Abstract
A transgenic mouse containing the complete human SLAM (hSLAM/CD150) gene, including its endogenous promoter for transcription, was generated by using human genomic DNA cloned into a bacterial artificial chromosome. hSLAM, the primary receptor for measles viruses (MV), was expressed on activated B, T, and dendritic cells with an expression profile equivalent to that of humans. We demonstrated that hSLAM+ cells obtained from the transgenic mouse, including activated B, T, and dendritic cells, were susceptible to MV infection in a receptor-dependent manner. Evidence was provided for transient infection in the nasal lymph nodes of hSLAM+ mice after intranasal inoculation. Virus was rapidly cleared without signs of secondary replication. To improve the efficiency of MV production, the hSLAM+ mice were bred with mice having a Stat1-deficient background. These mice were more susceptible to MV infection and produced more virus particles. After intranasal and intraperitoneal inoculation of these mice with MV, infections of the thymus, spleen, nasal, mesenteric, and leg lymph nodes were detected. Upon necropsy, enlarged lymph nodes and spleen were apparent. Flow cytometric analysis showed that abnormally large numbers of mature neutrophils and natural killer cells caused the splenomegaly. The hSLAM transgenic mouse constitutes an improved rodent model for studying the interaction of MV with immune cells that more accurately reflects the infection pattern found in humans.
Keywords: activated lymphocytes, transgenic mouse, dendritic cells
Measles viruses (MV) is one of the most contagious diseases known to man and is a major killer of children in the developing countries of Africa and South America. The high mortality rate associated with MV infection results from increased susceptibility to opportunistic infections. Although MV has been studied extensively, the mechanism for the immunosuppressive effects observed during MV infection remains unclear (1).
The development of a mouse model may lead to a better understanding of the immune suppression induced by MV. This was previously attempted by expressing CD46, one of the two receptors for MV, in mice (2-6). However, CD46 receptor, a ubiquitously expressed glycoprotein, is only used by vaccine or laboratory adapted strains of MV. Recently, a second receptor for MV was discovered: CD150 or signaling lymphocytic activation molecule (SLAM) (7-9). SLAM is a host cell receptor for both vaccine and WT MV strains of MV. It is a 70-kDa, type I transmembrane glycoprotein expressed on immune cells such as activated T cells, B cells, macrophages, and dendritic cells (DC) (10). The N-terminal V domain of SLAM interacts with the hemagglutinin protein of MV (9).
SLAM determines Th2 cytokine production such as IL-4 and it may be involved in production of IL-12, TNF, and NO by macrophages (11). In addition, SLAM may induce B cell proliferation and Ig synthesis. SLAM signal transduction in T cells is mediated through SAP (SLAM-associated protein). Mutations in SAP cause the lymphoproliferative disorder XLP, which results in an inappropriate response to Epstein-Barr virus infection (12). SAP is an SH2-containing adapter protein that binds with high affinity to tyrosines of SLAM's intracellular domain. SAP binds to the membrane proximal tyrosine (Tyr-281) of SLAM's cytoplasmic tail; it recruits and activates FynT (13). SAP interaction with the SH3 domain of FynT stabilizes the open, active conformation of FynT. This active form then phosphorylates Tyr-307 and Tyr-372 of SLAM resulting in the recruitment of multiple players involved in SLAM signaling (14).
To investigate MV receptor interactions and the effect of infection on SLAM function, we introduced a DNA fragment encompassing the entire human SLAM (hSLAM) gene into a mouse genome using a bacterial artificial chromosome (BAC). We showed that our hSLAM(+/+) mice express hSLAM in a regulated fashion and with an expression profile identical to that in humans. hSLAM+ cells obtained from the transgenic mice, including activated B, T, and dendritic cells, were susceptible to infection by MV in a receptor-dependent manner. To improve virus infection, the hSLAM transgenic mice were bred into a stat1-deficient background, which renders mice more susceptible to pathogens (15-18). Inoculation of the hSLAM(+/+)/stat1(-/-) mice yielded a productive infection of the lymphoid organs that resulted in enlarged lymph nodes and spleen. This mouse model should help researchers study receptor tropism and the immune dynamics of MV, and will provide a valuable tool for researchers studying the engineered vaccine and oncolytic potential of this virus (19-22).
Materials and Methods
Production of CD150 Mice. A BAC library from the Fondation Jean Dausset-Centre d'Etude Polymorphisme Humaine (Paris) was screened by using primers that recognized genomic sequence 10 kb upstream of the SLAM start codon and sequence 1.5 kb downstream of the SLAM stop codon. The BAC DNA was prepared by using a BAC purification kit (Clontech), and the DNA was resuspended in microinjection buffer (10 mM Tris·HCl, pH 7.5/0.1 mM EDTA, pH 8.0/100 mM NaCl/1× polyamines) at a concentration of 0.5-1.0 ng/μl. Microinjection of embryos was done as described in ref. 23. Offspring were weaned at 3-4 weeks of age, and DNA was prepared from a 1-cm portion of their tails. Heterozygous hSLAM mice were generated and bred to homozygosity and used in subsequent experiments. Mice deficient in stat1 were obtained from the laboratory of David Levy (New York University School of Medicine, New York).
Viruses and Infections. The GFP-expressing viruses MVedGFP, MVwtfGFP, and MVedGFP-SLAMblind are described in refs. 24-26. Activated lymphocytes and DC were infected at a multiplicity of infection of 5 unless otherwise stated. Intranasal (i.n.) infections were performed similarly to methods described in ref. 6. For i.n. and i.p. infections, respectively, 2.5 × 106 PFU and 1 × 107 PFU were used.
Isolation and Activation of Lymphocytes and DC. The lymph nodes and spleens of hSLAM or C57BL/6 mice were harvested through a 0.45-μm mesh in RPMI medium 1640 containing 10% FBS and 0.1% 2-mercaptoethanol. Both T and B cells were negatively selected for; purified T cells were activated by using anti-CD3 and IL-2 (50 units/ml), and purified B cells were activated by using 20 μg/ml LPS. Cell activation status was analyzed 48 h after harvest. DC were harvested as described in ref. 27. On day 9, the cells in suspension were harvested, counted, analyzed by FACS, and replated in activation media (RPMI medium 1640 + 10% FBS/0.1% 2-mercaptoethanol/antibiotics/5 ng/ml GM-CSF/100 ng/ml LPS). Activation status was analyzed 24 h later.
Immunoprecipitation Experiments. Tissues from dissected mice were sonicated by a polytron homogenizer in RIPA buffer (50 mM Hepes/150 mM NaCl/detergents and protease inhibitors). The lysed cells were centrifuged at 10,000 × g for 10-15 min at 4°C, and the supernatant was recovered. Protein concentrations were measured, 10 μl of anti-MV H monoclonal antibody (Chemicon) was added to equivalent amounts of protein, and the mixture was incubated overnight at 4°C, followed by incubation with protein G for 1-2 h at 4°C. The protein G-MV H complexes were centrifuged and washed with RIPA buffer five times. The beads were resuspended in 15 μl of reducing SDS/PAGE sample buffer and boiled for 5 min. After a 5-min spin, the supernatant was subjected to PAGE. The primary antibody was a rabbit polyclonal antibody directed to the C terminus of H protein.
Immunohistochemistry. Lymph nodes and spleens were snap-frozen in OCT embedding medium (EM Science) by using liquid N2, and 10-μm sections were made by using a cryoslicer. The sections were air-dried, fixed in cold acetone for 10 min, air-dried again, and rinsed in PBS. Endogenous peroxidase was blocked by using 0.3% hydrogen peroxide for 4 min. After protein blocking, the slides were incubated with a rabbit anti-MV H antibody at a dilution of 1/100 for 1 h at room temperature, washed well in PBS, and incubated with an anti-rabbit biotinylated linking antibody for 30 min at room temperature. They were then washed well in PBS, incubated with Ultra Streptavidin-Horseradish Peroxidase Complex (ID Labs, London, ON, Canada) for 30 min, and washed again in PBS. The slides were then developed with freshly prepared chromagen, washed in running tap water, and counterstained lightly with Mayer's hematoxylin. After another wash, they were dehydrated through alcohols, cleared in xylene, and mounted in Permount (Fisher Scientific).
Antibodies. Monoclonal antibodies specific for CD150 (clone A12) were purchased from BD Biosciences Pharmingen. Monoclonal antibodies recognizing H were purchased from Chemicon. A rabbit polyclonal antibody was generated against the C terminus of MV H protein. Antibodies that recognize CD11c, B220, CD4, CD8, B7.2, Iab, Gr-1, NK1.1, and CD11b were purchased from BD Biosciences.
Results
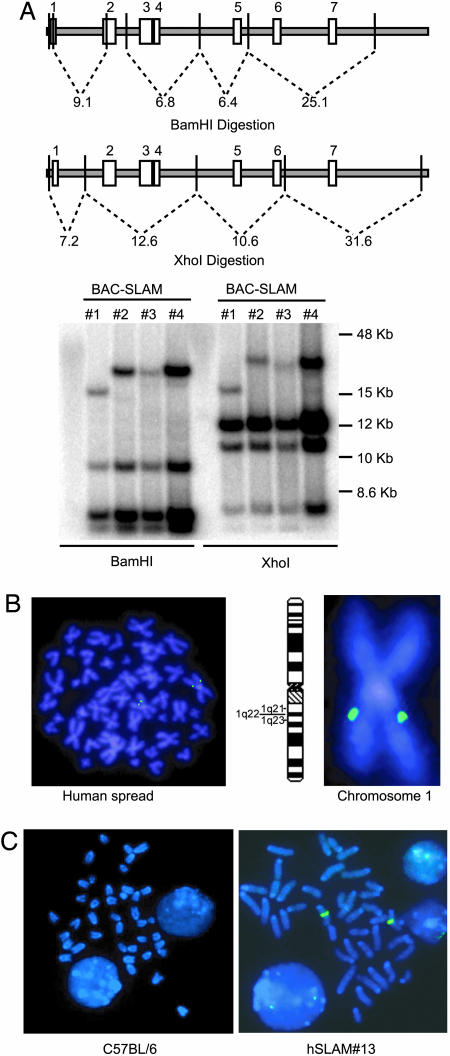
Generation of CD150 (SLAM) Transgenic Mice. To generate transgenic mice that express human SLAM (hSLAM), a BAC containing the hSLAM gene was used. Three of four BAC plasmids isolated from this library, #2-4, had the expected BamHI and XhoI DNA restriction digestion patterns in Southern blots hybridized to a radioactive hSLAM cDNA probe (Fig. 1B). By using BAC-hSLAM#2 as a probe, FISH was performed to confirm that the BAC genomic clone mapped to the expected CD150 gene location on chromosome #1 (1q22) (Fig. 1B).
Fig. 1.
Characterization of the BAC-hSLAM construct used to produce the SLAM transgenic mice. (A) Southern blot analysis of four BAC-hSLAM constructs obtained. The predicted fragments within the hSLAM gene after BamHI and XhoI digestion of human genomic DNA are indicated. The Southern blot was probed by using hSLAM cDNA. (B) FISH analysis of BAC-hSLAM#2 hybridized to a human chromosome spread. Chromosome #1 is enlarged to enhance visualization of the hybridization to locus 1q22. (C) FISH analysis of BAC-hSLAM#2 to mouse genomic spreads prepared from C57BL/6 or T cells from hSLAM #13 transgenic mouse.
Because the restriction map analysis of BAC-hSLAM#2 was consistent with complete coverage of the hSLAM genomic sequence, it was used to generate homozygous hSLAM transgenic mice. Three founder mice, two males and one female, were obtained, but only two mice were fertile: hSLAM#3 and hSLAM#13. hSLAM#13 has yielded homozygotes for the hSLAM transgene, whereas, to date, only hemizygotes have been obtained for hSLA M#3. Analysis of the homozygous hSLAM#13 transgenic mouse is presented in this article, but equivalent results with heterozygous hSLAM#3 mice have been obtained (data not shown). FISH analysis of mouse chromosomes from this line showed that the BAC integrated only at one locus (Fig. 1C).
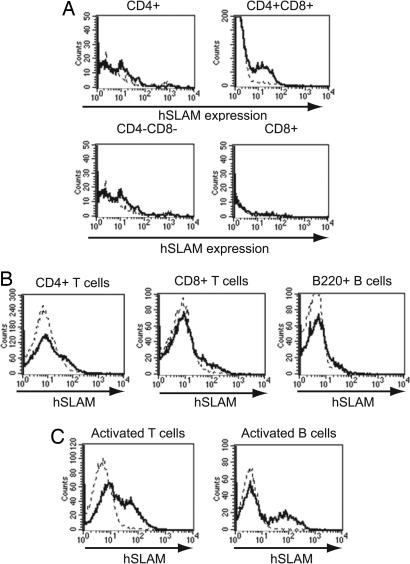
Profile of hSLAM Protein Expression in hSLAM Transgenic Mice. To determine the protein expression pattern of hSLAM in our hSLAM transgenic mice, RT-PCR (see the supporting information, which is published on the PNAS web site) and FACS analysis were performed. RT-PCR analysis of organs from the transgenic mice indicated that mRNA for hSLAM was synthesized in the thymus and in activated B and T cells (see the supporting information). Thymocytes were harvested from both C57BL/6 (WT) and hSLAM mice and stained with antibodies that recognize CD4, CD8, and hSLAM (Fig. 2A). In the hSLAM mice, hSLAM was strongly expressed only on CD4+ and CD8+ double-positive thymocytes. B and T cells harvested from the spleen or lymph nodes were stained for CD4, CD8, and hSLAM or B220 and hSLAM. As expected, expression of hSLAM on CD4+, CD8+, and B220+ cells was low (10) (Fig. 2B).
Fig. 2.
Profile of hSLAM protein expression. (A) Thymocytes harvested from hSLAM transgenic and C57BL/6 mice were stained with CD4, CD8, and hSLAM antibodies. SLAM expression was analyzed for the identified thymic populations. (B) Splenocytes were stained with CD4, CD8, B220, and hSLAM antibodies. hSLAM expression was analyzed in the indicated populations. Splenic B and T cells were purified, activated, and stained for hSLAM. (C) Activated B, activated T, and CD4+CD8+ T cells expressed hSLAM on their surfaces. The dotted line represents hSLAM staining of lymphocytes from C57BL/6 mice, and the solid line indicates hSLAM staining of lymphocytes from the transgenic mice.
To monitor hSLAM expression after activation, B and T cells were purified from the spleen and subsequently activated. FACS analysis indicated that 20% and 17% of B and T cells, respectively, could be activated to express hSLAM (Fig. 2C). Levels of hSLAM expression were comparable to those on activated PBMCs from humans (8).
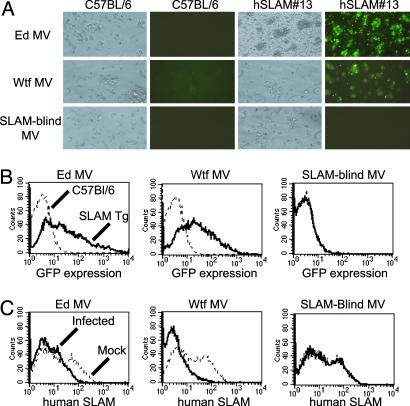
In Vitro Infection of hSLAM+ Cells from hSLAM Transgenic Mice. To determine whether hSLAM expression on T cells allowed for MV infection, activated T cells were infected with three different GFP-expressing MV: GFPedMV, GFPwtfMV, and MVedGFP-SLAMblind. All of these viruses are described in refs. 24-26. GFPedMV and GFPwtfMV can use hSLAM as a receptor, whereas the MVedGFP-SLAMblind cannot use hSLAM for infection and serves as a negative control.
Inoculation of activated T cells from hSLAM mice with GFPedMV and GFPwtfMV but not MVedGFP-SLAMblind resulted in GFP expression by 36 h postinfection (p.i.) (Fig. 3 A and B). Furthermore, hSLAM was down-regulated from the surface of infected T cells during GFPedMV and GFPwtfMV infection but not after inoculation with the SLAMblind virus (Fig. 3C). Down-regulation of hSLAM has previously been shown to occur in MV infections (28). C57BL/6 mice showed no signs of infection as monitored by GFP expression, confirming the importance of hSLAM expression for the infection of mouse T cells. Infection experiments were also performed with B cells and showed similar results (data not shown).
Fig. 3.
Activated T cells from hSLAM#13 transgenic mice can be infected in a receptor-dependent manner by MV. Activated T cells were infected with recombinant MV (multiplicities of infection of 5) that express either the Edmonston hemagglutinin (Ed MV), the WTF hemagglutinin (WTF), or a hemagglutinin that cannot use hSLAM for viral entry (SLAMblind), in addition to the GFP reporter gene. All experiments were performed 48 h p.i. (A) GFP expression in infected T cells detected by fluorescence microscope. (B) GFP expression in infected T cells determined by FACS analysis. (C) FACS analysis showing down-regulation of SLAM on activated T cells infected with Ed MV and WTF MV.
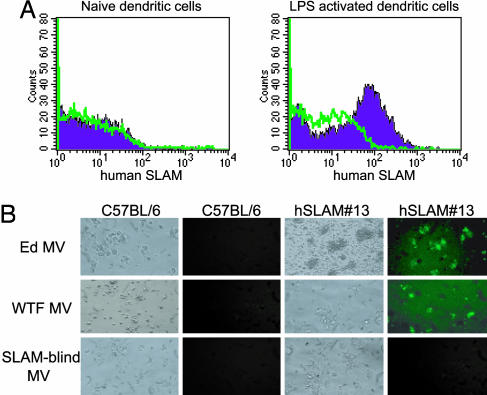
To analyze hSLAM expression on DC, bone marrow from the femurs of hSLAM transgenic mice was harvested and cultivated in the presence of GM-CSF for 9 days. At day 9, the cells were activated by using LPS for 24 h. FACS analysis demonstrated that hSLAM was expressed on LPS-activated DC but not on naïve DC (Fig. 4A). Inoculation of activated DC from hSLAM mice with GFPedMV, GFPwtfMV, or MVedGFP-SLAMblind resulted in receptor-dependent infection as determined by GFP expression (Fig. 4B).
Fig. 4.
Analysis of DC from hSLAM#13 transgenic mice. (A) Activated DC from bone marrow express hSLAM. DC were cultured from bone marrow in the presence of GM-CSF. On days 9 and 10 (after LPS activation), DC were harvested and stained for CD11c, MHC II, and hSLAM expression. For all FACS histograms, the blue peaks represent DC from hSLAM#13 transgenic mice stained for hSLAM. The green dotted lines represent dendritic cells from C57BL/6 mice stained for hSLAM. (B) Visualization of GFP expression in DC infected with GFP-expressing MV. DC were inoculated with Ed MV, WTF MV, or a SLAMblind MV (multiplicities of infection of 5). Photographs were taken at 48 h p.i.
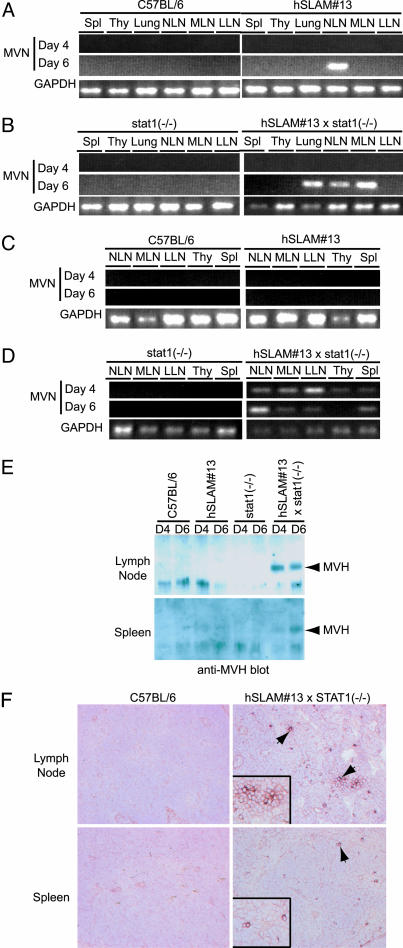
hSLAM Mice Are Transiently Infected by MV. To determine whether hSLAM mice can be infected in vivo, i.n. and i.p. inoculations of hSLAM mice were performed by using Montefiore MV, a WT isolate propagated in a primate B cell line (B95-8). Before infection, mice were primed by i.p. injection with 10 μg of LPS, which induces hSLAM expression in circulating lymphocytes, thereby increasing the pool of potential MV target cells (data not shown). RT-PCR for MV N mRNA with RNA derived from the lung, nasopharyngeal lymph nodes, mesenteric lymph nodes, thymus, and spleen from infected mice was performed to monitor levels of infection at 4 or 6 days p.i. (Fig. 5 A and C). After i.n. infection, MV N mRNA transcripts were observed in the nasopharyngeal lymph nodes of the hSLAM mice but not in other more distal nodes, thymus, spleen, or other organs of C57BL/6 mice. No N mRNA transcripts were detected after i.p. infections.
Fig. 5.
Evidence of MV infection in hSLAM#13/stat1(-/-) mice. (A and B) RT-PCR analysis of mice inoculated with WT Montefiore MV i.n. Primers specific for MV N and GAPDH cDNAs were used. (C and D) RT-PCR analysis of mice inoculated with Montefiore MV via i.p. injections. Primers specific for MV N and GAPDH cDNAs were used. (E) Western blots of immunoprecipitations for MV H protein. Arrows point to the MV H protein band. (F) Immunohistochemistry of sections from mice infected with Montefiore MV. Sections from lymph nodes and spleen were probed by using a rabbit polyclonal Ab specific for MV H. Cells positive for MV H have a deep reddish-brown stain on the cell surface as indicated by the arrows. Spl, spleen; Thy, thymus; NLN, nasal lymph node; MLN, mesenteric lymph node; LLN, leg lymph node.
STAT1 Deficiency Confers a Permissive Environment for Efficient Infection of the hSLAM Mice. Previous work with CD46 transgenic mice identified the innate immune response as a barrier to efficient MV replication in mice (6, 29). Therefore, the hSLAM mice were bred into a Stat1-deficient background, which renders mice extremely susceptible to pathogenic infections (15-18). To determine whether Stat1 deficiency could generate a permissive environment for MV infection, hSLAM(+/+)/stat1(-/-) mice were inoculated i.n. and i.p. by using Montefiore WT MV. In these mice, mRNA for MV N was detected in the lymph nodes, thymus, and spleen (Fig. 5 B and D). No signs of MV infection were observed in stat1-deficient mice inoculated with the virus (Fig. 5B) indicating that expression of hSLAM was required for MV infection. Immunoprecipitation of MV H from the cell lysates of infected organs and immunohistochemistry of infected lymph nodes and spleens with an anti-MV H antibody confirmed the RT-PCR results (Fig. 5 E and F).
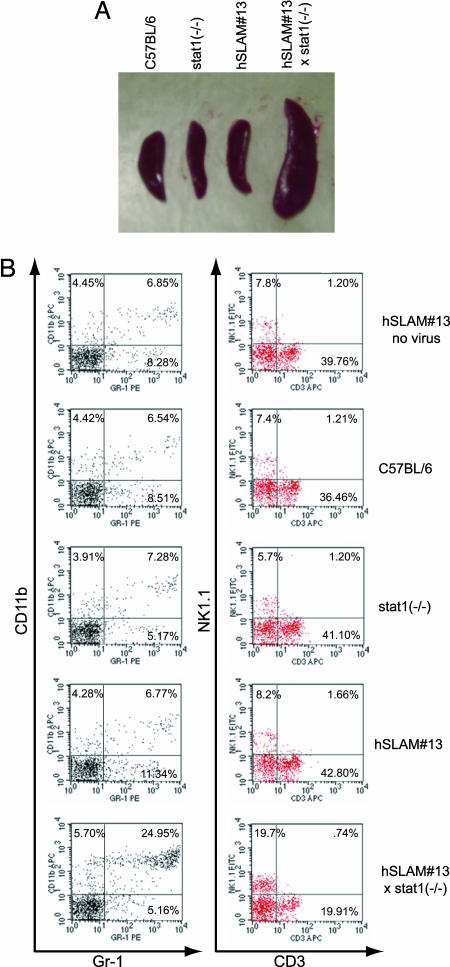
A striking effect of MV infection in the hSLAM(+/+)/stat1(-/-) mice was the presence of enlarged lymph nodes (not shown) and splenomegaly (Fig. 6A). The onset of these symptoms corresponded to the peak of viral infection and was consistently observed only in the hSLAM(+/+)/stat1(-/-) mice. To identify the cause for the enlarged spleens, FACS analysis of the splenic cell populations was performed at multiple time points p.i. This analysis revealed a difference in the numbers of mature neutrophils and natural killer (NK) cells in infected hSLAM(+/+)/stat1(-/-) mice (Fig. 6B). In the spleens with an enlarged phenotype, almost 20% of the splenic population consisted of Gr-1 and CD11b (MAC-1) double-positive cells, which are usually mature neutrophils (30). In infected C57BL/6, stat1(-/-) and hSLAM(+/+)/stat1(-/-) mice, these cells comprise only 4-8% of the cellular profile in the spleen. In addition to the increased levels of activated neutrophils, there was a substantial rise in the number of natural killer NK1.1+ cells (19%). Again, this was much higher than that seen in infected C57BL/6, stat1(-/-), and hSLAM mice. Combined, these two cell types make up almost 50% of the cellular compartment of the enlarged spleen in the infected hSLAM(+/+)/stat1(-/-) mice. Although their role in viral clearance remains to be explored, the increased level of these two cell types is the source of the observed splenomegaly after MV infection.
Fig. 6.
MV-infected hSLAM#13/stat1(-/-) mice develop splenomegaly due to immune cell infiltration. (A) Spleens from mice with and without hSLAM, and with and without STAT1 expression, were infected with WT Montefiore MV. Mice were dissected and photographed at 6 days p.i. Infections with WTF MV gave similar results. (B) FACS analysis of immune cells from spleens of infected mice. The presence of mature neutrophils was determined by staining for CD11b and Gr-1 surface markers. CD11b+Gr-1+ cells are mature neutrophils. The presence of NK cells was determined by staining for NK1.1 and CD3. NK1.1+CD3- cells are NK cells, and NK1.1+CD3+ cells are NK T cells. Percentages represent relative numbers of cells for each quadrant. The experiment was performed three separate times with similar results.
Discussion
The development of a small animal model that permits host cell entry and replication of MV should allow the assessment of the relevance of receptor usage by this virus. It should also have applications in the development of recombinant MV vaccines (19, 20, 31) and studying the oncolytic properties of this virus (21, 22, 32). Other groups have generated mice that express human CD46, which is the receptor used by vaccine and laboratory adapted strains of MV for virus entry (2-6, 33). These labs reported transient MV infections in these mice with limited virus production due to the induction of innate immune responses. Breeding of the CD46 mice into IFN-defective genetic backgrounds was used to overcome these difficulties. In a mouse strain deficient for IFN-α/β receptor, dissemination of MV to several tissues was observed (6, 29). Studies in mice lacking RAG2, which is a gene critical for the proper development of T and B cells, also resulted in more efficient spread of MV (34). However, it was not possible to analyze the role of T and B cells in MV infection in these experiments. Others studied the effect of MV infection in the brains of suckling mice, which lack innate immunity (4, 34). These studies did not recapitulate a normal course of infection or lead to the development of a neurological condition known as subacute sclerosing panencephalitis.
The most important limitation to the CD46 transgenic mouse models is the fact that CD46 supports efficient infection of only vaccine- or laboratory-adapted MV strains. Recently, however, the receptor for lymphocytic or WT strains of MV was determined to be hSLAM (8, 9, 35). Since this discovery, two transgenic mouse models, one that expresses hSLAM on T cells and one that expresses hSLAM on CD11c+ DC, were generated (36, 37). However, the utility of these mice as a small animal model for MV infection was hampered by the use of specific promoters that limited the expression of hSLAM to only certain types of cells. In contrast, we generated hSLAM transgenic mice that have a human-like expression profile through use of the human gene's endogenous promoter.
When hSLAM mice were inoculated with MV, infection was observed in the local lymph nodes, but the immune response cleared the virus before it could spread further. This clearance was expected because studies in CD46 transgenic mice indicated that MV is incapable of evading a strong innate immune response in mice. Because of the critical role of STAT1 in signaling downstream of types I and II IFN receptors, Stat1(-/-) mice are more susceptible to bacterial and viral infections (15-18). In stat1(-/-) mice, there is a reduction or absence of IFN-α- and IFN-γ-induced expression of major histocompatibility complex (MHC) class II protein, IFN regulatory factor 1 (IRF-1), guanylate-binding protein 1 (GBP-1), the MHC class II transactivating protein (CIITA), and the complement protein, C3.
When hSLAM(+/+)/stat1(-/-) mice were inoculated with MV, infected spleens were infiltrated by mature neutrophils and NK cells. The direct involvement of these two cell types in the clearance of the MV infection was not assessed, but it has been shown that NK cells do play a primary and immediate role in antiviral responses (38-40). In addition, the neutrophil response was shown to play a significant role in the oncolytic effect of replicating MV (41).
In conclusion, the present study reports the development of a small animal model for MV infection through transgenic expression of the primary receptor of this virus, hSLAM(CD150). This model will enable studies that relate immune responses to MV pathology but may also facilitate the assessment of recombinant MV-based vaccines and oncolytic vectors.
Supplementary Material
Acknowledgments
We thank the veterinarians and technicians of the University Health Network Toronto animal facility for housing and manipulating our mice. The Amgen Transgenic Mouse Facility (Thousand Oaks, CA) injected the hSLAM gene into oocytes for generation of the heterozygous transgenic mice. The immunology expertise of Gordon Duncan was much appreciated. The stat1-deficient mice were obtained from Dr. David Levy (New York University School of Medicine, New York). We also thank Dr. Wen-Chen Yeh and the entire Richardson laboratory for helpful discussions and support. This work was supported by Canadian Institutes of Health Research Grant MT-10638.
Author contributions: C.D.R. designed research; G.G.W., C.I., R.D., and J.B. performed research; J.B., J.S., S.V., and R.C. contributed new reagents/analytic tools; G.G.W., C.I., R.C., and C.D.R. analyzed data; and G.G.W. and C.D.R. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BAC, bacterial artificial chromosome; DC, dendritic cells; i.n., intranasal(ly); MV, measles viruses; NK, natural killer; p.i., postinfection.
References
- 1.Schneider-Schaulies, S. & ter Meulen, V. (2002) Springer Semin. Immunopathol. 24, 127-148. [DOI] [PubMed] [Google Scholar]
- 2.Horvat, B., Rivailler, P., Varior-Krishnan, G., Cardoso, A., Gerlier, D. & Rabourdin-Combe, C. (1996) J. Virol. 70, 6673-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorley, B. R., Milland, J., Christiansen, D., Lanteri, M. B., McInnes, B., Moeller, I., Rivailler, P., Horvat, B., Rabourdin-Combe, C., Gerlier, D., et al. (1997) Eur. J. Immunol. 27, 726-734. [DOI] [PubMed] [Google Scholar]
- 4.Manchester, M., Eto, D. S. & Oldstone, M. B. (1999) J. Neuroimmunol. 96, 207-217. [DOI] [PubMed] [Google Scholar]
- 5.Blixenkrone-Moller, M., Bernard, A., Bencsik, A., Sixt, N., Diamond, L. E., Logan, J. S. & Wild, T. F. (1998) Virology 249, 238-248. [DOI] [PubMed] [Google Scholar]
- 6.Mrkic, B., Pavlovic, J., Rulicke, T., Volpe, P., Buchholz, C. J., Hourcade, D., Atkinson, J. P., Aguzzi, A. & Cattaneo, R. (1998) J. Virol. 72, 7420-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatsuo, H., Ono, N. & Yanagi, Y. (2001) J. Virol. 75, 5842-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu, E. C., Iorio, C., Sarangi, F., Khine, A. A. & Richardson, C. D. (2001) Virology 279, 9-21. [DOI] [PubMed] [Google Scholar]
- 9.Erlenhoefer, C., Wurzer, W. J., Loffler, S., Schneider-Schaulies, S., ter Meulen, V. & Schneider-Schaulies, J. (2001) J. Virol. 75, 4499-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocks, B. G., Chang, C. C., Carballido, J. M., Yssel, H., de Vries, J. E. & Aversa, G. (1995) Nature 376, 260-263. [DOI] [PubMed] [Google Scholar]
- 11.Davidson, D., Shi, X., Zhang, S., Wang, H., Nemer, M., Ono, N., Ohno, S., Yanagi, Y. & Veillette, A. (2004) Immunity 21, 707-717. [DOI] [PubMed] [Google Scholar]
- 12.Sayos, J., Wu, C., Morra, M., Wang, N., Zhang, X., Allen, D., van Schaik, S., Notarangelo, L., Geha, R., Roncarolo, M. G., et al. (1998) Nature 395, 462-469. [DOI] [PubMed] [Google Scholar]
- 13.Latour, S., Roncagalli, R., Chen, R., Bakinowski, M., Shi, X., Schwartzberg, P. L., Davidson, D. & Veillette, A. (2003) Nat. Cell Biol. 5, 149-154. [DOI] [PubMed] [Google Scholar]
- 14.Latour, S., Gish, G., Helgason, C. D., Humphries, R. K., Pawson, T. & Veillette, A. (2001) Nat. Immunol. 2, 681-690. [DOI] [PubMed] [Google Scholar]
- 15.Sugawara, I., Yamada, H. & Mizuno, S. (2004) Tohoku J. Exp. Med. 202, 41-50. [DOI] [PubMed] [Google Scholar]
- 16.Meraz, M. A., White, J. M., Sheehan, K. C., Bach, E. A., Rodig, S. J., Dighe, A. S., Kaplan, D. H., Riley, J. K., Greenlund, A. C., Campbell, D., et al. (1996) Cell 84, 431-442. [DOI] [PubMed] [Google Scholar]
- 17.Lieberman, L. A., Banica, M., Reiner, S. L. & Hunter, C. A. (2004) J. Immunol. 172, 457-463. [DOI] [PubMed] [Google Scholar]
- 18.Durbin, J. E., Hackenmiller, R., Simon, M. C. & Levy, D. E. (1996) Cell 84, 443-450. [DOI] [PubMed] [Google Scholar]
- 19.Despres, P., Combredet, C., Frenkiel, M. P., Lorin, C., Brahic, M. & Tangy, F. (2005) J. Infect. Dis. 191, 207-214. [DOI] [PubMed] [Google Scholar]
- 20.Lorin, C., Delebecque, F., Labrousse, V., Da Silva, L., Lemonnier, F., Brahic, M. & Tangy, F. (2005) Vaccine 23, 4463-4472. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura, T. & Russell, S. J. (2004) Expert Opin. Biol. Ther. 4, 1685-1692. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura, T., Peng, K. W., Harvey, M., Greiner, S., Lorimer, I. A., James, C. D. & Russell, S. J. (2005) Nat. Biotechnol. 23, 209-214. [DOI] [PubMed] [Google Scholar]
- 23.Simonet, W. S., Hughes, T. M., Nguyen, H. Q., Trebasky, L. D., Danilenko, D. M. & Medlock, E. S. (1994) J. Clin. Invest. 94, 1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duprex, W. P., McQuaid, S., Hangartner, L., Billeter, M. A. & Rima, B. K. (1999) J. Virol. 73, 9568-9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider, U., von Messling, V., Devaux, P. & Cattaneo, R. (2002) J. Virol. 76, 7460-7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vongpunsawad, S., Oezgun, N., Braun, W. & Cattaneo, R. (2004) J. Virol. 78, 302-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutz, M. B., Kukutsch, N., Ogilvie, A. L., Rossner, S., Koch, F., Romani, N. & Schuler, G. (1999) J. Immunol. Methods 223, 77-92. [DOI] [PubMed] [Google Scholar]
- 28.Welstead, G. G., Hsu, E. C., Iorio, C., Bolotin, S. & Richardson, C. D. (2004) J. Virol. 78, 9666-9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mrkic, B., Odermatt, B., Klein, M. A., Billeter, M. A., Pavlovic, J. & Cattaneo, R. (2000) J. Virol. 74, 1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dumortier, A., Kirstetter, P., Kastner, P. & Chan, S. (2003) Blood 101, 2219-2226. [DOI] [PubMed] [Google Scholar]
- 31.Singh, M., Cattaneo, R. & Billeter, M. A. (1999) J. Virol. 73, 4823-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springfeld, C. F., Fielding, A. K., Pen, K. W., Galanis, E., Russell, S. J. & Cattaneo, R. (2005) in Virus Therapy of Human Cancers, eds. Horvath, J. C. S. & Sinkovics, J. G. (Dekker, New York), pp. 459-480.
- 33.Oldstone, M. B., Lewicki, H., Thomas, D., Tishon, A., Dales, S., Patterson, J., Manchester, M., Homann, D., Naniche, D. & Holz, A. (1999) Cell 98, 629-640. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence, D. M., Vaughn, M. M., Belman, A. R., Cole, J. S. & Rall, G. F. (1999) J. Virol. 73, 1795-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatsuo, H., Ono, N., Tanaka, K. & Yanagi, Y. (2000) Nature 406, 893-897. [DOI] [PubMed] [Google Scholar]
- 36.Hahm, B., Arbour, N., Naniche, D., Homann, D., Manchester, M. & Oldstone, M. B. (2003) J. Virol. 77, 3505-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hahm, B., Arbour, N. & Oldstone, M. B. (2004) Virology 323, 292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forthal, D. N., Landucci, G., Phan, T. B. & Becerra, J. (2005) J. Virol. 79, 2042-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta, N., Arthos, J., Khazanie, P., Steenbeke, T. D., Censoplano, N. M., Chung, E. A., Cruz, C. C., Chaikin, M. A., Daucher, M., Kottilil, S., et al. (2005) Virology 332, 491-497. [DOI] [PubMed] [Google Scholar]
- 40.Loh, J., Chu, D. T., O'Guin, A. K., Yokoyama, W. M. & Virgin, H. W., IV (2005) J. Virol. 79, 661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grote, D., Cattaneo, R. & Fielding, A. K. (2003) Cancer Res. 63, 6463-6468. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.