enhanceosome formation and gene activation. This report shows the importance of the histone methyltransferase, coactivator-associated arginine methyltransferase (CARM1/PRMT4), during IFN-γ-induced MHC-II gene activation. It also demonstrates the coordinated regulation of CIITA, CARM1, and the acetyltransferase cyclic-AMP response element binding (CREB)-binding protein (CBP) during this process. CARM1 synergizes with CIITA in activating MHC-II transcription and synergy is abrogated when an arginine methyltransferase-defective CARM1 mutant is used. Protein-arginine methyltransferase 1 has much less effect on MHC-II transcription. Specific RNA interference reduced CARM1 expression as well as MHC-II expression. The recruitment of CARM1 to the promoter requires endogenous CIITA and results in methylation of histone H3-R17; hence, CIITA is an upstream regulator of histone methylation. Previous work has shown that CARM1 can methylate CBP at three arginine residues. Using wild-type CBP and a mutant of CBP lacking the CARM1-targeted arginine residues (R3A), we show that arginine methylation of CBP is required for IFN-γ induction of MHC-II. A kinetic analysis shows that CIITA, CARM1, and H3-R17 methylation all precede CBP loading on the MHC-II promoter during IFN-γ treatment. These results suggest functional and temporal relationships among CIITA, CARM1, and CBP for IFN-γ induction of MHC-II.
Keywords: class II MHC, histone, interferon, methylation, transactivator
Class II major histocompatibility (MHC-II) proteins play a key role in the development of specific immune responses through the presentation of exogenously derived antigens to CD4+ T cells (1–5). Constitutive expression of MHC-II molecules is restricted to a subset of cells, but the induced expression of MHC-II by cytokines, particularly by IFN-γ, remains a key regulatory step. During an IFN-γ response, CIITA is transcriptionally activated by IRF-1 and STAT1 (1–3). Class II transactivator (CIITA) then leads to MHC-II promoter transcriptional complex assembly and histone acetylation, ultimately resulting in MHC-II activation (4).
CIITA is the master regulator of MHC-II and a member of the CATERPILLER family of genes (5, 6). Thus far, all CIITA-induced genes are in the MHC-II antigen presentation pathway except for plexin-A1, which nonetheless is important for the interaction of T cells and dendritic cells (7). CIITA is a scaffolding protein that bridges the MHC-II requisite transcription factors (RFX5, CREB, and NFY) (8) to various chromatin modifiers, including acetyltransferases [CREB-binding protein (CBP)/p300, P300/CBP-associated factor (PCAF), and steroid receptor coactivator-1 (SRC-1)] (9–13), ATP-dependent remodeling factors (BRG-1) (14, 15), and histone deacetylases (HDAC1) (16). These interactions are critical for the formation of a stable enhanceosome complex on the MHC-II promoter (17, 18).
Although roles for histone acetyltransferases and deacetylases in MHC-II regulation have been described, the possible implication of histone methylation has not been explored. Recent studies have demonstrated that arginine-specific methylation of histones H3 and H4 is an important modification modulating chromatin structure and gene transcription (19–21). In addition, methylation of nonhistone substrates involved in transcription, such as CBP/p300, has also emerged as a critical feature for transcriptional regulation (22, 23). There are seven arginine methyltransferases in mammals: protein-arginine methyltransferase (PRMT) 1, PRMT3, CARM1/PRMT4, PRMT5/JBP1, PRMT6, PRMT7, and PRMT8 (24, 25). The coactivator-associated arginine methyltransferase 1 (CARM1) preferentially methylates histone H3 (19, 26, 27) and cooperates with hormone receptor coactivators such as GRIP1 and SRC-1 and with the acetyltransferases CBP/p300 and P300/PCAF to enhance the ability of nuclear receptors to activate transcription (28–31). Recently, CARM1 was also shown to be a promoter-specific regulator of NF-κB-dependent gene expression (32). Mutations in the catalytic domain of CARM1 dramatically reduce both its methyltransferase and coactivator activities, suggesting that arginine methylation is the basis of transcriptional coactivation by CARM1 (26, 28). In addition to histones, CARM1 also methylates the transcriptional coactivator CBP/p300 (22, 23, 33, 34).
CIITA has been shown to synergize with CBP/p300, P300/PCAF, and most recently the steroid receptor coactivator SRC-1, thereby enhancing expression of MHC-II (12, 13, 35). CARM1 also cooperates with these same coactivators to enhance transcription by nuclear receptors (28, 30). These observations prompted us to examine the possible involvement of CARM1 and protein methylation in transcription of MHC-II genes by CIITA.
Materials and Methods
Tissue Culture, Cells, and Conditions. COS7, 293T, HeLa, Raji, and RJ2.2.5 cells were maintained as described in refs. 7 and 16.
Reagents. CARM1 and E267Q expression vectors were obtained from M. Stallcup (University of Southern California, Los Angeles). PRMT1 expression vector was obtained from Y. Zhang (University of North Carolina). GST-CIITA was obtained from J. Papamatheakis (University of Crete). FLAG-CIITA, MYC-CIITA, MHC-II-Luc, HA-CBP, HA-CBP-Δ685–774, and HA-CBPR3A have been described in refs. 8, 23, 36, and 37. Anti-hemagglutinin epitope (HA) was obtained from Roche. Anti-FLAG M5 was obtained from Sigma. Anti-MYC 9E10, anti-CARM1 and antidimethyl-H3-R17 (Me-R17) were obtained from Upstate Biotechnology (Lake Placid, NY). Anti-CBP was obtained from Santa Cruz Biotechnology; anti-CIITA was obtained from Rockland Immunochemicals (38); and IFN-γ was obtained from PeproTech (Rocky Hill, NJ).
Transient Transfections and Luciferase Assays. COS7 cells (5 × 104) were plated in six-well plates and then transfected 18–24 h later by using FuGene 6 transfection reagent (Roche). Cells were lysed and luciferase assays were performed 18–24 h after transfection as described in ref. 39.
Chromatin Immunoprecipitation (ChIP) Assays. Chromatin from 1 × 107 Raji, 5 × 106 HeLa, or 1 × 106 293 T cells was prepared as described in ref. 38, and ChIPs were performed by using the ChIP assay kit (Upstate Biotechnology) as described in ref. 16. The following antibodies were used: 5 μg of anti-HA, 10 μl of anti-CARM1, 10 μl of anti-Me-R17, 10 μg of anti-CBP, and 20 μg of anti-CIITA. Analysis of the immunoprecipitated products was done by real-time PCR.
Real-Time PCR. cDNA synthesis and real-time PCR were performed as described in ref. 40. Values were calculated based on standard curves generated for each gene. Samples were normalized by dividing copies of MHC-II RNA (HLA-DRA) or CIITA by copies of 18S rRNA. Analysis of chromatin immunoprecipitated products was performed by using MHC-II (HLA-DRA) promoter-specific primers and probe, and values were determined by subtracting values obtained from beads-only immunoprecipitations and normalizing to the total amount of MHC-II promoter DNA (input). All primer and probe sequences have been described in ref. 16.
Small Interfering RNA (siRNA) Transfections and Luciferase Assays. The siRNA sequence targeting CARM1 corresponded to the coding region 129–149 relative to the first nucleotide of the start codon. CARM1-specific siRNA or the control siRNA (200 nM) (Dharmacon RNA Technologies, Layfayette, CO) was transfected into U2OS by using Oligofectamine reagent (Invitrogen). Cells were trypsinized and seeded at a density of 5 × 104 cells in 12-well plates 24 h after transfection. Twelve hours later, U2OS cell lines were transfected by the calcium phosphate coprecipitation procedure with 1 μg of MHC-II-luc reporter vector and 50 ng of CIITA expression vector. Empty vector was included to keep the total amount of DNA per well constant. pCMV-lacZ was included in each experiment as a control for transfection efficiency. Cells were harvested 30 h after transfection. Luciferase and β-galactosidase activities were measured with Promega and Tropix (Bedford, MA) kits, respectively.
Results
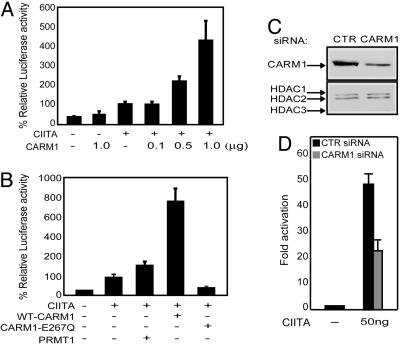
CARM1 Enhances CIITA-Mediated MHC-II Activation in a Methyltransferase-Dependent Manner. To investigate the possible role of histone methyltransferase in MHC-II transcription, we tested the effect of CARM1 on the function of CIITA in a transient transfection experiment in COS7 cells. Cotransfection of increasing amounts of CARM1 with CIITA resulted in the synergistic activation of an MHC-II promoter-driven reporter (Fig. 1A). A CARM1 mutant that lacks methyltransferase activity, E267Q (28), was also tested. The synergy between CIITA and CARM1 was abolished when the mutant E267Q was used (Fig. 1B). In contrast to CARM1, the arginine-specific methyltransferase PRMT1 exerted a modest effect on CIITA-mediated transcription, indicating an important role of CARM1 in MHC-II gene regulation (Fig. 1B). To further assess the role of CARM1 in CIITA-dependent transcription of MHC-II, we designed a siRNA duplex to specifically knock down endogenous CARM1 expression. As a control, we used a siRNA duplex, which does not anneal to any known mRNA. siRNA-mediated reduction of endogenous CARM1 by ≈50% relative to the control was verified by Western blot analysis (Fig. 1C Upper). The expression of several control proteins (HDACs 1–3) was not reduced by the CARM1 siRNA (Fig. 1C Lower). In control cells that express normal levels of CARM1, CIITA efficiently activated an MHC-II driven luciferase reporter, whereas in cells transfected with CARM1-specific siRNA, activation was reduced by half (Fig. 1D). This level of reduction correlates with the extent of CARM1 reduction seen in Fig. 1C.
Fig. 1.
CARM1 enhances CIITA transactivation in a methylation-dependent manner. (A) COS7 cells were cotransfected with 10 ng of CIITA, increasing amounts of CARM1, and 500 ng of the MHC-II-luc reporter construct. Cells were harvested 24 h later. Luciferase activity is reported as percent activation relative to that by CIITA alone. Values are shown as mean percent relative luciferase activity ± SEM for three experiments, each of which was repeated in triplicate. (B) COS7 cells were cotransfected with 10 ng of CIITA, 1 μg of CARM1, 1 μg of PRMT1 or the methylation-defective mutant E267Q, and 500 ng of the MHC-II-luc reporter construct. Luciferase activity was assayed and reported as in A.(C) Endogenous CARM1 expression in U20S cells transfected with control or CARM1 siRNA was detected by immunoblotting with anti-CARM1 (Upper). CARM1 siRNA successfully reduced the endogenous gene in U20S, hence this cell line was used in these experiments. Endogenous HDACs (1–3) served as controls (Lower). (D) U20S cells described in C were transfected with 50 ng of CIITA and 1 μg of the MHC-II-luc reporter construct. Luciferase activity is reported as fold activation relative to that of empty vector and was normalized to β-galactosidase.
CARM1 Enhances IFN-γ-Inducible and CIITA-Dependent Expression of Endogenous MHC-II. The experiments above used an artificial promoter–reporter system. To address the role of CARM1 in endogenous MHC-II gene expression, real-time PCR was used to measure endogenous MHC-II mRNA in HeLa cells. HeLa cells were used in all experiments measuring endogenous MHC-II expression because MHC-II is IFN-γ-inducible in these cells.
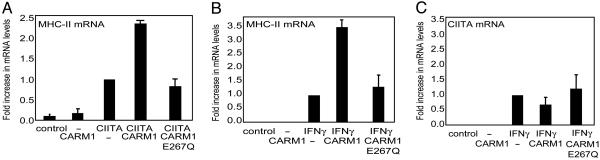
CARM1 significantly enhanced the expression of endogenous MHC-II but only in the presence of CIITA. The level of induction of MHC-II by CIITA is within the range of induction observed with physiologic inducers such as IFN-γ (41, 42). Furthermore, the coactivating effect of CARM1 is similar to that observed in another system (32). In contrast, the methyltransferase-deficient mutant, E267Q, did not enhance MHC-II expression (Fig. 2A). CIITA expression was not altered by CARM1 or the E267Q mutant as determined by immunoblot analysis (data not shown). To investigate the role of CARM1 with a physiologic stimulus of MHC-II gene, real-time PCR was used to measure endogenous MHC-II mRNA in HeLa cells after treatment with IFN-γ. CARM1 significantly enhanced endogenous MHC-II induced by IFN-γ (Fig. 2B). This enhancement was eliminated with the methyltransferase-deficient E267Q mutant. The level of endogenous CIITA transcript remained unaffected by CARM1 or the E267Q mutant (Fig. 2C).
Fig. 2.
Enhancement of CIITA-mediated and IFN-γ inducible expression of endogenous MHC-II depends on CARM1 methyltransferase activity. (A) HeLa cells were cotransfected with 0.5 μg of CIITA vector and 3 μg of CARM1 or CARM1-E267Q vector. MHC-II mRNA was measured by real-time PCR. Values are reported as fold increase in mRNA expression over a sample transfected with CIITA only. Samples were normalized to the number of 18S rRNA copies. (B) HeLa cells were induced with IFN-γ (25 ng/ml) for 24 h. Real-time PCR analysis was performed to measure endogenous mRNA levels of MHC-II in the presence of 3 μg of CARM1 or CARM1-E267Q vector. The value of the sample treated only with IFN-γ is set as 1. (C) CIITA mRNA levels from B were measured by real-time PCR. Values are reported as in B. All data shown are averages of three experiments.
One possible mechanism by which CARM1 enhances MHC-II transcription is by interaction with CIITA. To explore this interaction, coimmunoprecipitations of overexpressed CIITA and CARM1 were performed. CIITA efficiently associated with both wild-type CARM1 and E267Q (see Fig. 6 A and B, lanes 2 and 3, which is published as supporting information on the PNAS web site) but not to another nuclear MHC-II enhanceosome-associated protein, NF-YA (Fig. 6B, lane 4). This finding indicates that CARM1 interacts specifically with CIITA in vivo and that the loss of MHC-II transcriptional enhancement by E267Q is due not to the lack of its association with CIITA but to the lack of methyltransferase activity. However, thus far we have been unable to show the association of endogenous of CIITA and CARM1, which is likely due to the low level of endogenous proteins and to the lack of appropriate antibodies for endogenous CIITA. Nevertheless, in vitro evidence for a physical interaction between CIITA and CARM1 was obtained by GST pull-down experiments. In vitro-translated 35S-labeled CARM1 and E267Q mutant efficiently interacted with full-length CIITA immobilized on glutathione-Sepharose beads (Fig. 6C, lanes 3 and 6).
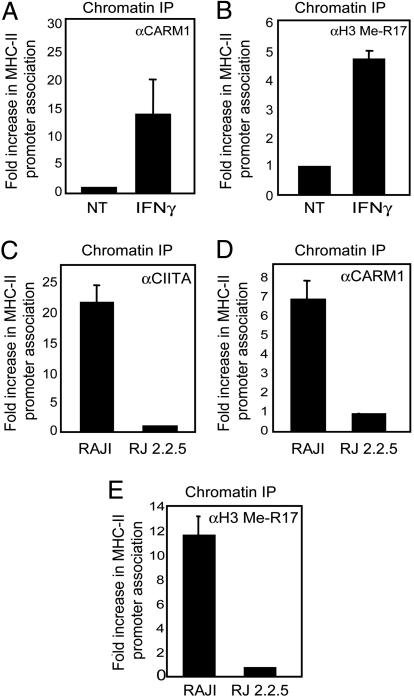
Recruitment of CARM1 to the Endogenous MHC-II Promoter Is Induced by IFN-γ. To test whether CARM1 is recruited to an endogenous MHC-II promoter, HLA-DRA, ChIP assay was used. HeLa cells were treated with IFN-γ (25 ng/ml) and endogenous CARM1 was immunoprecipitated. MHC-II promoter DNA sequences were detected by real-time PCR. IFN-γ significantly enhanced CARM1 association with the promoter (Fig. 3A) and the recruitment of CARM1 correlated with the methylation of histone H3-R17 (Me-R17), a known target of CARM1 (Fig. 3B) (19, 27), indicating that arginine methylation of histone H3 at the MHC-II promoter occurs during IFN-γ-induced MHC-II expression. To exclude the possibility that the results of Fig. 3A were due to altered expression of CARM1 after IFN-γ induction, extracts from cells untreated or treated with IFN-γ (25 ng/ml) for 24 h were immunoblotted with an anti-CARM1 antibody. No dramatic change in CARM1 expression was observed in these conditions (see Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 3.
CARM1 association with the MHC-II promoter requires CIITA. (A and B) Chromatin was prepared from HeLa cells that were untreated (NT) or treated with IFN-γ (25 ng/ml). ChIP assays were performed using anti-CARM1 or anti-Me-R17 as indicated. MHC-II promoter DNA was detected by real-time PCR. Data are presented as fold increases in relative promoter association of the immunoprecipitated protein compared with untreated cells. Real-time PCR values were determined by subtracting values obtained from bead-only immunoprecipitates and normalizing to the total amount of MHC-II promoter DNA (input). (C–E) Chromatin from Raji or RJ2.2.5 cells was prepared for ChIP assays using anti-CIITA, anti-CARM1, or anti-Me-R17 as indicated. MHC-II promoter DNA was detected by quantitative real-time PCR and values were determined as for A and B. Data are presented as fold change compared with the negative control RJ2.2.5. All data shown are representatives of three to five experiments.
CIITA Mediates CARM1 Promoter Association in a B Cell Line. The Raji and RJ2.2.5 cells lines respectively represent a CIITA-expressing, MHC-II+ B cell line and its CIITA-defective and MHC-II-negative variant (43). These cell lines provide another biologic system to analyze the effect of endogenous CIITA on CARM1. To investigate the mode of CARM1 recruitment to the MHC-II promoter in B cells, ChIP assays were performed. Both CIITA and CARM1 were associated with MHC-II promoter in Raji cells (Fig. 3C, left bar, and Fig. 3D, left bar). In the CIITA-deficient cell line RJ2.2.5, CIITA (Fig. 3C, right bar) and CARM1 (Fig. 3D, right bar) were not detected by ChIP on the promoter, indicating that a functional CIITA is required for optimal association of CARM1 with the MHC-II promoter. Me-R17 was also found at the MHC-II promoter in Raji cells but not in RJ2.2.5 cells (Fig. 3E). As a control for specificity, ChIP analysis of the β-actin promoter showed that neither CARM1 nor CIITA interacted with the β-actin promoter, and methylation that produces Me-R17 was not observed (data not shown). These data demonstrate that CARM1 is specifically recruited to the promoter in a CIITA-dependent fashion, resulting in histone methylation at the HLA-DRA promoter.
Methylation of CBP by CARM1 Is Required for CIITA-Dependent Transcription. The above data strongly suggest that CARM1 enhances the transcription of MHC-II genes in a methyltransferase-dependent manner. One possibility is that CARM1 regulates MHC-II transcription by methylating CIITA. To test this hypothesis, in vitro methylation assays were performed in which GST-CIITA was incubated with recombinant CARM1 in the presence of radiolabeled S-adenosyl-l-[methyl-3H]methionine (SAM). In contrast to the predicted result, CIITA was not methylated by CARM1 in vitro (see Fig. 8A, which is published as supporting information on the PNAS web site). In contrast, residues 685–774 of the histone acetyltransferase CBP were methylated by CARM1. The full-length CBP was also methylated by CARM1, and this methylation was not affected by including CIITA (Fig. 8B). This observation prompted us to examine whether CARM1-mediated methylation enhances MHC-II transcription by targeting CBP. CBP is known to enhance CIITA-dependent transcription and has been shown to interact and cooperate with CARM1 in nuclear receptor-dependent transcription (23, 28). However, the importance of CARM1-mediated methylation of CBP has not been explored in IFN-γ-regulated transcription.
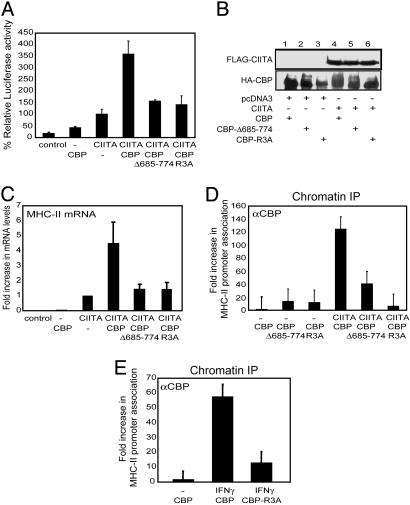
To address the potential role of CARM1-mediated methylation of CBP in MHC-II transcription, we took advantage of previous studies that showed that CARM1 methylates CBP in a region that spans residues 685–774 and that specifically targets arginines 714, 742, and 768 (23). Luciferase reporter assays using a mutant of CBP (CBP-Δ685–774) that lacks the region spanning residues 685–774 (Fig. 4A) were performed. Cotransfection of CIITA with wild-type CBP resulted in a synergistic enhancement of MHC-II activation. However, CBP-Δ685–774 failed to enhance MHC-II luciferase activity. The methylation-deficient CBP mutant R3A, in which arginines R714, R742, and R768 are replaced by alanines, also failed to enhance CIITA-dependent MHC-II reporter activation. CBP-R3A and wild-type CBP were expressed to a similar extent (Fig. 4B), indicating that these differences were not due to uneven expression. Furthermore, mutant and wild-type CBP had the same histone acetyltransferase activity using histones as a substrate as determined both in vivo and in vitro (see Fig. 9 A and B, which is published as supporting information on the PNAS web site). Moreover, both had similar stability and displayed a similar nuclear localization (Fig. 9 C and D). Finally, CBP mutant interacted with CIITA equally as well as the wild type (see Fig. 10, which is published as supporting information on the PNAS web site). These observations indicate that CBP mutant R3A is comparable to wild-type CBP except in its ability to be methylated by CARM1. Together, these results suggest that methylation of CBP by CARM1 at these arginine residues may be critical for enhanced MHC-II transcription. To extend these studies to the regulation of endogenous MHC-II genes, we cotransfected HeLa cells with CIITA and wild-type CBP or methylation-defective mutants and measured endogenous MHC-II (HLA-DRA) expression by real-time PCR (Fig. 4C). Whereas CBP enhanced CIITA-mediated expression of endogenous HLA-DRA transcripts, both CBP methylation mutants Δ685–774 and R3A failed to do so.
Fig. 4.
methylation sites within CBP are required for enhanced MHC-II transcription by CIITA. (A) COS7 cells were cotransfected with 10 ng of CIITA, 1 μg of CBP, CBP-Δ685–774, or CBP-R3A and 500 ng of the MHC-II-luc reporter construct. Cells were harvested 24 h later. Luciferase activity is reported as percent activation relative to that by CIITA alone. Values are shown as mean percent relative luciferase activity ± SEM for three experiments, each of which was repeated in triplicate. (B) 293T cells were cotransfected with FLAG-CIITA and HA-CBP, HA-Δ685–774 or HA-R3A. Whole cell lysates were prepared and expression of CIITA or CBP was confirmed by immunoblotting with anti-FLAG (Upper) or anti-HA (Lower) antibodies respectively. Data shown are representative of two experiments. (C) HeLa cells were cotransfected with 500 ng of CIITA and 3 μg of CBP, CBP-Δ685–774, or CBP-R3A. MHC-II mRNA levels were measured by real-time PCR. Values are reported as fold increase in mRNA expression. Samples were normalized to the number of 18S rRNA copies. Data shown are representative of three experiments. (D) 293T cells were cotransfected with FLAG-CIITA and HA-CBP, HA-Δ685–774, or HA-R3A. Chromatin was prepared 24 h after transfection, and ChIPs were performed with anti-HA to detect CBP. MHC-II promoter DNA was analyzed by real-time PCR. Data shown are representative of four experiments. (E) HeLa cells were transfected with HA-CBP or HA-R3A and treated with IFN-γ (25 ng/ml) where indicated. Chromatin was prepared 24 h after induction, and ChIPs were performed with anti-HA to detect CBP. MHC-II promoter DNA was analyzed by real-time PCR. Data shown are representative of three experiments.
The Triple Arginine Residues of CBP Are Necessary for Its Stable Association with the HLA-DRA Promoter. To investigate whether the methylation target sites within CBP are important for promoter association, we cotransfected 293T cells with FLAG-CIITA and HA-CBP or HA-tagged methylation-defective mutants (Δ685–774 and R3A) and performed ChIP assays using anti-HA antibody to detect CBP (Fig. 4D). MHC-II promoter DNA that immunoprecipitated with CBP was detected by real-time PCR. Wild-type CBP efficiently associated with the promoter in the presence of CIITA but not in its absence. In contrast, promoter association of both CBP-Δ685–774 and R3A, which lacked the CARM1 methylation sites, was significantly decreased, suggesting that methylation of one or more of the arginines R714, R742, and R768 may be critical for the stable association of CBP with the endogenous HLA-DRA promoter. To extend these studies to a more physiological inducer, we transfected HeLa cells with either wild-type HA-tagged CBP or the mutant R3A and performed ChIP assays with anti-HA antibody after induction with IFN-γ. MHC-II promoter DNA immunoprecipitated with anti-HA was detected by real-time PCR (Fig. 4E). Wild-type CBP efficiently associated with the promoter after IFN-γ induction, but promoter association of R3A was significantly decreased, indicating the importance of these three arginine residues in CBP association with the MHC-II promoter during an IFN-γ response.
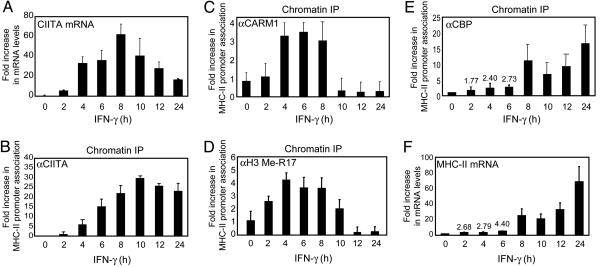
IFN-γ Causes the Coordinated Recruitment of CIITA, CARM1, and CBP to the MHC-II Promoter. The results thus far suggest that CARM1-mediated methylation of CBP at specific arginines within residues 685–774 may be critical for its stable association with the MHC-II promoter (Fig. 4D). This observation suggests that the activities of chromatin-modifying coactivators, such as CBP and CARM1, are likely coordinated during MHC-II gene induction. To investigate the kinetic profile of coactivator recruitment to the endogenous MHC-II promoter during a physiologic stimulus, we treated HeLa cells with IFN-γ over a 24-h time course and performed ChIP assays. CIITA association with the MHC-II promoter was detected 2–4 h after IFN-γ induction and was subsequently enhanced and sustained throughout the time course (Fig. 5B). This induction pattern during the first 8 h matches the profile of CIITA induction by IFN-γ as measured by real-time PCR (Fig. 5A). A basal level of CARM1 was detected at the MHC-II promoter before induction and was later enhanced 4 h after exposure to IFN-γ and maintained until 8 h after exposure (Fig. 5C). At later time points (10 h and later), promoter association of CARM1 with the promoter was not detected, indicating the dissociation of CARM1. The possibility exists that the antibody no longer recognizes CARM1; however, the correlative loss of Me-R17 (a specific modification of CARM1) (see below) suggests that this possibility is less likely. The association of CARM1 with the MHC-II promoter is correlated with enhanced methylation of H3 (Me-R17) at all time points except the 10-h time point. At the 10 h time point, methylated histone was still detected after CARM1 had already reverted to a basal level (Fig. 5D). This result suggests that methylated H3 remained associated with MHC-II promoter shortly after the dissociation of CARM1. However, beyond this time point, Me-R17 was also very low.
Fig. 5.
IFN-γ inducible recruitment of CARM1 to the MHC-II promoter correlates with CIITA but precedes CBP. (A) Expression profile of endogenous CIITA mRNA was determined by real-time PCR in HeLa cells after treatment with IFN-γ (25 ng/ml) at the indicated time points. Values are reported as fold increase in mRNA levels. Data shown are representative of three experiments. (B) Chromatin was prepared from HeLa cells after treatment with IFN-γ (25 ng/ml) at the indicated time points and ChIPs were performed using an anti-CIITA antibody. Data shown are averages of three experiments. (C) Chromatin was prepared as indicated in B, and ChIPs were performed with an anti-CARM1 antibody. Data shown are averages of seven experiments for CARM1. (D) Chromatin was prepared as described in B and immunoprecipitations were performed with anti-Me-R17 antibody. Data shown are averages of four experiments. (E) Chromatin was prepared as described in B, and ChIPs were performed with anti-CBP antibody. Data shown are averages of three experiments. (F) Expression profile of endogenous MHC-II mRNA was determined by real-time PCR in HeLa cells after treatment with IFN-γ (25 ng/ml) at the indicated time points. Values are reported as fold increase in mRNA levels. Data shown are representative of three experiments.
Low levels of CBP were detected at the promoter during early time points (2–6 h), but association peaked at 8 h (Fig. 5E), which was preceded by the association of CIITA and CARM1 with the promoter (Fig. 5 B and C). CBP association with the promoter was sustained for the duration of the time course. significantly, CBP recruitment correlated with the onset of MHC-II expression, which was also significantly enhanced at 8 h after IFN-γ induction (Fig. 5F), suggesting that recruitment of CBP to the promoter is a key step in the initiation of MHC-II gene transcription. Collectively, these results indicate that the association of transcription factors and modifying enzyme at the MHC-II promoter is a highly coordinated and dynamic process.
Discussion
Interactions of CIITA with the MHC-II requisite transcription factors, the basal transcription machinery, and chromatin modifiers play a fundamental role in MHC-II promoter assembly (4). However, the role of arginine methylation, a modification recently shown to be an important player in the regulation of chromatin structure and transcription (24, 44), has not yet been explored. Here, we showed that the arginine methyltransferase CARM1 enhances CIITA-dependent and IFN-γ-induced MHC-II transcription, and this enhancement depends on an intact methyltransferase activity. CARM1 may have two functions, the methylation of H3 at arginine-17 and the methylation of CBP. The latter is supported by the requirement for intact methylation sites within CBP for its association with the endogenous MHC-II promoter and for its activation of MHC-II genes. Whereas CARM1 has been shown to be an important component of nuclear receptor-dependent transcription, this report specifically indicates its role in IFN-γ-induced transcription.
This study shows a dynamic series of events involving CIITA, CARM1, and CBP. CARM1 is not observed at the MHC-II promoter in the CIITA-negative RJ2.2.5 cells, indicating a role of CIITA in recruiting CARM1 to the endogenous MHC-II promoter. In turn, the molecular mechanism by which CARM1 enhances the expression of MHC-II genes depends on an intact methyltransferase domain. Me-R17 is highly enriched at the MHC-II promoter shortly after IFN-γ induction, correlating with the presence of CARM1 at the promoter. Although the functional consequence of Me-R17 on the localized chromatin structure is not yet fully understood, a prevailing view is that this modification could facilitate chromatin alterations necessary for transcription (45, 46). In addition to histone methylation, CARM1 also methylates CBP within a region containing arginines R714, R742, and R768. These residues are required for the association of CBP with the MHC-II promoter, suggesting that the methylation of CBP by CARM1 is likely important for promoter association. In addition, the kinetics study shows that whereas a maximal level of CARM1 was found at the MHC-II promoter 4 h after IFN-γ treatment, CBP–promoter association trailed that of CARM1.
Notably, methylation of CBP at arginines 714, 742, and 768 is not known to affect transcription by CREB, an important transcription factor for MHC-II expression (23, 47). CBP methylation within the kinase-inducible (KIX) domain has been previously demonstrated to inhibit the interaction between CBP and CREB, and thus decrease CREB-mediated transcription (22). However, the methylation sites within the KIX domain are entirely different from the ones investigated here and do not affect CREB transactivation (23).
A recent paper by Gomez et al. (48) examined IFN-γ-induced chromatin changes, albeit at a later time point (16, 24, and 48 h). A host of modifications involving histone acetylation were documented. Whether these later modifications are predicated on CARM1-mediated methylation of H3-R17 to produce Me-R17 or whether they influence the function and release of CARM1 will be of future interest.
In summary, our results are consistent with the following model. Shortly after IFN-γ stimulation, CIITA expression is induced and is recruited to the MHC-II promoter. CIITA then enhances the recruitment of CARM1, which mediates methylation that produces Me-R17. CARM1 also methylates CBP, thereby facilitating its association with the promoter. Finally, CARM1 dissociates from the promoter, whereas promoter association with CIITA and CBP is more sustained. These results indicate a coordinated sequence of events involving the interplay of CIITA, CARM1, and CBP during IFN-γ-induced gene activation.
Supplementary Material
Acknowledgments
We thank Drs. Michael Stallcup, Yi Zhang, and Joseph Papamatheakis for reagents; Susanna Greer for careful review of the manuscript; Brian Strahl for helpful discussion; and Brian O'Connor and Zhengmao ye for assistance with β-actin promoter PCR and real-time PCR. We also thank John Lich for the interaction of endogenous CARM1 and CIITA. This work was supported by National Institutes of Health Grants AI29564, AI45580, and AI41751 (to J.P.-Y.T.).
Author contributions: E.Z., L.V., and J.P.-Y.T. designed research; E.Z., L.F., and L.V. performed research; L.V. contributed new reagents/analytic tools; E.Z., L.F., L.V., and J.P.-Y.T. analyzed data; E.Z., L.V., and J.P.-Y.T. wrote the paper; and J.P.-Y.T. supervised the study.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CIITA, class II transactivator; CARM1, coactivator-associated arginine methyltransferase 1; CBP, CREB-binding protein; ChIP, chromatin immunoprecipitation; CREB, acetyltransferase cyclic-AMP response element binding; HA, hemagglutinin epitope; HDAC, histone deacetylase; Me-R17, dimethyl-H3-R17; MHC-II, major histocompatibility class II; PRMT, protein arginine methyltransferase; P/CAF, P300/CBP-associated factor; siRNA, small interfering RNA.
References
- 1.Ramana, C. V., Chatterjee-Kishore, M., Nguyen, H. & Stark, G. R. (2000) Oncogene 19, 2619-2627. [DOI] [PubMed] [Google Scholar]
- 2.Williams, B. R. (1991) Eur. J. Biochem. 200, 1-11. [DOI] [PubMed] [Google Scholar]
- 3.Ransohoff, R. M. (1998) N. Engl. J. Med. 338, 616-618. [DOI] [PubMed] [Google Scholar]
- 4.Ting, J. P. & Trowsdale, J. (2002) Cell 109, Suppl. 1, S21-S33. [DOI] [PubMed] [Google Scholar]
- 5.Harton, J. A., Linhoff, M. W., Zhang, J. & Ting, J. P. (2002) J. Immunol. 169, 4088-4093. [DOI] [PubMed] [Google Scholar]
- 6.Ting, J. P. & Davis, B. K. (2004) Annu. Rev. Immunol. 23, 387-414. [DOI] [PubMed] [Google Scholar]
- 7.Wong, A. W., Brickey, W. J., Taxman, D. J., van Deventer, H. W., Reed, W., Gao, J. X., Zheng, P., Liu, Y., Li, P., Blum, J. S., et al. (2003) Nat. Immunol. 4, 891-898. [DOI] [PubMed] [Google Scholar]
- 8.Zhu, X. S., Linhoff, M. W., Li, G., Chin, K. C., Maity, S. N. & Ting, J. P. (2000) Mol. Cell. Biol. 20, 6051-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spilianakis, C., Papamatheakis, J. & Kretsovali, A. (2000) Mol. Cell. Biol. 20, 8489-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sisk, T. J., Gourley, T., Roys, S. & Chang, C. H. (2000) J. Immunol. 165, 2511-2517. [DOI] [PubMed] [Google Scholar]
- 11.Zhu, X. S. & Ting, J. P. (2001) Mol. Cell. Biol. 21, 7078-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kretsovali, A., Agalioti, T., Spilianakis, C., Tzortzakaki, E., Merika, M. & Papamatheakis, J. (1998) Mol. Cell. Biol. 18, 6777-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzortzakaki, E., Spilianakis, C., Zika, E., Kretsovali, A. & Papamatheakis, J. (2003) Mol. Endocrinol. 17, 2509-2518. [DOI] [PubMed] [Google Scholar]
- 14.Mudhasani, R. & Fontes, J. D. (2002) Mol. Cell. Biol. 22, 5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattenden, S. G., Klose, R., Karaskov, E. & Bremner, R. (2002) EMBO J. 21, 1978-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zika, E., Greer, S. F., Zhu, X. S. & Ting, J. P. (2003) Mol. Cell. Biol. 23, 3091-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masternak, K., Muhlethaler-Mottet, A., Villard, J., Zufferey, M., Steimle, V. & Reith, W. (2000) Genes Dev. 14, 1156-1166. [PMC free article] [PubMed] [Google Scholar]
- 18.Spilianakis, C., Kretsovali, A., Agalioti, T., Makatounakis, T., Thanos, D. & Papamatheakis, J. (2003) EMBO J. 22, 5125-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer, U. M., Daujat, S., Nielsen, S. J., Nightingale, K. & Kouzarides, T. (2002) EMBO Rep. 3, 39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strahl, B. D., Briggs, S. D., Brame, C. J., Caldwell, J. A., Koh, S. S., Ma, H., Cook, R. G., Shabanowitz, J., Hunt, D. F., Stallcup, M. R. & Allis, C. D. (2001) Curr. Biol. 11, 996-1000. [DOI] [PubMed] [Google Scholar]
- 21.Wang, H., Huang, Z. Q., Xia, L., Feng, Q., Erdjument-Bromage, H., Strahl, B. D., Briggs, S. D., Allis, C. D., Wong, J., Tempst, P. & Zhang, Y. (2001) Science 293, 853-857. [DOI] [PubMed] [Google Scholar]
- 22.Xu, W., Chen, H., Du, K., Asahara, H., Tini, M., Emerson, B. M., Montminy, M. & Evans, R. M. (2001) Science 294, 2507-2511. [DOI] [PubMed] [Google Scholar]
- 23.Chevillard-Briet, M., Trouche, D. & Vandel, L. (2002) EMBO J. 21, 5457-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides, T. (2002) Curr. Opin. Genet. Dev. 12, 198-209. [DOI] [PubMed] [Google Scholar]
- 25.Miranda, T. B., Miranda, M., Frankel, A. & Clarke, S. (2004) J. Biol. Chem. 279, 22902-22907. [DOI] [PubMed] [Google Scholar]
- 26.Chen, D., Ma, H., Hong, H., Koh, S. S., Huang, S. M., Schurter, B. T., Aswad, D. W. & Stallcup, M. R. (1999) Science 284, 2174-2177. [DOI] [PubMed] [Google Scholar]
- 27.Ma, H., Baumann, C. T., Li, H., Strahl, B. D., Rice, R., Jelinek, M. A., Aswad, D. W., Allis, C. D., Hager, G. L. & Stallcup, M. R. (2001) Curr. Biol. 11, 1981-1985. [DOI] [PubMed] [Google Scholar]
- 28.Lee, Y. H., Koh, S. S., Zhang, X., Cheng, X. & Stallcup, M. R. (2002) Mol. Cell. Biol. 22, 3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, D., Huang, S. M. & Stallcup, M. R. (2000) J. Biol. Chem. 275, 40810-40816. [DOI] [PubMed] [Google Scholar]
- 30.Koh, S. S., Chen, D., Lee, Y. H. & Stallcup, M. R. (2001) J. Biol. Chem. 276, 1089-1098. [DOI] [PubMed] [Google Scholar]
- 31.Stallcup, M. R., Chen, D., Koh, S. S., Ma, H., Lee, Y. H., Li, H., Schurter, B. T. & Aswad, D. W. (2000) Biochem. Soc. Trans. 28, 415-418. [PubMed] [Google Scholar]
- 32.Covic, M., Hassa, P. O., Saccani, S., Buerki, C., Meier, N. I., Lombardi, C., Imhof, R., Bedford, M. T., Natoli, G. & Hottiger, M. O. (2005) EMBO J. 24, 85-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, H., Park, S., Kilburn, B., Jelinek, M. A., Henschen-Edman, A., Aswad, D. W., Stallcup, M. R. & Laird-Offringa, I. A. (2002) J. Biol. Chem. 277, 44623-44630. [DOI] [PubMed] [Google Scholar]
- 34.Lee, J. & Bedford, M. T. (2002) EMBO Rep. 3, 268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontes, J. D., Kanazawa, S., Jean, D. & Peterlin, B. M. (1999) Mol. Cell. Biol. 19, 941-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cressman, D. E., Chin, K. C., Taxman, D. J. & Ting, J. P. (1999) Immunity 10, 163-171. [DOI] [PubMed] [Google Scholar]
- 37.Cressman, D. E., O'Connor, W. J., Greer, S. F., Zhu, X. S. & Ting, J. P. (2001) J. Immunol. 167, 3626-3634. [DOI] [PubMed] [Google Scholar]
- 38.Greer, S. F., Zika, E., Conti, B., Zhu, X. S. & Ting, J. P. (2003) Nat. Immunol. 4, 1074-1082. [DOI] [PubMed] [Google Scholar]
- 39.Piskurich, J. F., Linhoff, M. W., Wang, Y. & Ting, J. P. (1999) Mol. Cell. Biol. 19, 431-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong, A., Ghosh, N., McKinnon, K. P., Reed, W., Piskurich, J. F., Wright, K. L. & Ting, J. P. (2002) J. Immunol. 169, 3112-3119. [DOI] [PubMed] [Google Scholar]
- 41.Most, J., Schaweble, W., Drach, J., Sommerauer, A. & Dierich, M. P. (1992) J. Immunol. 148, 1635-1642. [PubMed] [Google Scholar]
- 42.Littman, B. H., Dastvan, F. F., Carlson, P. L. & Sanders, K. M. (1989) J. Immunol. 142, 520-525. [PubMed] [Google Scholar]
- 43.Accolla, R. S. (1983) J. Exp. Med. 157, 1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y. & Reinberg, D. (2001) Genes Dev. 15, 2343-2360. [DOI] [PubMed] [Google Scholar]
- 45.Trievel, R. C. (2004) Crit. Rev. Eukaryotic Gene Expression 14, 147-169. [DOI] [PubMed] [Google Scholar]
- 46.Bedford, M. T. & Richard, S. (2005) Mol. Cell 18, 263-272. [DOI] [PubMed] [Google Scholar]
- 47.Moreno, C. S., Beresford, G. W., Louis-Plence, P., Morris, A. C. & Boss, J. M. (1999) Immunity 10, 143-151. [DOI] [PubMed] [Google Scholar]
- 48.Gomez, J. A., Majumder, P., Nagarajan, U. M. & Boss, J. M. (2005) J. Immunol. 175, 1030-1040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.