Abstract
Every residue of the 101-aa Escherichia coli toxin CcdB was substituted with Ala, Asp, Glu, Lys, and Arg by using site-directed mutagenesis. The activity of each mutant in vivo was characterized as a function of Controller of Cell Division or Death B protein (CcdB) transcriptional level. The mutation data suggest that an accessibility value of 5% is an appropriate cutoff for definition of buried residues. At all buried positions, introduction of Asp results in an inactive phenotype at all CcdB transcriptional levels. The average amount of destabilization upon substitution at buried positions decreases in the order Asp>Glu>Lys>Arg>Ala. Asp substitutions at buried sites in two other proteins, maltose-binding protein and thioredoxin, also were shown to be severely destabilizing. Ala and Asp scanning mutagenesis, in combination with dose-dependent expression phenotypes, was shown to yield important information on protein structure and activity. These results also suggest that such scanning mutagenesis data can be used to rank order sequence alignments and their corresponding homology models, as well as to distinguish between correct and incorrect structural alignments. With continuous reductions in oligonucleotide costs and increasingly efficient site-directed mutagenesis procedures, comprehensive scanning mutagenesis experiments for small proteins/domains are quite feasible.
Keywords: accessibility, aspartate, scanning mutagenesis, phenotype
It is well known that buried residues in a protein are important determinants of protein stability while surface residues are involved in protein function (1). Residue burial in a protein structure is typically quantitated in terms of accessible surface area and percent side-chain accessibility of a residue in a protein (ACC) (2). ACC is the accessible surface area of the residue relative to that found for the same residue in a Gly-X-Gly tripeptide of extended conformation, and burial is often equated to 100-ACC (3). There is no universally accepted definition of what ACC cutoff should be used to distinguish buried from surface residues, although a variety of values ranging from 5% to 25% have been used (4, 5).
In the absence of a 3D structure, it is not always straightforward to determine the extent of residue burial experimentally. For residues with chemically reactive functional groups (such as Lys or Cys), it is possible to probe burial by examining the reactivity toward residue-specific labeling reagents. Solvent accessibilities of residues also can be characterized by using site-directed fluorescence labeling monitored by fluorescent anisotropy and lifetime measurements (6). However, labeling rates are often influenced by local geometries, the nature of surrounding residues, and other features besides side-chain burial. Hydrogen exchange is another powerful tool for studying protein structure and dynamics. Unfortunately, hydrogen-exchange rates are poorly correlated with residue burial (7). Recently, a combination of synchrotron radiolysis and mass spectrometry (MS) has been used to provide qualitative information on residue burial and residues involved in protein:protein or protein:DNA interaction (8). In the present work, we examine the feasibility of using scanning mutagenesis to distinguish between buried and exposed positions and also to arrive at an experimental definition of the appropriate ACC cutoff to distinguish between buried and exposed residues. The experimental system, Controller of Cell Division or Death B protein (CcdB), is a 101-residue, homodimeric protein encoded by F plasmid. The protein is an inhibitor of DNA gyrase and is a potent cytotoxin in Escherichia coli (9). Crystallographic structures of CcdB in the free and gyrase bound forms (10, 11) are also available. Transformation of normal E. coli cells with plasmid expressing the wild-type (WT) ccdb gene results in cell death. If the protein is inactivated through mutation, cells transformed with such mutant genes will survive. Each residue of CcdB was replaced with Ala, Asp, Glu, Lys, and Arg. All mutants were expressed under the control of the PBAD promoter, which allows for dose-dependent protein expression by varying the amount of inducer added (12). CcdB phenotype was assayed as a function of expression level and residue burial by monitoring the presence or absence of cell growth.
Methods
Plasmids and Host Strains. The ccdb gene was cloned under the control of the arabinose PBAD promoter in the vector pBAD24 to yield the construct pBAD24CcdB (13). Three E. coli host strains were used, TOP10, XL1-Blue, and CSH501. TOP10 is sensitive to the action of CcdB and used for screening the phenotype. XL1-Blue is able to tolerate low levels of CcdB protein expression because of the presence of the antidote CcdA, which is encoded by the resident F plasmid and was used for plasmid propagation. CSH501 is completely resistant to the action of CcdB because the strain harbors the GyrA462 mutation in its chromosomal DNA and prevents gyrase from binding to CcdB. CSH501 was kindly provided by M. Couturier (Universite Libre de Bruxelles, Brussels) and was used for monitoring expression of mutant proteins. Maltose-binding protein (MBP) and thioredoxin (Trx) genes also were cloned in pBAD24. The strains A307 and Pop6590 deleted for chromosomal trxA and malE genes were used for monitoring expression of Trx and MBP mutants. Pop6590 was kindly provided by M. Hofnung (Institut Pasteur, Paris), and A307 was received from the E. coli Genetic Stock Center at Yale University.
Mutagenesis. Thirty-nucleotide-long primers to generate CcdB mutants were designed by using oligo (Version 6.0, Molecular Biology Insights, Cascade, CO) and were obtained in 96-well format from the PAN Oligo facility at Stanford University. Each residue in CcdB was replaced with Ala, Asp, Glu, Arg, and Lys by using a mega-primer-based method of site-directed mutagenesis. The first PCR reaction was carried out in 96-well format by using PCR strips (each having eight tubes). Each tube contained a specific internal mutagenic primer and a vector-specific primer. Twenty cycles of PCR were carried out by using a low annealing temperature of 50°C to generate a short product, which serves as a mega-primer in the next amplification to obtain full-length mutant plasmid. Subsequently, 20 cycles of a PCR-like reaction were carried out without any annealing step to prevent further formation of the mega-primer. WT template was digested with DpnI, leaving only mutant plasmid. This product was transformed into E. coli strain XL1-Blue (transformation efficiency 107 to 108 per μg of pUC18 DNA). A similar procedure was used to generate mutants of MBP and Trx.
Sequencing. Templates for sequencing to confirm mutations in CcdB were isolated directly from either a colony or culture of mutant plasmid transformed in XL1-Blue using the Templiphi reaction kit (Amersham Pharmacia Bisociences). The entire coding region of CcdB was subjected to automated DNA sequencing. After sequence confirmation, plasmids were isolated from XL1-Blue grown in 96-deep-well plates. MBP and Trx mutations were confirmed by sequencing of the plasmids.
Screening of Phenotype of CcdB Mutants. Mutant CcdB plasmids were transformed in TOP10 in 96-well format by using PCR strips, and activity was assayed by plating 5 μl of transformation mix on square LB-amp plates (120 × 120 mm) placed on 96-well grids in the absence of arabinose at 37°C. Because active CcdB is toxic to E. coli, only cells transformed with inactive mutants will survive. The phenotype of all mutants that were inactive at 0% arabinose also was examined at 0.001%, 0.01%, and 0.1% arabinose inducer. Expression level was monitored for all inactive mutants in CSH501 in the presence of 0.1% arabinose. Cultures were grown in 96-deep-well plates. After cell lysis by a freeze-thaw method (14), expression and solubility of all aspartate mutants of CcdB in CSH501 were monitored using SDS/PAGE. MBP and Trx WT and aspartate mutant expression was monitored in Pop6590 and A307, respectively. Expression and solubility of these mutants also were monitored in DH5α after detergent-based lysis (Bugbuster, Novagen).
Data Analysis. The phenotypes of the various mutants were determined as a function of inducer concentration. The data were analyzed by using Microsoft access database management systems software by generating various queries as a function of residue ACC and inducer concentration.
Trx Purification and in Vitro Insulin Aggregation Assay. WT-Trx and Trx78D mutant were purified to homogeneity from DH5α by using chloroform shock followed by anion exchange chromatography as described in ref. 15. Masses were confirmed by electron spray ionization MS. Far- and near-UV circular dichroism (CD) spectra for WT and mutant Trx were acquired in 10 mM phosphate buffer (pH 7.0) on a J715A spectropolarimeter (Jasco, Tokyo). Protein concentration used was 9 μM for far-UV CD and 20 μM for near-UV CD. Fluorescence spectra were acquired on a SPEX-Fluoromax 3 instrument by using 1 μM protein concentration in the absence and presence of a 10-fold excess of DTT. Insulin aggregation assay was performed by using 1 μM protein and 0.14 mM insulin in 100 mM phosphate buffer (pH 6.5) and 3 mM DTT. Insulin aggregation was monitored by the absorbance at 650 nm as a function of time (16). Time scans were taken for 3,600 sec.
Homology Modeling Studies. Structural homologs of CcdB were identified, and corresponding structure-based sequence alignments of CcdB with these homologs were carried out by using dali (www.ebi.ac.uk/dali). The CcdB sequence also was threaded onto its structural homologs by using 3dpssm (www.sbg.bio.ic.ac.uk/~3dpssm), 123d (http://123D.ncifcrf.gov), and fugue (www-cryst.bioc.cam.ac.uk/fugue) online threading softwares. In a separate series of studies, decoy templates with approximately the same length as CcdB and dimeric in nature, but of unrelated structure, were selected from Protein Quaternary Server database at European Bioinformatics Institute (http://pqs.ebi.ac.uk). Sequence alignments of CcdB with these decoys were generated by fugue online threading software as indicated above. In all of the cases, homology models of CcdB were generated from the sequence alignments by using modeller 7v7 (17).
Results and Discussion
CcdB Mutagenesis Data. A total of 390 of the 454 possible site-directed mutants were isolated at all 101 positions of CcdB. All mutants at position 1 (the initiator Met) were inactive and are not considered in the subsequent analysis. Tables 5 and 6, which are published as supporting information on the PNAS web site, show the phenotypes for each mutant at low (0% arabinose) and high (0.1% arabinose) levels of expression. Table 7, which is published as supporting information on the PNAS web site, summarizes the level of tolerance for each of the six substitutions averaged over all positions of the protein. The level of tolerance is quite sensitive to expression level. At low levels of expression (0% arabinose), an appreciably smaller percentage of mutants show an active phenotype (Fig. 1) for the following reason: at a given inducer concentration, destabilization of the protein through mutation results in a decrease in the amount of soluble protein in vivo. If this amount falls below the threshold required for activity, the mutant shows an inactive phenotype. However, at a higher inducer concentration, there is an increase in the total amount of soluble protein, and the same mutant is more likely to show an active phenotype (Fig. 1). As has been shown previously (13), phenotype is a function of expression level, and overexpression often results in the rescue of inactive phenotypes. The PBAD promoter has been shown to have an induction/repression ratio of ≈250-fold (12). Hence, any mutant that shows an inactive phenotype even upon induction has an activity 250-fold lower than that of WT. At higher expression levels, a remarkably high fraction of the mutants are active, given the drastic nature of most of the amino acid substitutions described above. However, Asp is clearly less well tolerated than the remaining amino acids. Table 1 shows mutational effects on activity as a function of residue burial and expression level. Residues were subdivided into four categories based on the ACC of the WT residue. ACC cutoffs were chosen to have approximately equal numbers of residues in each category. As expected, there is an increase in the fraction of active mutants with increase in ACC. However, residues with 0–5% ACC behave in a markedly different fashion from other residues. Indeed, there is insignificant difference between residues in the other three burial categories, especially at high expression levels of the protein. Residues can therefore be classified into two primary categories, buried ones with ACC of <5% and exposed ones with an ACC of >5%. There does not appear to be a strong case for having a third class of intermediately buried residues that have been used in some prior analyses (6, 18, 19); however, see below for additional discussion.
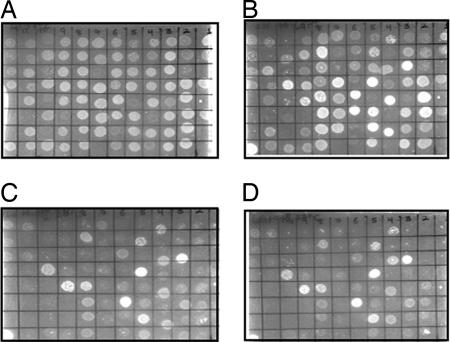
Fig. 1.
Ninety-six-well screening to obtain phenotypes of CcdB mutants as a function of inducer arabinose concentration at 37°C. Mutants were transformed into E. coli strain Top10 and plated at 0% (A), 0.001% (B), 0.01% (C), and 0.1% (D) arabinose. Identical grid positions on different plates correspond to the same mutant. Cells transformed with active mutants do not grow, whereas those transformed with inactive mutants will grow. The fraction of mutants showing an active phenotype clearly increases with increasing inducer concentration.
Table 1. Fraction of active mutants as a function of residue burial and expression level.
| Ala
|
Asp
|
Glu
|
Lys
|
Arg
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Active, %
|
Active, %
|
Active, %
|
Active, %
|
Active, %
|
|||||||||||
| ACC,* % | Total | 0% | 0.1% | Total | 0% | 0.1% | Total | 0% | 0.1% | Total | 0% | 0.1% | Total | 0% | 0.1% |
| 0–5 (22) | 18 | 44 | 77 | 18 | 0 | 0 | 20 | 5 | 35 | 18 | 5.5 | 61 | 20 | 16 | 68 |
| 5–15 (19) | 17 | 70 | 100 | 16 | 56 | 87 | 15 | 66 | 100 | 16 | 50 | 93 | 15 | 73 | 86 |
| 15–40 (20) | 11 | 91 | 100 | 13 | 61 | 76 | 14 | 64 | 71 | 14 | 71 | 92 | 15 | 80 | 86 |
| >40 (40) | 33 | 93 | 93 | 29 | 90 | 90 | 29 | 90 | 90 | 35 | 86 | 89 | 24 | 88 | 100 |
| Total | 79 | 76 | 78 | 83 | 74 | ||||||||||
Total refers to the total number of substitutions made in this ACC class; Active refers to the fraction of active mutants; 0% and 0.1% refer to the arabinose inducer concentrations at the lowest and highest expression levels of CcdB, respectively.
Values in parentheses represent the number of residues in this ACC class in WT CcdB
Introduction of charged residues at buried (<5% ACC) positions typically results in an inactive phenotype at low expression levels. In a large fraction of cases, overexpression of such mutants results in an active phenotype (Table 1). However, at all buried positions, substitution of the WT residue by Asp invariably leads to an inactive phenotype at all expression levels. Hence, introduction of Asp at buried positions appears to be more destabilizing than introduction of larger, charged amino acids. Because Asp is a relatively small and rigid amino acid, the carboxylate side chain is likely to remain buried in the mutant protein. For longer and more flexible side chains, it is possible (although we have no structural data to support this hypothesis) that the charged groups may find a way to reach the protein surface, thereby minimizing the potentially destabilizing effects of the substitution. In addition, the longer charged side chains contain several hydrophobic methylene groups that can remain stably buried in the protein interior. In support of such an explanation, the amount of destabilization, upon substitution at buried positions, appears to decrease in the order Asp>Glu>Lys>Arg.
Correlation of Residue Burial with Mutagenesis Data. Because introduction of Asp at buried positions invariably leads to loss of activity, this result suggests that Asp scanning mutagenesis may be a useful method of identifying buried positions in a protein of unknown structure. For such an approach to be useful, it should generate only a small number of “false positives,” i.e., Asp substitutions at nonburied positions should not lead to loss of activity, except at surface positions important for activity of the protein. Such surface positions can be identified by Ala scanning mutagenesis (20). Thus, a combination of Asp and Ala scanning mutagenesis, when combined with dose-dependent expression phenotypes, can yield important information on protein structure and activity. Active-site residues (involved in DNA gyrase binding) were identified from the recent CcdB:DNA gyrase crystal structure (10) using the “Structure Analysis” module of ccp4 (21) using a contact distance of 4 Å. This set consisted of residues 24, 25, 26, 87, 88, 91, 92, 95, 99, 100, and 101. Table 2 summarizes the kind of information that can be obtained from such studies. Essentially, phenotypes of Ala mutants help to identify many functionally important residues, which are typically solvent accessible. Phenotypes of Asp mutants provide information about residue burial. The combination of these two scanning mutagenesis strategies provides valuable additional information. Because Ala and Asp substitution phenotypes are monitored at two different expression levels, there is a maximum of 16 different combinations of phenotypes. However, in a simple bacterial system, some of these combinations are unlikely to occur, for example mutants that show an active phenotype at low expression levels and an inactive phenotype at higher expression levels. Indeed, only six different combinations of phenotypes were observed (Table 2). The observed phenotypic combinations were used to predict residue burial and active-site proximity. Also shown are the number of positions where a given phenotype occurs and the accuracy of the burial/activity predictions. For example, in class 1, 90% of the residues that showed the indicated phenotype were not active-site residues and were exposed (ACC > 5%). Without input from Ala scanning mutagenesis, active-site residues (which are typically exposed) would be assigned as buried based on inactivity of the corresponding Asp mutants at all arabinose concentrations (compare classes 3 and 6, Table 2). Partially buried residues also can be identified because they show activity at high, but not at low, expression levels (classes 2 and 4 in Table 2). One drawback of Ala scanning mutagenesis is that it may fail to identify exposed hydrophobic active-site residues because substitution of a hydrophobic by Ala will result only in the loss of a few van der Waals interactions. However, Asp substitutions at such exposed hydrophobic active sites are likely to be inactive because complex formation will result in energetically unfavorable burial of the Asp. This hypothesis was indeed the case: for example, at residues 25 and 95, which are part of the active site, the Ala mutant was active whereas the Asp mutant was inactive. A similar combination of mutant phenotypes also may occur at buried residue positions. However, at such positions, protein expression level and/or solubility will be substantially lowered for the Asp mutant as demonstrated in the next section. In contrast, for exposed, active-site residues, expression level and solubility will be unaffected. The data in Table 2 suggest that valuable information on residue burial and contribution to activity can be obtained from such scanning mutagenesis experiments. Mutational data for the remaining charged residue substitutions (Glu, Lys, Arg) are less informative than Ala and Asp scanning mutagenesis data, although they do provide useful confirmatory evidence in some cases. Examples include positions where the Asp mutation was either not available or showed a phenotype discordant with phenotypes of other charged substitutions at that position, e.g., residues 14, 51, 55, 80, and 96 (Tables 5 and 6). There has been recent interest in determining the fraction of single or multiple site mutants of a protein that show an inactive phenotype. Efforts have been made to measure this parameter through mutagenesis and also to correlate it with the protein fold (22, 23). The present work clearly demonstrates that this parameter is a function of both expression level as well as the nature of the mutation and hence cannot be simply correlated with the protein fold.
Table 2. Prediction of residue burial and active-site proximity from Ala and Asp scanning mutagenesis.
| Ala*
|
Asp*
|
||||||
|---|---|---|---|---|---|---|---|
| Class | 0% | 0.1% | 0% | 0.1% | Burial prediction | No. of positions | Accuracy, % |
| 1 | A | A | A | A | Exposed,† not active site | 40 | 90 |
| 2 | A | A | I | A | Partially exposed,‡ not active site | 5 | 80 |
| 3 | A | A | I | I | Buried, not active site or exposed, active site§ | 7 | 86 |
| 4 | I | A | I | A | Partially exposed‡ | 1 | 100 |
| 5 | I | A | I | I | Buried, not active site | 9 | 77 |
| 6 | I | I | I | I | Active site proximal | 4 | 100 |
0% and 0.1% refer to phenotypes of Ala or Asp mutants at 0% and 0.1% arabinose, respectively. A and I refer to active and inactive phenotype, respectively
Represents exposed residues with an ACC > 5%
Represents partially exposed residues with an ACC of 5–15%. Exposed residues can be subdivided into partially (ACC 5–15%) and completely (ACC > 15%) exposed categories
For buried positions, the corresponding Asp mutant will show substantial reductions in protein expression and/or solubility. If the residue is an exposed hydrophobic that is part of the active site, expression and solubility will not be affected
Correlation of Solubility of Asp Mutants with Residue Burial. Destabilizing substitutions often result in low or undetectable levels of the mutant protein in the aqueous fraction of cell lysates. Such mutants are either found in insoluble inclusion bodies or subjected to proteolysis in vivo (24, 25). It was therefore of interest to examine the solubilities of the various Asp mutants. After induction, cells were lysed, either by a freeze–thaw protocol or by using detergent-based lysis. Both methods gave similar results. Lysates were centrifuged, and the supernatant and pellet fractions were isolated. Relative amounts of CcdB in the two fractions were quantitated by SDS/PAGE. The data are summarized in Table 3. Consistent with the activity data in Table 1, Asp substitutions of residues with ACC of <5% were typically insoluble (see Fig. 2, which is published as supporting information on the PNAS web site). In contrast, for ACC of >5%, most Asp mutants were soluble. Most partially accessible (ACC of 5–15%) and virtually all fully exposed (ACC > 15%) mutants were soluble. Thus, solubility data can provide useful additional information to predict residue burial and to distinguish buried hydrophobic residues from surface hydrophobics that are part of the active site as discussed in the preceding section.
Table 3. Solubility and activity of aspartate mutants of CcdB in CSH501 as a function of ACC.
| ACC, % | Total no. of residues | No. soluble | No. insoluble | No. active (0.1% arabinose) |
|---|---|---|---|---|
| 0–5 | 16 | 1 | 15 | 0 |
| 5–15 | 15 | 12 | 3 | 13 |
| 15–40 | 38 | 38 | 0 | 34 |
Soluble and insoluble indicate whether protein is present in supernatant or pellet, respectively, after cell lysis.
Use of Scanning Mutagenesis Data to Validate Homology Models and Sequence Alignments. Genome sequencing has led to an explosion in the number of known protein sequences. Obtaining structural information for these proteins would help to elucidate protein function. Experimental determination of protein structures by either NMR or crystallography is a laborious and difficult process. Homology modeling is widely used to generate protein models at the atomic level. The primary inputs required are sequence alignments with protein(s) of known 3D structure (17). An alternative approach to generate protein models is through threading procedures wherein the sequence of interest is threaded through different template structures from the Protein Data Bank (PDB) and energetic criteria are used to evaluate the best model from the library of generated structures (26). The most difficult steps in both homology modeling and threading are to identify the correct template and obtain the correct alignment of query and template sequences. When the sequence identity drops to <30%, it is difficult to distinguish correct from incorrect sequence alignments. It was therefore of interest to examine whether the residue burial predictions from scanning mutagenesis experiments could be used for this purpose. Decoy templates consisting of nonhomologous, homodimeric structures of similar size as CcdB but belonging to different protein folds were selected from the Protein Quaternary Server Macromolecular structure database (http://pqs.ebi.ac.uk) and alignments were generated by using fugue online threading software (www-cryst.bioc.cam.ac.uk/fugue). Homology models of CcdB were generated from each sequence alignment by using the program modeller (Version 7v7; ref. 17). CcdB also was modeled by using structure-based sequence alignments generated from dali (www.ebi.ac.uk/dali) between CcdB and its structural homologs, namely, Kid toxin of plasmid R1 (PDB ID code 1m1f), YdcE protein from Bacillus subtilis (PDB ID code 1ne8), and MazF toxin from E. coli (PDB ID code 1ub4). The CcdB sequence also was threaded onto the above structural homologs as well as other homologs belonging to the same Structural Classification of Proteins family by using 3dpssm (www.sbg.bio.ic.ac.uk/~3dpssm), 123d (http://123D.ncifcrf.gov), and fugue, and sequence alignments were obtained. These sequence alignments were also input into modeller. The program generated models of CcdB using homology modeling based on the specified template structure and sequence alignment. Twenty-three different models of CcdB were generated by using modeller. A comparison was performed between ACC predicted from aspartate scanning mutagenesis of CcdB and ACC obtained in various homology models of CcdB. Residues of CcdB were divided into three different classes depending on the phenotype of the aspartate mutant at low and high arabinose: class 1 (predicted ACC of 0–5%) at positions where Asp mutants were inactive at both low and high arabinose; class 2 (predicted ACC of 5–15%) where mutants are inactive at low and active at high arabinose; and class 3 (predicted ACC of >5%) where mutants are active at both arabinose concentrations. If a given residue position was predicted to be in the same ACC class using either the modeled structure of CcdB or mutagenesis data, the position was assigned a value of 1; otherwise, it was assigned a value of 0. The overall score was obtained by summing up the values over all positions of CcdB for each model. Finally, Z score, a normalized score (27), was calculated for the comparison. Significant correlations (Z ≈ 2) were only observed with CcdB models generated from structural homologs and using structure-based sequence alignments (see Table 8, which is published as supporting information on the PNAS web site). Poor correlations were seen with models generated by threading as well as decoy alignments (see Table 8). We also compared ACC in models generated by homology modeling with the corresponding ACC in the CcdB crystal structure (see Table 9, which is published as supporting information on the PNAS web site). Here, too, ACC correlations between the homology model and crystallographic structure of CcdB are best when structural homologs of CcdB are used as templates in combination with structure-based sequence alignments. However, if the same structural homologs were used as templates for threading, the resultant CcdB model showed poor ACC correlation with the crystal structure primarily because of poor alignments generated by threading. Thus, the current accuracies of threading procedures are insufficient to generate models with ACC values close to the true ones, even when the appropriate template is used. Similar results were obtained even if mutational data or ACC correlations solely at hydrophobic residue positions were used (Tables 8 and 9). Exposed hydrophobic sites are often mistakenly assigned as buried ones in modeling and protein-structure prediction studies. These results suggest that scanning mutagenesis data can be used to rank order sequence alignments and their corresponding homology models, as well as to distinguish between correct and incorrect structural alignments. With continuous reductions in oligonucleotide costs and increasingly efficient site-directed mutagenesis procedures, comprehensive scanning mutagenesis experiments for small proteins/domains are quite feasible.
Characterization of Asp Mutants of MBP and Trx. CcdB is a moderately stable protein [Tm = 61°C, ΔGou (298 K) = 21 kcal/mol (1 cal ≈ 4.184 J) of dimer] (28). In this system, Asp substitution at all buried (ACC < 5%) positions leads to loss of activity at all expression levels of the protein. In all of the cases, such mutants are found in inclusion bodies when overexpressed. To assess the effects of introduction of Asp at buried positions in other small and large proteins, a total of four buried sites in two proteins were selected for mutagenesis. Asp was introduced at residues 25 and 78 (ACC of 0% in each case) in E. coli Trx and at residues 224 and 264 (ACC of 0 and 0.5%, respectively) in E. coli MBP. Trx is a 108-aa, monomeric single-domain disulfide oxidoreductase protein. It is extremely stable with a ΔGou of 9.1 kcal/mol at 298 K and Tm of 84°C (29). MBP is a 370-aa, monomeric, two-domain periplasmic protein involved in maltose uptake and chemotaxis. It has ΔGou of 8.9 kcal/mol at 298 K and a Tm of 63.4°C (30). All proteins were expressed by using the PBAD promoter in strains deleted for chromosomal MBP or Trx (Pop6590 and A307, respectively). In contrast to the WT proteins, none of these mutants showed expression in the appropriate deleted strains, even when induced with arabinose (see Figs. 3 and 4, which are published as supporting information on the PNAS web site). Subsequently these mutants were expressed in E. coli strain DH5α. Three of the four mutants showed detectable expression in this strain. After growth and induction in LB, solubilities of mutants in whole-cell lysates were assessed by SDS/PAGE as described above. MBP224D was partly soluble, whereas MBP 264D was found exclusively in the pellet fraction. No expression for Trx 25D could be detected in whole-cell lysates, suggesting that it is proteolyzed. Trx 78D was soluble, like WT, and was purified to homogeneity. The protein showed similar CD and fluorescence spectra to WT (see Fig. 5 A–C, which is published as supporting information on the PNAS web site) but did not show any fluorescence increase after reduction with DTT, characteristic of WT (Fig. 5C). The protein also was found to be incapable of catalyzing the reduction of insulin, suggesting that it is misfolded in some way (Fig. 6, which is published as supporting information on the PNAS web site). In all four cases, expression of mutants was not detectable in the strains deleted for chromosomal copies of Trx or MBP. In the normal E. coli strain, DH5α, the amount of expressed protein in the soluble fraction decreased appreciably for three of the four mutants. The only mutant that was soluble to an appreciable extent, Trx 78D, did not show any activity. Hence, by using a combination of activity and expression data in all four cases, it would have been possible to predict that all four sites were at buried positions.
Suggested Method to Determine Residue Burial. The data in the present study as well as earlier studies (described in the next section) clearly indicate that charged residue substitutions at buried positions typically lead to protein misfolding and targeting to inclusion bodies or greatly reduced levels of expression. Both of these effects typically lead to an inactive phenotype. Asp is the most destabilizing of all of the charged residue substitutions examined. To probe burial of a specific residue, it should be mutated to Ala and Asp, and the activity of the mutants should be characterized in vivo, preferably at both low and high transcriptional levels. The residue burial as well as contribution to activity can be inferred by using the results established for CcdB, which are summarized in Tables 2 and 3.
Analysis of Previous Mutational Studies. To confirm the generality of our results, we have analyzed data from several previous mutagenesis studies on a number of different proteins. Systematic large-scale mutagenesis has been reported on two proteins previously, namely, T4 lysozyme (31) and lac repressor (32) using suppressor strains of Salmonella typhimurium and E. coli, respectively. Single-site mutants of T4 lysozyme were generated by introducing amber mutations into every codon of the 164-aa bacteriophage T4 lysozyme gene (31). The amber alleles were introduced into a bacteriophage P22 hybrid, and the 2,015 resulting mutant phenotypes were scored depending on their ability to form plaques in 13 different suppressor strains (31). Each suppressor results in substitution of a different amino acid at an amber codon. We have analyzed the results of the T4 lysozyme studies in a similar fashion as that for CcdB (Table 4). In similar studies on lac repressor (32), amber codons were introduced from positions 2 to 329 of lac repressor. The resulting mutants were transformed into 13 different suppressor strains to yield ≈4,000 mutant phenotypes. The phenotype was scored on the basis of the ability to repress β-galactosidase activity and to respond to inducer. The 4,000 individual phenotypes were not explicitly listed in the publication. Instead, the overall results were summarized in table 1 of ref. 32. Residues were classified into 16 groups depending on their location in the protein structure and the level of tolerance to different substitutions. Only 6 of a total of 123 buried sites (group III, table 1 of ref. 32) were found to be tolerant of nonconservative substitutions. Of these six sites, only three were hydrophobic and able to tolerate Glu, Lys, or Arg substitution. No aspartate suppressor strain was available for the T4 lysozyme and lac repressor studies, and hence the phenotypes of Asp mutants could not be studied. The suppression efficiencies of different suppressor strains used in these studies depend on the DNA sequence surrounding the mutated codon. Also, the studies were performed only at a single expression level, and protein expression was not directly monitored. Despite these caveats, the overall results of these studies are quite consistent with those observed with CcdB.
Table 4. Correlation of residue burial with activity for mutants of T4 lysozyme.
| Active mutants,* %
|
|||||
|---|---|---|---|---|---|
| ACC, % | Total no. of sites | Ala | Glu | Lys | Arg |
| 0–5 | 32 | 89 | 23 | 8 | 14 |
| 5–15 | 28 | 96 | 72 | 68 | 84 |
| 15–40 | 32 | 96 | 86 | 61 | 68 |
| >40 | 59 | 100 | 98 | 92 | 100 |
Data taken from ref. 31
There also have been extensive, but more limited, mutagenesis studies on other proteins using random mutagenesis. In the case of Gene V protein, an 87-residue dimeric protein, 371 mutants at 86 positions were characterized for activity (33). For HIV-1 protease, a 99-residue dimer, 330 mutants at 99 sites were characterized for activity (34). In no case was a charged substitution at a buried site found to be active. A similar result was seen in the case of lambda repressor, wherein seven core residues were randomized (35). Charged residue substitutions at these sites were always found to inactivate the protein. Six additional examples of effects of Asp substitutions of buried, noncharged residues were obtained from the Protherm database (http://gibk26.bse.kyutech.ac.jp/jouhou/protherm/protherm_search.html): A52D and I53D in RNaseH (36, 37), L133D in case of T4 lysozyme (38), S91D in hen egg white lysozyme (39), Y282D in case of Lac repressor (40), and Y78D in U1A protein (41). In all of the cases, there was a drastic decrease in protein thermodynamic stability relative to WT. The average amount of destabilization, ΔΔGou, was found to be ≈4.5 kcal/mol, and the average decrease in Tm was 10°C. Spectroscopic studies for all mutants (except L133D of T4 lysozyme where no data were reported) showed the mutants to have similar conformations to WT. A52D RNaseH showed lowered protein expression, and I53D RNaseH was targeted to inclusion bodies. L133D (T4 lysozyme), Y78D (U1A), and Y282D (lac repressor) were all inactive. Introduction of charged amino acids into the protein interior results in appreciable destabilization of the protein. Consequently, when expressed in E. coli, such mutants either form inclusion bodies or are expressed in soluble form at much lower levels than the WT. We anticipate that exceptions may occur with unusually stable proteins such as those from thermophiles or hyperthermophiles. For such proteins with high values of ΔGou, the destabilization caused by introduction of buried charges may be insufficient to trigger inclusion body formation or proteolysis in vivo.
Another potential concern with using Asp mutants as probes of residue burial is that protein conformational change upon mutation may allow the charge to be solvent exposed and hence allow the mutation to be tolerated. However, the observation that the vast majority of charged residue substitutions at buried positions result in loss of activity indicates that such conformational changes occur very rarely, if at all. There are only a few structural studies on mutants with buried, charged amino acids. In the case of T4 lysozyme, the crystal structure of the M102K mutant shows that the Lys side chain is accommodated without major structural rearrangement (38). A similar result was observed for the V66K mutant of staphylococcal nuclease (42). Two-dimensional NMR studies of the V68D and V68E mutants of human myoglobin also showed that the mutant residues were accommodated without significant conformational changes (43). In summary, the data from the present study as well as earlier studies consistently suggest that <5% ACC is a reliable cutoff for buried residues and that phenotypes and solubilites of Asp mutants are reliable indicators of residue burial for most proteins.
Supplementary Material
Acknowledgments
We thank Dr. M. S. Madhusudhan and Rajan Prabu for helpful suggestions regarding homology modeling and Ravindra Babu Ch. and Ankita Roy for carrying out some of the initial mutagenesis experiments. K.B. and P.C. are Council of Scientific and Industrial Research Fellows. R.V. is an International Senior Research Fellow of The Wellcome Trust. This work was supported by grants from The Wellcome Trust and Department of Biotechnology, Government of India (to R.V.).
Author contributions: R.V. designed research; K.B. and P.C. performed research and analyzed data; and K.B. and R.V. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ACC, percent side-chain accessibility of a residue in protein; CcdB, Controller of Cell Division or Death B protein; MBP, maltose-binding protein; Trx, thioredoxin.
References
- 1.Ponder, J. W. & Richards, F. M. (1987) J. Mol. Biol. 193, 775-791. [DOI] [PubMed] [Google Scholar]
- 2.Lee, B. & Richards, F. M. (1971) J. Mol. Biol. 55, 379-400. [DOI] [PubMed] [Google Scholar]
- 3.Shrake, A. & Rupley, J. A. (1973) J. Mol. Biol. 79, 351-371. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad, S. & Gromiha, M. M. (2002) Bioinformatics 18, 819-824. [DOI] [PubMed] [Google Scholar]
- 5.Miller, S., Janin, J., Lesk, A. M. & Chothia, C. (1987) J. Mol. Biol. 196, 641-656. [DOI] [PubMed] [Google Scholar]
- 6.Mansoor, S. E., McHaourab, H. S. & Farrens, D. L. (1999) Biochemistry 38, 16383-16393. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarty, S. & Varadarajan, R. (1999) Struct. Folding Des. 7, 723-732. [DOI] [PubMed] [Google Scholar]
- 8.Guan, J. Q., Almo, S. C. & Chance, M. R. (2004) Acc. Chem. Res. 37, 221-229. [DOI] [PubMed] [Google Scholar]
- 9.Bernard, P., Kezdy, K. E., Van Melderen, L., Steyaert, J., Wyns, L., Pato, M. L., Higgins, P. N. & Couturier, M. (1993) J. Mol. Biol. 234, 534-541. [DOI] [PubMed] [Google Scholar]
- 10.Dao-Thi, M. H., Van Melderen, L., De Genst, E., Afif, H., Buts, L., Wyns, L. & Loris, R. (2005) J. Mol. Biol. 348, 1091-1102. [DOI] [PubMed] [Google Scholar]
- 11.Loris, R., Dao-Thi, M. H., Bahassi, E. M., Van Melderen, L., Poortmans, F., Liddington, R., Couturier, M. & Wyns, L. (1999) J. Mol. Biol. 285, 1667-1677. [DOI] [PubMed] [Google Scholar]
- 12.Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995) J. Bacteriol. 177, 4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakshusmathi, G., Mondal, K., Lakshmi, G. S., Singh, G., Roy, A., Ch., R. B., Madhusudhanan, S. & Varadarajan, R. (2004) Proc. Natl. Acad. Sci. USA 101, 7925-7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, B. H. & Hecht, M. H. (1994) Biotechnology 12, 1357-1360. [DOI] [PubMed] [Google Scholar]
- 15.Ghoshal, A. K., Swaminathan, C. P., Thomas, C. J., Surolia, A. & Varadarajan, R. (1999) Biochem. J. 339, 721-727. [PMC free article] [PubMed] [Google Scholar]
- 16.Holmgren, A. (1979) J. Biol. Chem. 254, 9627-9632. [PubMed] [Google Scholar]
- 17.Sanchez, R. & Sali, A. (1997) Proteins, Suppl. 1, 50-58. [DOI] [PubMed]
- 18.Russell, R. B. & Barton, G. J. (1994) J. Mol. Biol. 244, 332-350. [DOI] [PubMed] [Google Scholar]
- 19.Rose, G. D., Geselowitz, A. R., Lesser, G. J., Lee, R. H. & Zehfus, M. H. (1985) Science 229, 834-838. [DOI] [PubMed] [Google Scholar]
- 20.Dickinson, C. D., Kelly, C. R. & Ruf, W. (1996) Proc. Natl. Acad. Sci. USA 93, 14379-14384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 22.Guo, H. H., Choe, J. & Loeb, L. A. (2004) Proc. Natl. Acad. Sci. USA 101, 9205-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloom, J. D., Silberg, J. J., Wilke, C. O., Drummond, D. A., Adami, C. & Arnold, F. H. (2005) Proc. Natl. Acad. Sci. USA 102, 606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pakula, A. A., Young, V. B. & Sauer, R. T. (1986) Proc. Natl. Acad. Sci. USA 83, 8829-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vershon, A. K., Bowie, J. U., Karplus, T. M. & Sauer, R. T. (1986) Proteins 1, 302-311. [DOI] [PubMed] [Google Scholar]
- 26.Hendlich, M., Lackner, P., Weitckus, S., Floeckner, H., Froschauer, R., Gottsbacher, K., Casari, G. & Sippl, M. J. (1990) J. Mol. Biol. 216, 167-180. [DOI] [PubMed] [Google Scholar]
- 27.Murray, R. & Spiegel, L. J. S. (2000) Statistics (Tata McGraw–Hill, New York).
- 28.Bajaj, K., Chakshusmathi, G., Bachhawat-Sikder, K., Surolia, A. & Varadarajan, R. (2004) Biochem. J. 380, 409-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakrabarti, A., Srivastava, S., Swaminathan, C. P., Surolia, A. & Varadarajan, R. (1999) Protein Sci. 8, 2455-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beena, K., Udgaonkar, J. B. & Varadarajan, R. (2004) Biochemistry 43, 3608-3619. [DOI] [PubMed] [Google Scholar]
- 31.Rennell, D., Bouvier, S. E., Hardy, L. W. & Poteete, A. R. (1991) J. Mol. Biol. 222, 67-88. [DOI] [PubMed] [Google Scholar]
- 32.Suckow, J., Markiewicz, P., Kleina, L. G., Miller, J., Kisters-Woike, B. & Muller-Hill, B. (1996) J. Mol. Biol. 261, 509-523. [DOI] [PubMed] [Google Scholar]
- 33.Terwilliger, T. C., Zabin, H. B., Horvath, M. P., Sandberg, W. S. & Schlunk, P. M. (1994) J. Mol. Biol. 236, 556-571. [DOI] [PubMed] [Google Scholar]
- 34.Loeb, D. D., Swanstrom, R., Everitt, L., Manchester, M., Stamper, S. E. & Hutchison, C. A., III (1989) Nature 340, 397-400. [DOI] [PubMed] [Google Scholar]
- 35.Lim, W. A. & Sauer, R. T. (1989) Nature 339, 31-36. [DOI] [PubMed] [Google Scholar]
- 36.Akasako, A., Haruki, M., Oobatake, M. & Kanaya, S. (1997) J. Biol. Chem. 272, 18686-18693. [DOI] [PubMed] [Google Scholar]
- 37.Spudich, G. M., Miller, E. J. & Marqusee, S. (2004) J. Mol. Biol. 335, 609-618. [DOI] [PubMed] [Google Scholar]
- 38.Dao-pin, S., Anderson, D. E., Baase, W. A., Dahlquist, F. W. & Matthews, B. W. (1991) Biochemistry 30, 11521-11529. [DOI] [PubMed] [Google Scholar]
- 39.Shih, P., Holland, D. R. & Kirsch, J. F. (1995) Protein Sci. 4, 2050-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, J. & Matthews, K. S. (1994) Biochemistry 33, 8728-8735. [DOI] [PubMed] [Google Scholar]
- 41.Kranz, J. K., Lu, J. & Hall, K. B. (1996) Protein Sci. 5, 1567-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sondek, J. & Shortle, D. (1990) Proteins 7, 299-305. [DOI] [PubMed] [Google Scholar]
- 43.Varadarajan, R., Lambright, D. G. & Boxer, S. G. (1989) Biochemistry 28, 3771-3781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.