Abstract
Selenoproteins are a diverse group of proteins that contain selenocysteine (Sec), the 21st amino acid. In the genetic code, UGA serves as a termination signal and a Sec codon. This dual role has precluded the automatic annotation of selenoproteins. Recent advances in the computational identification of selenoprotein genes have provided a first glimpse of the size, functions, and phylogenetic diversity of eukaryotic selenoproteomes. Here, we describe the identification of a selenoprotein family named SelJ. In contrast to known selenoproteins, SelJ appears to be restricted to actinopterygian fishes and sea urchin, with Cys homologues only found in cnidarians. SelJ shows significant similarity to the jellyfish J1-crystallins and with them constitutes a distinct subfamily within the large family of ADP-ribosylation enzymes. Consistent with its potential role as a structural crystallin, SelJ has preferential and homogeneous expression in the eye lens in early stages of zebrafish development. A structural role for SelJ would be in contrast to the majority of known selenoenzymes. The unusually highly restricted phylogenetic distribution of SelJ, its specialization, and the comparative analysis of eukaryotic selenoproteomes reveal the diversity and functional plasticity of selenoproteins and point to a mosaic evolution of the use of Sec in proteins.
Keywords: ADP-ribosylation, J1-crystallins, selenocysteine, selenium
Selenocysteine (Sec) residues are found only in a small group of proteins called selenoproteins. Selenoproteins with characterized functions are enzymes involved in redox reactions. Sec is inserted by dynamic recoding of an in-frame UGA stop codon located upstream of the actual termination codon (1). Selenoproteins are present in eukaryotes and prokaryotes and, although their sets of selenoproteins (selenoproteomes) do not completely overlap (2), their intersection appears to be larger than previously thought (3). In eukaryotes, the Sec insertion sequence (SECIS), an RNA stem-loop located in the 3′ UTR of selenoprotein genes, recruits several transacting factors to recode UGA from termination to Sec insertion (4). The dual function of this codon confounds gene-finding programs and human curators, making selenoprotein identification a challenge (5–10).
In addition, Cys and Sec are structurally related amino acids, with Sec incorporating selenium in the position of sulfur in Cys. The functional resemblance of Sec and Cys is supported by the observation that their residues occupy equivalent positions in homologous proteins. As expected, mutation of the Sec residue to Cys results in variants that are active but have a lower catalytic efficiency (11–13), although they may have an enhanced translation (13). Furthermore, natural Cys-containing counterparts are usually inferior catalysts (14, 15), but similar reactivity has also been reported (16). Hence, Sec, per se, does not possess an essential role in protein function and these two residues seem to be partially interchangeable. Accordingly, Sec and Cys residues are alternatively found in orthologous proteins in different species raising the issue of the evolutionary direction (if any) of Sec/Cys interconversion. This distribution of Sec and Cys residues across genomes hinders the identification of true Sec-containing proteins.
In consequence, the description of eukaryotic selenoproteomes is incomplete. The number, functional diversity, and phylogenetic distribution of eukaryotic selenoproteins are poorly known and, thus, the importance of Sec and selenium in protein function and evolution remains unclear, despite an increasing body of evidence linking selenium deficiency to a number of pathologies (reviewed in ref. 17). Recently, computational approaches have been developed that have greatly contributed to the characterization of selenoproteins in many eukaryotic genomes. Specific methods have been developed to predict SECIS elements in nucleotide sequences (5, 6). SECIS predictions, in turn, have been used to instruct modified gene prediction algorithms to ignore UGA codons as terminators in putative exon sequences when SECIS elements are predicted at the appropriate distance (7–9). In addition, comparative sequence analysis methods have proven to be very powerful in uncovering novel selenoproteins. Indeed, protein sequence conservation across a TGA codon between sequences from species at large phylogenetic distances is strongly indicative of the Sec coding function (10).
Here, we report the discovery by computational and experimental means of a selenoprotein family designated SelJ in the genome of Tetraodon nigroviridis. SelJ has a very restricted phylogenetic distribution and, in contrast to all known eukaryotic selenoproteins, does not exist in mammalian genomes, not even as a Cys homologue. In addition, although the majority of selenoproteins are assumed to have enzymatic functions, computational and experimental data suggest that SelJ could have a structural role. Comparison of eukaryotic selenoproteomes highlights the scattered phylogenetic distribution of selenoproteins, arguing for a mosaic evolution of specialized proteins in which the differential use of Sec and Cys shapes the size and function of eukaryotic selenoproteomes.
Methods
Gene and SECIS Prediction. The T. nigroviridis sequence data (18) was screened for previously uncharacterized selenoprotein genes using three independent computational methods (see Supporting Methods, which is published as supporting information on the PNAS web site). The SelJ (Fig. 1A) selenoprotein mRNA was found through a coordinated prediction of ORFs interrupted by in-frame UGA codons in cDNA sequences with the gene prediction program geneid (19) and SECIS elements (Fig. 1B) with secisearch (8, 9).
Fig. 1.
SelJ gene and SECIS structure. (A) SelJ gene structure in T. nigroviridis. Coding exon regions are shown in blue. The annotated gene in Ensembl (GSTENG00015130001) has the frame changed and uses a different termination codon. Therefore, SelJ is reannotated with the Sec codon (TGA in the genomic sequence) and the SECIS element in the noncoding region of the last exon. (B) SelJ SECIS element.
75Se Labeling. The partial sequence of the zebrafish SelJ cDNA that included a region coding for a 18-kDa C-terminal portion of the protein and the 3′ UTR with a predicted SECIS element was amplified with 5′-AGTCGCTCGAGGTTGAAGAAGCAGTCCGTGTCAC-3′ and 5′-GCTTGGGATCCATTTTCCGCATGTCATGCTG-3′ primers and cloned into pEGFP-C3 (Clontech) vector by using XhoI and BamHI restriction sites to generate the GFP–SelJ construct, which codes for a fusion protein containing GFP followed by a C-terminal region of SelJ. The plasmid was purified with an EndoFree Maxi kit (Qiagen, Valencia, CA). NIH 3T3 cells were transfected with the GFP–SelJ construct using Lipofectamine and Plus reagents (Invitrogen). Transfected cells were supplemented with 5–10 mCi (1 Ci = 37 GBq) of [75Se]selenite (University of Missouri Research Reactor, Columbia) per 60-mm cell culture dish. After 24 h, cells were collected, and protein samples were prepared, subjected to SDS/PAGE, and transferred onto a polyvinylidene difluoride membrane. Radioactive bands were detected by using a phosphorimager (Amersham Biosciences, which is now GE Healthcare). See Fig. 2.
Fig. 2.
75Se labeling. The left lane shows cells transfected with a GFP–SelJ construct, and the right lane shows cells transfected with a control vector. Migration of endogenous selenoproteins TR1 and GPx1 is indicated by arrows on the right. Migration of the 45-kDa GFP–SelJ fusion selenoprotein is shown by an arrow on the left.
Subcellular Localization. The Sec-encoding TGA codon was mutated to TGC, a Cys codon, by using 5′-CTGTGTTTCCAAATACCTGCGGTTTGCCTGGTGC-3′ and 5′-GGAATGCACCAGGCAAACCGCAGGTATTTGGAAA-3′ primers and a QuikChange Mutagenesis kit (Stratagene). The coding region of the resulting Cys mutant of SelJ was amplified with 5′-TCGATCTCGAGGATTTCAAGATGGCTCTTGC-3′ and 5′-CTAAGGAATCCCTTGTTTTTCCAGGAAGGTGGAATC-3′ and cloned upstream of GFP in pEGFP-N2 vector by using EcoR I and XhoI restriction sites. NIH 3T3 cells were transfected as described above, and fluorescence was detected with an Olympus FV500 confocal microscope at the Microscopy Core Facility at the University of Nebraska, Lincoln (see Fig. 3).
Fig. 3.
Subcellular localization of SelJ. (Upper) The SelJ–GFP fusion protein, in which Sec was mutated to Cys, was expressed in NIH 3T3 cells. (Lower) A control in which the cells were transfected with a construct expressing GFP alone.
Homology Searches. tblastn (20) was used to query the following for SelJ homologues: the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov) collection of 62 eukaryotic (partial and complete), 25 archaeal, and 354 bacterial genomes, the NCBI eukaryotic ESTs, and sets of eukaryotic transcripts (81 species) for SelJ homologues from The Institute for Genomic Research (www.tigr.org). Additional homologues were identified by using iterated psi-blast (20) searches in the NCBI and UniProt (Universal Protein Resource, www.pir.uniprot.org) databases.
Analysis of Promoter Regions. The available jellyfish J1A (289 nt), J1B (265 nt), and J1C (178 nt) promoter sequences were obtained from NCBI entries L05524, L05523, and L05522, respectively. SelJ promoter regions (1,000 nt upstream of the most 5′ transcript) were extracted for T. nigroviridis (chromosome 15), Tetraodon rubripes (scaffold_182) and Danio rerio (Zv4_scaffold126.1) fishes. bl2seq was used to compare these promoter regions at the sequence level. The PROMO server with transfac 8.3 (21, 22) was used to predict promoter elements (see Supporting Methods).
In Situ Hybridization in Zebrafish. A tblastn search in EST databases identified eight zebrafish sequences encoding homologues to the T. nigroviridis SelJ protein. These ESTs generated a 1,292-bp contiguous sequence containing the entire ORF and the 3′ UTR with the SECIS motif. A 1,300-bp DNA fragment encompassing positions 12–1313 of the zebrafish SelJ cDNA was obtained by SmaI–EcoRI digestion and cloned into pBluescriptSK(–). The antisense probe was synthesized with T7 RNA polymerase, and whole-mount in situ hybridizations were performed (23). The fully detailed protocol is available from the authors upon request (see Fig. 4 and Supporting Methods; see also Fig. 6, which is published as supporting information on the PNAS web site).
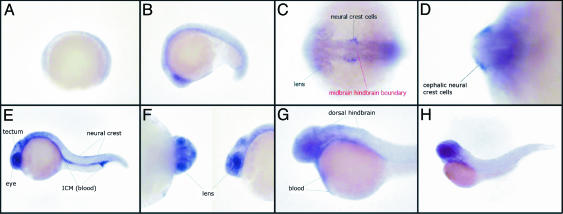
Fig. 4.
Expression pattern of the SelJ gene during development in zebrafish embryos. Shown are a gastrula (A), an embryo at middle somitogenesis (B), dorsoventral (C), or transversal (D) views of the 20-somite stage, a whole embryo 24 h after fertilization (E) and a closer view of the eyes (F), a whole embryo 36 h after fertilization (G), and an embryo 48 h after fertilization (H). All views are lateral, except C and D.(C) Territories in which SelJ is expressed are indicated in black, and the position of the midbrain–hindbrain boundary is shown in red.
EST Expression Pattern. Tissue distribution of SelJ in different fishes was obtained from ESTs (indicated here with each species and the respective database accession nos.) in Ensembl (T. nigroviridis, GSTENG00015130001), UniGene (D. rerio, Dr.21881), and the Institute for Genomic Research databases [D. rerio, TC292667; Salmo salar, TC23857 (SelJa), TC23525 (SelJb), and TC33491 (SelJb); Oryzias latipes, TC38404 (SelJa) and TC38404 (SelJb); Fundulus heteroclitus, TC1043 and CN987014; Oncorhynchus mykiss, TC56113 and TC64467].
Structural and Functional Analysis. Protein domain architectures were obtained from the Pfam (Protein Families Database of Alignments and HMMs) and SCOP (Structural Classification of Proteins) databases. The protein structure of the ADP-ribosylglycohydrolase (ARH) from Methanococcus jannaschii (mjARH) was obtained from the Protein Data Bank (PBD ID code 1T5J), and its secondary structure was obtained from the DSSP (Definition of Secondary Structure of Proteins) database (24). Highly conserved regions among ARHs were mapped onto the 3D structure of mjARH to identify their locations with respect to putative binding sites (see Table 1 and Fig. 7, which are published as supporting information on the PNAS web site). The NCBI accession numbers and species of depicted protein sequences in Fig. 7 are listed in Table 2, which is published as supporting information on the PNAS web site. See Supporting Methods for details.
Distribution of Eukaryotic Selenoproteins. Human selenoproteins were compared (tblastn) to selected eukaryotic genomes and transcriptomes (E < 10–3) with different blosum matrices (see Fig. 5).
Fig. 5.
Distribution of eukaryotic selenoproteins and Cys homologues in eukaryotes. Sec proteins are shown in red, Cys proteins are shown in green, and unknown proteins are shown in yellow. Open boxes represent absent proteins in the genome. S. purpuratus, Xenopus laevis, Sus scrofa, and Bos taurus are incomplete genomes. The protein homologous to known disulfide isomerases with Sec instead of Cys in the haptophyte alga Emiliana huxleyi (39) is not depicted. See Table 3, which is published as supporting information on the PNAS web site, for full scientific and common names.
Results
T. nigroviridis Selenoproteome. The T. nigroviridis selenoproteome consists of 19 known selenoprotein families (18): 15kDa, DI, GPx, SelH, SelI, SelK, SelM, SelN, SelO, SelP, SelR, SelS, SelT, SelU, SelV, SelW, SPS2, and TR. Thus, all known vertebrate selenoproteins exist in true Sec form in this fish. In addition, this genome encodes SelJ on chromosome 15. SelJ has 9 exons, with a single Sec residue lying in exon 7 (Fig. 1A). The SelJ gene encodes a protein of 341 residues and has a type I SECIS (Fig. 1B). Type I SECIS, in contrast to type II, have no additional small-stem loop at the end of the apical loop (25).
SelJ Experimental Validation. To verify that SelJ is a true selenoprotein, 75Se-labeling of NIH 3T3 cells was undertaken. A 75Se-labeled band (Fig. 2) of the expected size for the protein product containing the in-frame UGA encoding Sec was observed. This band was absent in the control transfection experiments, whereas the endogenous mammalian selenoproteins were expressed in both samples. Thus, SelJ is a selenoprotein and has a functional SECIS element, which is used to insert Sec at an in-frame UGA codon. To determine the cellular location of SelJ, a SelJ–GFP fusion protein was expressed in NIH 3T3 cells in which Sec was mutated to Cys. The fusion protein, which was detected by GFP fluorescence, was distributed uniformly in the cell (Fig. 3).
SelJ Relationship to J1-Crystallins. A psi-blast search with the zebrafish SelJ protein returned J1-crystallins (J1A, J1B, and J1C of the jellyfish Tripedalia cystophora) as the closest homologues (the best E value was 3 × 10–57 for J1C-crystallin, and sequence identity was 36% for J1A-, J1B-, and J1C-crystallins). These proteins have Cys in place of Sec (Fig. 8, which is published as supporting information on the PNAS web site; see also Fig. 7) and are encoded by three intronless genes (26). Crystallins maintain the transparency and proper light diffraction in the eye lens across the animal taxa, but unrelated crystallins are found in vertebrates, cephalopods, and jellyfish. At the base of this polyphyletic origin lies the mechanism known to produce crystallin-acting proteins, that is, the independent recruitment of metabolic enzymes and stress-related proteins in the eye lens. There are two different pathways for crystallin recruitment, including gene duplication and subsequent sequence divergence with or without loss of the original function (27) and gene sharing, which consists of the recruitment of a gene product to serve additional functions with neither duplication nor loss of the primary function (28). Gene sharing may, however, require a change of tissue specificity, usually a dramatic increase of expression in the eye lens or a change in developmental timing. J1-crystallins are of unknown origin.
To further investigate the potential role of SelJ as a crystallin, we have analyzed the promoter region of the fish SelJ and jellyfish J1-crystallins. As reported in ref. 26, the J1A-, J1B-, and J1C-crystallins have completely different promoter regions. Furthermore, the fish promoters do not show sequence similarity to the jellyfish promoter regions. However, T. nigroviridis and T. rubripes, two puffer fishes, have stretches of significant sequence similarity. We searched for binding sites in common in all promoter regions and, specifically, shared by T. nigroviridis and T. rubripes. Among hundreds of potential binding sites, we searched for motifs that could be indicative of eye function, for example, activator protein 1, musculoaponeurotic fibrosarcoma, and those in the Pax family. Pax-6 has been recognized as a master control gene for eye morphogenesis, and other members of this family also are related to eye differentiation (28). Pax-2 (T01823), Pax-2a (T00678), Pax-4a (T02983), Pax-6 (T00682), Pax-9a (T03593), Pax-9b (T03594) transfac hits were found in all sequences. However, all hits had a high random expectation (>0.25). Therefore, the evidence for these elements is inconclusive. Furthermore, the hits in the T. nigroviridis and T. rubripes sequences did not correlate with their conserved regions, suggesting a high false-positive rate.
In addition, to assess whether SelJ in fishes has significant eye lens expression, we analyzed the tissue and temporal expression of SelJ in situ during zebrafish embryogenesis. An RNA probe complementary to the zebrafish SelJ cDNA was synthesized, and in situ hybridization was performed on whole zebrafish embryos from different developmental stages. The hybridization sites were revealed by a chromogenic reaction. The SelJ gene was not expressed during the primary developmental stages, up to middle somitogenesis (Fig. 4 A and B). The first transcripts were detected at the stage of 20 somites and were observed within the forming lens and the anterior neural crest cells flanking the midbrain–hindbrain boundary (Fig. 4 C and D). Twenty-four hours after fertilization, SelJ was predominantly expressed within different territories: the eye; the tectum, a highly proliferating tissue of the brain; the inner cell mass, which is the tissue of hematopoiesis; and the dorsal neural crest (Fig. 4). Neural crest cells are highly migrating cells with a large spectrum of differentiation potential, such as pigment cells, peripheral nervous system, or parts of facial cartilage. Within the eye, SelJ was expressed in the lens where the labeling was homogenous (Fig. 4F). Later in development, the expression turned less restricted and spread to the whole embryo with higher levels detected in the blood and dorsal hindbrain (Fig. 4G). At 48 h of development, the gene was widely expressed in all embryonic tissues (Fig. 4H). Additionally, tissue and temporal expression was explored through the identification of ESTs from various tissue origins in several actinopterygian fishes. SelJ showed no preferential expression pattern in later embryonic or adult stages and was observed in liver, kidney, heart, muscle, ovary, gut, spleen, eyes, testis, and brain in adult fishes. However, the EST data (76 sequences) might be too scarce to infer a reliable expression pattern. In conclusion, SelJ is widely expressed in advanced embryo and adult stages, but a more restricted pattern of expression in the eye lens is detected in early embryogenesis.
SelJ Relationship to ADP-Ribosylation Enzymes. The first psi-blast iteration also retrieved significant hits to bacterial proteins (best E value, 5 × 10–19), all of which belong to the family of ADP-ribosylation enzymes (termed ADP_ribosyl_GH in Pfam). This search converged at the fifth iteration after retrieving >170 potential ADP-ribosylation enzymes from all domains of life, that is, bacteria, archaea, eukarya, and viruses, including bacteriophages and the mimivirus. We conclude that SelJ and J1-crystallins form a distinct subfamily within this large family of ADP-ribosylation enzymes. ADP-ribosylation is a reversible posttranslational modification of proteins involving the addition of an ADP-ribose moiety from nicotinamide adenine dinucleotide to acceptor amino acid(s). In particular, ARHs cleave ADP-ribose-l-Arg (29, 30) (EC 3.2.2.19). Other well studied family members are dinitrogenase reductase-activating glycohydrolases (EC 3.2.2.24) of the photosynthetic Rhodospirillum rubrum and related bacteria, which regulate nitrogen fixation by removing the ADP-ribosyl group of inactivated dinitrogenase reductases to recover their activity (31, 32).
To assess whether SelJ or the J1-crystallins may have an ADP-ribosylation function similar to the previously described ARHs, we first identified functionally relevant amino acids of these enzymes and then compared them to the corresponding residues of SelJ and J1-crystallin proteins (Table 1 and Fig. 7). The analysis of mjARH provided by Gogos et al. in the Protein Data Bank file identifies a catalytic pocket with two alternative adjacent locations of modeled Mg2+ ions, but the presence of different metals cannot be ruled out. Indeed, the activity of mammalian ARHs depends on DTT and Mg2+ (29, 30). It has also been shown experimentally that dinitrogenase reductase ADP-ribosyltransferases require Mg-ATP and a free divalent metal for their activity. Furthermore, a binuclear Mn2+ center, also found in arginases (33), has been detected in the active site of dinitrogenase reductase-activating glycohydrolases (31–33). Therefore, the modeled Mg2+ ions may really point to the approximate positions of Mg2+ or Mn2+ in vivo.
In general, Mg2+ and Mn2+ ions are frequently bound to and coordinated through Asp and Glu (33). Indeed, six highly conserved Asp residues and one Glu are present within the putative binding pocket of mjARH and are modeled to be involved in metal binding and coordination (Table 1 and Fig. 9, which is published as supporting information on the PNAS web site). This catalytic cleft is also clearly marked by large, negatively charged, protein-surface patches computed by grasp2 for the crystal structure of mjARH (Fig. 10, which is published as supporting information on the PNAS web site). Visual inspection of this structure suggested further functional and structural roles for certain amino acids (Table 1). The functional relevance of Asp and other amino acids in and near the active site has also been demonstrated experimentally by inactivating mutations of dinitrogenase reductase-activating glycohydrolase in human or rat ADP-ribosylarginine hydrolase (29–32). Interestingly, most functionally relevant Asp residues are not conserved in SelJ and J1-crystallin homologues or in six other bacterial proteins (Fig. 7, sequences in violet and brown; see also Table 1). However, these proteins conserve other residues (Cys/Ser and His) corresponding to active Asp residues in ARHs (Table 1 and Fig. 7). This high degree of conservation would not be expected if the catalytic site of Asp-lacking proteins was inactive. Therefore, SelJ, J1-crystallins, and other Asp-lacking proteins may still be involved in catalytic functions related to ADP-ribosylation, but these functions may be different from well known ARHs.
SelJ Phylogenetic Distribution. SelJ is also present in other fishes in either Sec or Cys form (see Figs. 7 and 8). However, no SelJ homologues were found in mammals, birds, amphibians, or other vertebrates. Interestingly, SelJ is also present in Strongylocentrotus purpuratus (sea urchin). In addition, Cys homologues are present in two cnidaria, the jellyfish T. cystophora and Hydra magnipapillata. SelJ appears to have a very restricted phylogenetic distribution and is known in vertebrates but is absent, even as a Cys homologue, in mammals.
Distribution of Eukaryotic Selenoproteins. The mapping of known selenoproteins across eukaryotes showed a scattered pattern of Sec/Cys distribution among proteins and taxa (Fig. 5). Sec-containing proteins seemed to accumulate in vertebrate or mammalian genomes (Fig. 5, unframed families); however, the recently identified families (Fig. 5, framed families) no longer followed this previously observed trend of Sec/Cys usage in eukaryotes. A greater diversity among selenoproteomes is thus observed.
Discussion
As new genome sequences become available and improved computational methods are being developed, a more accurate picture of the use and roles of Sec in eukaryotic proteins is emerging. Indeed, a much richer and more complex history of eukaryotic selenoproteins than suspected is taking shape. Recent findings have uncovered taxa-specific selenoproteins with unexpected functions as well as Sec-dependent homologues of well known Cys-containing genes.
Here, we have used computational methods to analyze the recently released genomic sequence of the actinopterygian fish T. nigroviridis and predicted a previously uncharacterized selenoprotein family, which we designated SelJ. We have shown SelJ to be a bona fide selenoprotein that possesses a functional SECIS element by 75Se labeling (Fig. 2). SelJ is widely distributed in actinopterygian fishes and present in at least one species of sea urchin. It is also found among cnidarians in J1-crystallins with Cys (Figs. 7 and 8). In contrast to all known eukaryotic selenoproteins, SelJ is absent, even as a Cys homologue, in mammals.
Our study indicates that SelJ and J1-crystallins may have been derived from ancestral ADP-ribosylation enzymes, suggesting that taxa-specific selenoproteins may have evolved specialized functions from them. Within the family of ADP-ribosylation enzymes, SelJ is more closely related to the J1-crystallins. Analyses of SelJ, J1-crystallin, and several bacterial proteins with Cys in place of Sec show common functional residues that are conserved but different from those found in the active sites of ADP-ribosylation enzymes (Table 1 and Figs. 7 and 9). This observation argues for the acquisition of a different protein function, although possibly related to ADP-ribosylation. Like the majority of eukaryotic selenoproteins, the function of SelJ is not fully clear. Because the Sec insertion mechanisms differ between bacteria and eukaryotes, the recombinant expression of selenoproteins is difficult and often results in a low amount of pure protein, which complicates biochemical analyses. However, whereas in most recently identified selenoproteins there are no clues of which assays should be used to look for function, we expect the work presented here to motivate and lay the ground for the experimental test of ADP-ribosylation-related activity in SelJ.
In addition, the close relationship between SelJ (with Sec) and J1-crystallins (with Cys) suggests the possibility of SelJ also being a crystallin. Most selenoproteins and Cys-containing homologues are thought to have an exclusively enzymatic role, and only GPx4 (34) has been previously predicted to have a structural function. If confirmed, the structural role of SelJ would strongly support a greater functional plasticity of Sec-containing proteins. In this regard, in situ hybridization of SelJ in zebrafish shows high expression almost exclusively in the eye lens in early embryogenesis but a wide expression (including in the eye lens) at later developmental stages. Analyses of EST distribution in several fishes support such expression in adult tissues. This pattern, the lack of SelJ paralogous genes in most fish genomes, and its similarity to bacterial proteins would be consistent with a crystallin role for SelJ in fishes with neither gene duplication nor the loss of a previous enzymatic function. Interestingly, in jellyfish, J1-crystallins are highly expressed in the eye lens and weakly expressed elsewhere, which is consistent also with crystallin and noncrystallin roles (35). In this regard, the analysis of the promoter regions of the fish SelJ is inconclusive. However, it cannot be ruled out that the presence of SelJ in the eye lens may be related exclusively to its original enzymatic function. Indeed, several antioxidant selenoproteins are typically expressed in the eye lens and, to our knowledge, perform no crystallin function (36).
The recent analysis of nonmammalian or nonvertebrate lineages has resulted in the identification of several selenoprotein families (Fig. 5, framed families) with restricted taxa distribution in Sec-containing form and a more widespread distribution in Cys form (10, 37–39). These results argue for a scattered mosaic-like evolution of selenoprotein genes rather than for an accumulation of them in a sort of evolutionary cul-de-sac taxon (10), because it was thought to be the mammalian lineage from the Sec/Cys distribution of the first identified selenoprotein families (Fig. 5, unframed families). SelJ distribution is indeed more extreme because it does not exist, not even in Cys form, in mammals. This result further supports the hypothesis of a distinct evolutionary history of each selenoprotein gene.
The ad hoc use of selenium and Sec among taxa is further observed in the range of selenoproteome sizes. Well studied selenoproteomes, mainly in mammals (9), consist of small sets of selenoproteins (possibly <30 genes), which is in accordance with a limited supply of selenium in nature. Studies of the fly genome suggest the presence of three selenoproteins (7, 8), and a recent analysis of model nematodes limit their selenoproteome to a single protein (40). Furthermore, known yeast and land plants do not possess selenoproteins (nor the necessary selenoprotein machinery) but only alternative Cys variants. Thus, Sec is a rarely used amino acid in extant proteins and, although the selenoprotein sets are unknown for most species, it seems reasonable to expect them to be variable and rather small.
The presence of specialized Sec-containing proteins among lineages and the scattered distribution of Sec and Cys homologues of all selenoproteins are consistent with the highly diverse nature of selenoproteomes (Fig. 5). However, whether there is a general gain/loss trend of Sec underlying the observed mosaic distribution of extant selenoproteins in eukaryotes (Fig. 5) remains unknown. Furthermore, the dynamics of Sec/Cys interconversion for each selenoprotein family and lineage and whether the evolutionary forces driving it can be correlated to biological constraints (e.g., selenium availability, Sec incorporation efficiency, Sec reactivity, and Sec/Cys exchangeability) also are uncertain. Preliminary conclusions can be drawn from the study of specific taxa. In nematodes, the distribution of Sec-containing enzymes suggests that the sizes of selenoproteomes were reduced during evolution and, although some nematodes have few Sec enzymes, in an extreme reduction, Caenorhabditis elegans and Caenorhabditis briggsae have only one selenoprotein left (40).
In summary, although the distribution of selenoproteins we know is only a static snapshot of the current use of Sec among a subset of eukaryotes, the small but different selenoproteome sizes, their protein diversity and functional plasticity, with SelJ as an example, and the uneven distribution of Sec among proteins and taxa further supports a landscape of mosaic evolution in the use of the 21st amino acid in proteins.
Supplementary Material
Acknowledgments
We thank R. J. Stillwell, M. Coromines, and M. Oertel for helpful discussions; C. and B. Thisse for expertise with zebrafish in situ hybridization. This work was supported by National Institutes of Health Grant GM061603 (to V.N.G.) and Plan Nacional de Investigación Científica Desarrollo e Innovación Tecnológica Grant BIO2000-1358-C02-02 from the Ministerio de Educación y Ciencia of Spain (to R.G.). The use of 75Se was supported by U.S. Department of Energy Grant DE-FG07-02ID14380. The work conducted at the Max Planck Institute for Informatics and, partially, at the Institut Municipal d'Investigació Mèdica was performed in the context of the BioSapiens Network of Excellence funded by the European Commission.
Author contributions: S.C., A.V.L., M.A., A.L., T.L., A.K., V.N.G., and R.G. designed research; S.C., A.V.L., C.C., S.V.N., M.A., D.H., and A.L. performed research; S.C. and M.A. analyzed data; and S.C., M.A., V.N.G., and R.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Sec, selenocysteine; SECIS, Sec insertion sequence; ARH, ADP-ribosylglycohydrolase; mjARH, ARH from Methanococcus jannaschii.
See Commentary on page 16123.
References
- 1.Atkins, J. F. & Gesteland, R. F. (2000) Nature 407, 463. [DOI] [PubMed] [Google Scholar]
- 2.Kryukov, G. V. & Gladyshev, V. N. (2004) EMBO Rep. 5, 538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, Y., Fomenko, D. E. & Gladyshev, V. N. (2005) Genome Biol. 6, R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driscoll, D. M. & Copeland, P. R. (2003) Annu. Rev. Nutr. 23, 17–40. [DOI] [PubMed] [Google Scholar]
- 5.Kryukov, G. V., Kryukov, V. M. & Gladyshev, V. N. (1999) J. Biol. Chem. 274, 33888–33897. [DOI] [PubMed] [Google Scholar]
- 6.Lescure, A., Gautheret, D., Carbon, P. & Krol, A. (1999) J. Biol. Chem. 274, 38147–38154. [DOI] [PubMed] [Google Scholar]
- 7.Castellano, S., Morozova, N., Morey, M., Berry, M. J., Serras, F., Corominas, M. & Guigó, R. (2001) EMBO Rep. 2, 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Romero, F. J., Kryukov, G. V., Lobanov, A. V., Carlson, B. A., Lee, B. J., Gladyshev, V. N. & Hatfield, D. L. (2001) J. Biol. Chem. 276, 29798–29804. [DOI] [PubMed] [Google Scholar]
- 9.Kryukov, G. V., Castellano, S., Novoselov, S. V., Lobanov, A. V., Zehtab, O., Guigó, R. & Gladyshev, V. N. (2003) Science 300, 1439–1443. [DOI] [PubMed] [Google Scholar]
- 10.Castellano, S., Novoselov, S. V., Kryukov, G. V., Lescure, A., Blanco, E., Krol, A., Gladyshev, V. N. & Guigó, R. (2004) EMBO Rep. 5, 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Axley, M. J., Böck, A. & Stadman, T. C. (1991) Proc. Natl. Acad. Sci. USA 88, 8450–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berry, M. J., Kieffer, J. D., Harney, J. W. & Larsen, P. R. (1991) J. Biol. Chem. 266, 14155–14158. [PubMed] [Google Scholar]
- 13.Berry, M. J., Mai, A. L., Kieffer, J., Harney, J. W. & Larsen, P. (1992) Endocrinology 131, 1448–1852. [DOI] [PubMed] [Google Scholar]
- 14.Stadtman, T. C. (1996) Annu. Rev. Biochem. 65, 83–100. [DOI] [PubMed] [Google Scholar]
- 15.Hatfield, D. L., Gladyshev, V. N., Park, J., Park, S. I. Chittum, H. S., Baek, H. J., Carlson, B. A., Yang, E. S., Moustafa, M. E. & Lee, B. J. (1999) Comp. Nat. Prod. Chem. 4, 353–380. [Google Scholar]
- 16.Gromer, S., Johansson, L., Bauer, H., Arscott, L. D., Rauch, S., Ballou, D. P., Williams, C. H., Jr., Schirmer, R. H. & Arner, E. S. (2003) Proc. Natl. Acad. Sci. USA 100, 12618–12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatfield, D. L. (2001) Selenium: Its Molecular Biology And Role In Human Health (Kluwer, New York).
- 18.Jaillon, O., Aury, J.-M., Brunet, F., Petit, J.-L., Stange-Thomann, N., Mauceli, E., Bouneau, L., Fischer, C., Ozouf-Costaz, C., Bernot, A., et al. (2004) Nature 431, 946–957. [DOI] [PubMed] [Google Scholar]
- 19.Parra, G., Blanco, E. & Guigó, R. (2000) Genome Res. 10, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul, S. F., Madden, T., Schaffer, A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messeguer, X., Escudero, R., Farré, D., Nuñez, O., Martínez, J. & Albà, M. (2002) Bioinformatics 18, 333–334. [DOI] [PubMed] [Google Scholar]
- 22.Farré, D., Roset, R., Huerta, M., Adsuara, J. E., Roselló, L., Albà, M. & Messeguer, X. (2003) Nucleic Acids Res. 31, 3651–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thisse, C., Thisse, B., Schilling, T. F. & Postlethwait, J. H. (1993) Development (Cambridge, U.K.) 119, 1203–1215. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 25.Grundner-Culemann, E., Martin, G. W., III, Harney, J. W. & Berry, M. J. (1999) RNA 5, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piatigorsky, J., Horwitz, J. & Norman, B. L. (1993) J. Biol. Chem. 268, 11894–11901. [PubMed] [Google Scholar]
- 27.Piatigorsky, J. (2003) J. Struct. Funct. Genomics 3, 131–137. [PubMed] [Google Scholar]
- 28.Nilsson, D.-E. (2004) Curr. Opin. Neurobiol. 14, 407–414. [DOI] [PubMed] [Google Scholar]
- 29.Takada, T., Iida, K. & Moss, J. (1993) J. Biol. Chem. 268, 17837–17843. [PubMed] [Google Scholar]
- 30.Konczalik, P. & Moss, J. (1999) J. Biol. Chem. 274, 16736–16740. [DOI] [PubMed] [Google Scholar]
- 31.Antharavally, B. S., Poyner, R. R., Zhang, Y., Roberts, G. P. & Ludden, P. W. (2001) J. Bacteriol. 183, 5743–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, K., Zhang, Y. & Roberts, G. P. (2004) FEBS Lett. 559, 84–88. [DOI] [PubMed] [Google Scholar]
- 33.Christianson, D. W. & Cox, J. D. (1999) Annu. Rev. Biochem. 68, 33–57. [DOI] [PubMed] [Google Scholar]
- 34.Ursini, F., Heim, S., Kiess, M., Maiorino, M., Roveri, A., Wissing, J. & Flohe, L. (1999) Science 285, 1393–1396. [DOI] [PubMed] [Google Scholar]
- 35.Piatigorsky, J., Norman, B., Dishaw, L. J., Kos, L., Horwitz, J., Steinbach, P. J. & Kozmik, Z. (2001) Proc. Natl. Acad. Sci. USA 98, 12362–12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flohe, L. (2005) Dev. Ophthalmol. 38, 89–102. [DOI] [PubMed] [Google Scholar]
- 37.Fu, L. H., Wang, X. F., Eyal, Y., She, Y. M., Donald, L. J., Standing, K. G. & Ben-Hayyim, G. (2002) J. Biol. Chem. 277, 25983–25991. [DOI] [PubMed] [Google Scholar]
- 38.Novoselov, S. V., Rao, M., Onoshko, N. V., Zhi, H., Kryukov, G. V., Xiang, Y., Weeks, D. P., Hatfield, D. L. & Gladyshev, V. N. (2002) EMBO J. 21, 3681–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obata, T. & Shiraiwa, Y. (2005) J. Biol. Chem. 280, 18462–18468. [DOI] [PubMed] [Google Scholar]
- 40.Taskov, T., Chapple, C., Kryukov, G. V., Castellano, S., Lobanov, A. V., Korotkov, K. V., Guigó, R. & Gladyshev, V. N. (2005) Nucleic Acids Res. 33, 2227–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.