Abstract
The Lactococcus lactis Ll.LtrB group II intron encodes a reverse transcriptase (LtrA protein) that binds the intron RNA to promote RNA splicing and intron mobility. Here, we used LtrA–GFP fusions and immunofluorescence microscopy to show that LtrA localizes to cellular poles in Escherichia coli and Lactococcus lactis. This polar localization occurs with or without coexpression of Ll.LtrB intron RNA, is observed over a wide range of cellular growth rates and expression levels, and is independent of replication origin function. The same localization pattern was found for three nonoverlapping LtrA subsegments, possibly reflecting dependence on common redundant signals and/or protein physical properties. When coexpressed in E. coli, LtrA interferes with the polar localization of the Shigella IcsA protein, which mediates polarized actin tail assembly, suggesting competition for a common localization determinant. The polar localization of LtrA could account for the preferential insertion of the Ll.LtrB intron in the origin and terminus regions of the E. coli chromosome, may facilitate access to exposed DNA in these regions, and could potentially link group II intron mobility to the host DNA replication and/or cell division machinery.
Keywords: catalytic RNA, intron mobility, protein localization, retrotransposon, ribozyme
Mobile group II introns are retroelements that insert at high frequencies into unoccupied target sites in intronless alleles (“retrohoming”) and retrotranspose at low frequencies into ectopic sites that resemble the normal homing site (reviewed in refs. 1 and 2). These mobility processes are mediated by a ribonucleoprotein (RNP) complex that is formed during RNA splicing and contains the intron-encoded protein and the excised intron lariat RNA. For mobility, the excised intron RNA in the RNPs uses its ribozyme activity to reverse splice directly into a DNA target site and is then reverse transcribed by the associated intron-encoded protein. The resulting intron cDNA is integrated into the recipient genome by cellular DNA recombination or repair mechanisms. The primer for reverse transcription can either be generated by cleavage of the opposite DNA strand or can be a nascent strand at a DNA replication fork (reviewed in ref. 2). Although group II intron mobility mechanisms have now been characterized extensively, there is little information about how mobility occurs in the context of cellular structures or how it might be coordinated with cellular processes, such as DNA replication or cell division.
The Lactococcus lactis Ll.LtrB intron, which has been studied as a model system, encodes a protein (LtrA) with four conserved domains: reverse transcriptase (RT), which corresponds to the fingers and palm regions of retroviral RTs; X, which corresponds to the RT thumb; DNA-binding (D); and DNA endonuclease (En). The RT and X domains function together to bind the intron RNA and stabilize its active structure for RNA splicing and reverse splicing (3, 4). Domain D is required for efficient reverse splicing into dsDNA, whereas En cleaves the opposite strand to generate the primer for reverse transcription (5). Although En-dependent retrohoming is favored, when En cleavage is blocked by mutation, Ll.LtrB can still retrohome by using nascent strands at DNA replication forks to prime reverse transcription (6, 7). Analogous En-independent mechanisms are also used for retrotransposition of Ll.LtrB to ectopic sites (8, 9).
Ll.LtrB RNPs initiate mobility by binding DNA nonspecifically and searching for relatively long (30–35 bp) target sites, which are recognized by a combination of intron-encoded protein interactions and base pairing of the intron RNA (10, 11). The region of the DNA target site recognized by intron RNA base pairing extends from positions –12 to +3 (relative to the intron insertion site) and consists of three sequence elements, denoted intron-binding sites 1 and 2 in the 5′ exon and δ′ in the 3′ exon; the complementary intron RNA sequences are denoted exon-binding sites 1 and 2 and δ. The intron-encoded protein recognizes only a few specific nucleotide residues in the distal regions of the target site and facilitates local DNA melting, enabling the intron RNA to base pair to the intron-binding site and δ′ sequences (11). Because most of the DNA target site is recognized by base pairing of the intron RNA, it is possible to retarget the Ll.LtrB intron to insert into desired DNA sites simply by modifying the intron RNA, enabling its development into a gene targeting vector (“targetron”) (reviewed in ref. 12).
In addition to targeted disruption, by incorporating a genetic marker for selection, an Ll.LtrB intron with randomized exon-binding site and δ sequences was used to obtain insertions at sites distributed throughout the Escherichia coli genome, analogous to global transposon mutagenesis (13). Surprisingly, however, the Ll.LtrB insertion sites obtained by using this approach were strongly clustered around the bidirectional replication origin (oriC), with 57% of the sites found within 5% of the genome on either side of oriC. Coros et al. (9) studying retrotransposition of the wild-type Ll.LtrB intron to ectopic sites in E. coli also observed clustering of insertion sites but in both the origin region (Ori region) and terminus region (Ter region), with the clustering most pronounced under conditions in which cells were growing slowly. In E. coli, the newly replicated Ori regions migrate rapidly to opposite cell poles, whereas the Ter region remains at mid-cell (14, 15). Under slow-growth conditions, daughter cells have Ori and Ter regions at opposite poles, whereas, under rapid growth conditions, daughter cells have Ori regions at opposite poles (16). Thus, the clustering of Ll.LtrB insertion sites in the Ori and Ter regions could potentially reflect the intracellular localization of LtrA and/or the interaction of Ll.LtrB RNPs with proteins bound to the origin or terminus of DNA replication. These possibilities were intriguing in light of recently discovered links between group II intron mobility and DNA replication, including the ability of group II introns to use nascent strands at DNA replication forks to prime reverse transcription and that Ll.LtrB mobility in E. coli requires actively replicating DNA and likely uses the host replicative polymerase Pol III for second-strand DNA synthesis (see above and ref. 17).
Here, we directly investigated the intracellular localization of LtrA in E. coli and L. lactis by using LtrA/GFP fusions and immunofluorescence microscopy. We found that LtrA shows a polar localization pattern similar to that of E. coli oriC-linked sequences. Furthermore, when coexpressed in E. coli, LtrA interferes with the pole localization of the Shigella IcsA protein, which mediates polarized actin tail assembly, suggesting competition for a common localization determinant. The polar localization of LtrA could account for the preferential insertion of Ll.LtrB in the Ori and Ter regions of the E. coli chromosome and could potentially link group II intron mobility to the host DNA replication or cell division machinery.
Materials and Methods
Bacterial Strains and Growth Conditions. E. coli strains were HMS174(DE3) (F– recA hsdR rifR) (Novagen), BL21(DE3) (F– ompT hsdSB gal dcm) (Stratagene), DH5α, and AQ10033 (oriC+) and AQ10060 (oriC–) (obtained from Jim Walker, University of Texas, Austin) (18). DE3-derivatives of AQ10033 and AQ10060 containing an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible phage T7 RNA polymerase were constructed with a λDE3 lysogenization kit (Novagen). Standard growth conditions were LB medium at 37°C, except for AQ10033(DE3) and AQ10060(DE3), which were grown in minimal medium (19) supplemented with 0.5% casamino acids; 1% glucose; 5 μg/ml thiamine-HCl; and thymine, tryptophan, and asparagine at 50 μg/ml each. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; spectinomycin, 100 μg/ml; and tetracycline, 25 μg/ml. For plasmid induction, overnight cultures were diluted 1:100 into fresh medium, grown 2–3 h at 37°C to an OD600 of 0.2–0.3, and induced with IPTG and/or arabinose the under conditions indicated for individual experiments.
L. lactis strain NZ9800 and its ΔltrB derivative were grown overnight without aeration in M17 plus 1% glucose medium at 30°C (plus 10 μg/ml tetracycline for NZ9800ΔltrB). For cells transformed with pLE-RIG-GFP/LtrA (RIG, retrotransposition indicator gene), chloramphenicol was added at 10 μg/ml. For induction of plasmid expression, overnight cultures were diluted 1:100 and grown at 30°C until the OD600 was 0.3, then 1 ml of cells was induced with 25 ng/ml nisin for 3 h at 30°C.
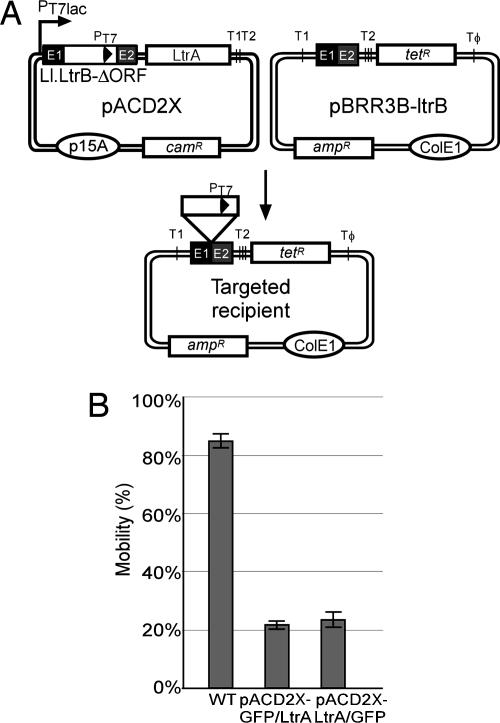
Recombinant Plasmids. The intron-donor plasmid pACD2X contains a 0.9-kb Ll.LtrB-ΔORF intron and short flanking exons cloned downstream of a T7lac promoter in a pACYC184-based vector with chloramphenicol resistance gene camR; the intron contains an additional T7 promoter inserted in DIV, and the LtrA protein is expressed from a position just downstream of the 3′ exon (5, 20). The recipient plasmid pBRR3-ltrB contains a 45-bp Ll.LtrB intron target site (position –30 to +15 from the intron insertion site) cloned upstream of a promoterless tetracycline resistance gene tetR in a pBR322-based vector carrying ampicillin resistance gene ampR (20).
pACD2X–GFP/LtrA and pACD2X–LtrA/GFP are derivatives of pACD2X in which GFPuv (an enhanced GFP variant) (21) is fused in-frame to the N or C terminus of the LtrA ORF, respectively. pACSD2–GFP/LtrA has the N-terminal GFP fusion in intron-donor plasmid pACSD2, which carries a spectinomycin resistance gene spcR instead of camR (17). pAC–GFP/LtrA is a derivative of pACD2X–GFP/LtrA that expresses the LtrA protein without the Ll.LtrB-ΔORF intron, and pAC–GFP is a matched construct that expresses GFPuv. pAC–GFP/LtrA(2–200), pAC–GFP/LtrA(201–400), pAC–GFP/RT(70–361), and pAC–GFP/En(543–599) express N-terminal GFP fusions with the indicated LtrA subsegments.
pLE-LtrA/RIG contains the Ll.LtrB intron and short flanking exons cloned downstream of an inducible nisA promoter in the E. coli/L. lactis shuttle vector pSKH1 (8). The intron carries a kanamycin-resistant (kanR)-RIG marker in intron domain IV for use in intron mobility assays. pLE-RIG–GFP/LtrA is a derivative expressing an N-terminal GFP/LtrA fusion made with GFPrft. GFPrft is a derivative of GFPuv with improved folding and solubility properties (G. Georgiou, unpublished data), which we found performs better in L. lactis than does GFPuv. pLE-RIG–GFP is a matched plasmid that expresses GFPrft from within intron domain IV.
pBAD24-icsA507–620::gfp (22), which expresses IcsA507–620–GFP from the pBAD (arabinose) promoter, was obtained from Marcia Goldberg (Massachusetts General Hospital, Boston). pBAD24-icsA507–620::gfpt expresses IcsA507–620 linked to a truncated, nonfluorescent GFP. pACD2X-IcsA and pACD2X-IcsAΔSP are derivatives of pACD2X expressing full-length IcsA and IcsA with its signal peptide deleted, respectively.
Details of plasmid constructions are given in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. In all constructs, regions subjected to PCR were sequenced to ensure that no adventitious mutations had been introduced.
Intron Mobility Assays. Intron mobility was assayed by using an E. coli two-plasmid system (Fig. 1A) (20, 23). The CamR intron-donor plasmid pACD2X and AmpR recipient plasmid pBRR3-ltrB were cotransformed into E. coli HMS174 (DE3), and cells were grown overnight at 37°C in LB medium containing chloramphenicol and ampicillin. The overnight culture was then diluted 1:100 into fresh LB with the same antibiotics, grown 2–3 h at 37°C to an OD600 of 0.2–0.3, and induced with IPTG for 1–3 h at 37°C. After induction, cells were washed, resuspended in fresh medium, and plated at different dilutions on LB containing tetracycline plus ampicillin or ampicillin alone. Colonies were counted after overnight incubation at 37°C, and mobility frequencies were calculated as the ratio of (TetR + AmpR)/AmpR colonies.
Fig. 1.
LtrA–GFP fusions are active in intron mobility. (A) Intron mobility assay. The CamR intron-donor plasmid pACD2X, which expresses a 0.9-kb Ll.LtrB-ΔORF intron with a phage T7 promoter near its 3′ end plus LtrA protein downstream of the 3′ exon, is cotransformed into E. coli HMS174(DE3) with the AmpR recipient plasmid pBRR3-ltrB. The latter contains the Ll.LtrB target site (positions –30 to +15) cloned upstream of a promoterless tetR gene. After induction with IPTG, insertion of the intron carrying the T7 promoter into the target site activates the tetR gene, and mobility frequencies are measured from the ratio of (TetR + AmpR)/AmpR colonies. PT7lac is the T7lac promoter used for donor intron expression, and T1, T2, and TΦ are transcription terminators for E. coli RNA polymerase (T1 and T2) and phage T7 RNA polymerase (TΦ). (B) Mobility assays with donor plasmids in which GFP is fused in-frame to LtrA's N terminus (GFP/LtrA) or C terminus (LtrA/GFP). The bar graphs show the mean ± SD for three experiments.
Localization of LtrA–GFP Fusions. Cells from 1 ml of culture were washed and resuspended in 0.1 ml of fresh growth medium without IPTG or antibiotics, and ≈1 μl was placed on a glass slide and examined by fluorescence microscopy with a Zeiss Axioplan2 with a ×63/1.40 oil differential interference contrast lens and a GFP filter. Photographs were taken with a Hamamatsu c4742-95 digital camera and processed with photoshop (Adobe Systems, Palo Alto, CA). DNA was stained with 10 μg/ml DAPI (Molecular Probes) for 15 min to 1 h before the end of the induction period, and cell membranes were stained with 2 μg/ml FM4–64 (Molecular Probes) for 15 min before the end of the induction period. L. lactis cells were chilled at 4°C for 5–6 h before microscopy to enhance fluorescence (24).
Immunofluorescence Microscopy. Cells from 10 ml of culture were fixed directly in growth medium by adding paraformaldehyde, glutaraldehyde, and NaPO4 buffer, pH 7.4, to final concentrations of 2.4% (vol/vol), 0.04% (vol/vol), and 30 mM, respectively, and then incubating at room temperature for 10 min and on ice for 50 min. The cells were washed three times by centrifugation in PBS at room temperature, resuspended in 50 mM glucose/10 mM EDTA/20 mM Tris·HCl, pH 7.5, and incubated with freshly prepared 5 μg/ml lysozyme (Sigma) for 30 min at room temperature. After washing twice with PBS, the cells were air-dried, rehydrated with PBS, incubated for 4 min, recentrifuged, incubated in blocking solution [2% (wt/vol) BSA in PBS] for 30 min at room temperature, and centrifuged again. The cells were then incubated overnight at 4°C in blocking solution containing a 1:100 dilution of rabbit polyclonal anti-LtrA antibody preparation [obtained from Gary Dunny (University of Minnesota, Minneapolis) and preadsorbed to fixed untransformed HMS174(DE3)], washed 10 times with blocking solution, and incubated in the dark for 2 h at room temperature in blocking solution with a 1:1,000 dilution of secondary antibody (anti-rabbit IgG-FITC, The Jackson Laboratory). Cells were examined microscopically after 10 washes with PBS.
SDS/PAGE and Immunoblotting. SDS/PAGE and immunoblotting were as described (3). Samples containing protein from 0.1 OD600 units of cells were analyzed in 7.5% polyacrylamide/1% SDS (GFP/LtrA) or 10% polyacrylamide/1% SDS (IcsA/GFP) gels. Immunoblots were probed with a 1:1,000 dilution of anti-LtrA antibody (see above) or anti-GFP antibody (BD Biosciences, Franklin Lakes, NJ), followed in both cases by a 1:100,000 dilution of goat anti-rabbit secondary antibody (Pierce). Blots were developed with SuperSignal West Pico Chemilluminescent substrate (Pierce). Equal loading was confirmed by Coomassie blue staining of a parallel gel.
Results
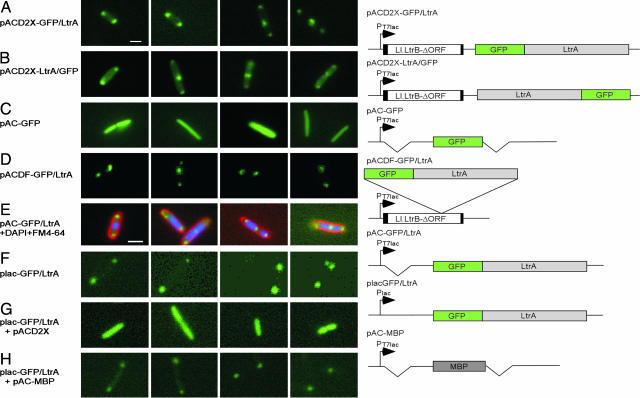
Construction of Active LtrA–GFP Fusions. To study the intracellular localization of the LtrA protein, we constructed N- and C-terminal LtrA–GFP fusions in the intron-donor plasmid pACD2X (Fig. 1A). This plasmid contains a 0.9-kb Ll.LtrB-ΔORF intron with short flanking exons cloned behind a T7lac promoter, with the LtrA ORF expressed from a position just downstream of the 3′ exon. The intron has an additional T7 promoter inserted near its 3′ end for use in intron mobility assays described below. The LtrA protein expressed from the downstream cis position promotes RNA splicing and then remains tightly bound to the excised intron RNA in RNPs that promote intron mobility. In the intact Ll.LtrB intron and pACD2X, the synthesis of LtrA is autoregulated by binding to its own Shine–Dalgarno sequence, thereby limiting the accumulation of excess unbound protein (25). SDS/PAGE and immunoblotting with an anti-LtrA antibody showed that the N- and C-terminal LtrA–GFP fusions (denoted GFP/LtrA and LtrA/GFP, respectively) were expressed at somewhat reduced levels (33–50% wild type), but the proportion of expressed protein recovered in RNPs was essentially the same as for wild-type LtrA (data not shown).
To determine whether the LtrA/GFP fusions are active, we carried out intron mobility assays in which CamR donor plasmids expressing wild-type LtrA or the LtrA–GFP fusions were cotransformed into E. coli HMS174(DE3) with a compatible AmpR recipient plasmid containing the Ll.LtrB target site cloned upstream of a promoterless tetR gene (Fig. 1A). After induction of donor plasmid expression with IPTG, insertion of the intron carrying the T7 promoter into the target sites activates the expression of the tetR gene, and mobility frequencies are measured as the ratio of (TetR + AmpR)/AmpR colonies. The N- and C-terminal LtrA–GFP fusions supported mobility at frequencies that were roughly proportional to their somewhat lower expression levels (Fig. 1B; mobility frequencies 22 ± 1% and 23 ± 3% for pACD2X–GFP/LtrA and pACD2X-LtrA/GFP, respectively, compared with 85 ± 3% for wild-type LtrA).
Intracellular Localization of LtrA–GFP Fusions. We next examined the localization of the LtrA–GFP fusions by fluorescence microscopy in E. coli HMS174(DE3) grown and induced under the same conditions as in the mobility assay (250 μM IPTG for 1 h at 37°C) (Fig. 2 A and B and Table 1). Most of the cells expressing the N- or C-terminal LtrA–GFP fusions showed a localization pattern with two foci at the poles or two foci at the poles plus an additional focus elsewhere (82.3% and 53% for pACD2X–GFP/LtrA and pACD2X–LtrA/GFP, respectively). Cells with two foci were smaller (3.1 ± 0.47 μm) than those with three foci (5.2 ± 0.57 μm), suggesting that the appearance of the third focus is correlated with incipient cell division. Lower proportions of the cells showed only one focus at a pole (1.3% and 2.0% for pACD2X–GFP/LtrA and pACD2X–LtrA/GFP, respectively), or four or more foci, with two at the poles and the remainder distributed throughout the cell (16.4% and 35.0% for pACD2X–GFP/LtrA and pACD2X–LtrA/GFP, respectively). Controls showed that GFP expressed under the same conditions from a parallel construct was uniformly distributed throughout the cell, as expected (Fig. 2C, pAC–GFP).
Fig. 2.
LtrA–GFP fusions are pole-localized in E. coli. Images show fluorescence microscopy. E. coli HMS174(DE3) containing the indicated plasmids was grown and induced with 250 μM IPTG for 1 h at 30°C. In E, DAPI and FM4–64 were added to stain DNA and cell membranes, respectively. Constructs are diagrammed to the right. The Ll.LtrB-ΔORF intron is indicated by an open rectangle with flanking exons shaded black. Deletions relative to the pACD2X–GFP/LtrA parent construct are indicated by breaks. MBP, maltose-binding protein. (Scale bar ≈ 2 μm; in E, magnification is ×1.3 that in other images.)
Table 1. Localization of GFP fusion proteins.
| Conditions, °C/μM IPTG/min
|
Percentage of cells with n foci
|
|||||
|---|---|---|---|---|---|---|
| Strains and constructs | 1 | 2 | 3 | ≥4 | Diffuse | |
| E. coli HMS174(DE3) | ||||||
| pAC—GFP/LtrA | 25/50/180 | 12.1 | 65.5 | 19.6 | 1.4 | 1.4 |
| 25/250/180 | 7.2 | 67.1 | 23.9 | 1.2 | 0.6 | |
| 30/50/150 | 12.8 | 65.3 | 20.7 | 0.4 | 0.8 | |
| 30/250/120 | 9.1 | 69.0 | 21.0 | 0.9 | 0.0 | |
| 37/50/90 | 16.2 | 60.6 | 19.4 | 1.5 | 2.3 | |
| 37/250/60 | 7.6 | 69.1 | 20.0 | 1.8 | 1.5 | |
| plac—GFP/LtrA | 37/0/120 | 3.2 | 56.3 | 31.5 | 4.0 | 5.0 |
| 37/250/120 | 3.0 | 60.2 | 34.8 | 0.8 | 1.2 | |
| pACD2X—GFP/LtrA | 37/50/60 | 17.9 | 45.3 | 26.4 | 9.4 | 1.0 |
| 37/100/60 | 3.6 | 47.2 | 41.1 | 6.7 | 1.4 | |
| 37/250/60 | 1.3 | 29.6 | 52.7 | 16.4 | 0.0 | |
| 37/500/60 | 1.9 | 30.7 | 46.2 | 20.3 | 0.9 | |
| 37/1,000/60 | 2.4 | 33.3 | 45.5 | 17.2 | 1.6 | |
| pACD2X—LtrA/GFP | 37/250/60 | 2.0 | 19.0 | 44.0 | 35.0 | 0.0 |
| pACDF—GFP/LtrA | 37/250/60 | 2.4 | 40.6 | 34.7 | 22.3 | 0.0 |
| pAC—GFP/RT | 37/250/60 | 7.4 | 53.5 | 28.0 | 9.4 | 1.6 |
| pAC—GFP/En | 37/250/60 | 7.6 | 60.1 | 29.6 | 2.7 | 0.0 |
| pAC—GFP/LtrA(2-200) | 37/250/60 | 3.0 | 44.0 | 39.7 | 12.3 | 1.0 |
| pAC—GFP/LtrA(201-400) | 37/250/60 | 0.4 | 21.1 | 21.1 | 55.4 | 2.0 |
| pBAD24-icsA507-620::gfp* | 37/250/60 | 62.4 | 31.8 | 5.8 | 0 | 0 |
| pBAD24-icsA507-620 + pACD2X::gfp | 37/250/60 | 14.6† | 7.1 | 1.1 | 0 | 70.5 |
| E. coli AQ10033(DE3) (oriC+) | ||||||
| pACSD2—GFP/LtrA | 37/250/60 | 6.1 | 61.4 | 26.5 | 3.6 | 2.4 |
| E. coli AQ10060(DE3) (oriC-)‡ | ||||||
| pACSD2—GFP/LtrA | 37/250/60 | 6.0 | 40.8 | 25.3 | 27.9 | 0.0 |
| L. lactis NZ9800 | ||||||
| pLE—GFP/LtrA-RIG | 30/25§/180 | 35/5¶ | 55 | 5 | 0 | 0 |
Indicated are the number of foci of LtrA—GFP fusion proteins detected by fluorescence microscopy. In cells with one or two foci, the foci were always at the poles. In cells with three foci, two were at the poles and the third was elsewhere. In cells with four or more foci, two were at the poles and the remainder were elsewhere. At least 200 cells were counted in each experiment.
Data are for the localization of IcsA507-620/GFP.
Another 6.7% showed one fluorescent focus outside the pole area.
Of the AQ10060(DE3) oriC- cells, 27.9% formed filaments, most of which had more than four fluorescent foci.
Value indicates 25 ng/ml nisin.
Values indicate that 35% of cells showed one focus apposed or near the membrane at a putative pole in elongated or linked cells, and 5% showed one focus elsewhere.
In other experiments, we also observed pole localization of the N-terminal LtrA–GFP fusion expressed from the normal location within the intron (pACDF–GFP/LtrA) (Fig. 2D and Table 1) and for the “protein only” construct, which lacks the Ll.LtrB intron (pAC–GFP/LtrA) (Fig. 2E and Table 1). The GFP/LtrA fusion was also pole localized in E. coli strains BL21(DE3) and DH5α (data not shown). Additionally, we found that pole-localized GFP/LtrA expressed at lower levels from the lac promoter (plac–GFP/LtrA) (Fig. 2F and Table 1) could be competed from the poles by the full-length LtrA protein expressed from pACD2X (Fig. 2G) but not by maltose-binding protein expressed at similar levels and confined intracellularly by deletion of its signal peptide (pAC–maltose-binding protein) (Fig. 2H and SDS/PAGE data not shown).
Staining of DNA with DAPI and cell membranes with FM 4–64 showed that pole-localized GFP/LtrA is located at the edge of, but mostly excluded from the nucleoid region and generally apposed to the inside of the inner membrane (shown in Fig. 2E for pAC–GFP/LtrA).
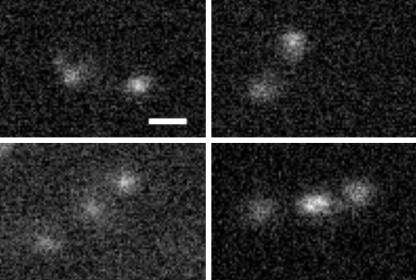
Immunofluorescence Microscopy. To exclude the possibility that the pole localization of the GFP/LtrA fusion protein is an artifact resulting from the disruption of normal localization signals by the GFP fusions, we examined the localization of native LtrA by immunof luorescence microscopy. These experiments used HMS174(DE3) and the RNP expression construct pACD2X (Fig. 1A). After IPTG induction, the cells were fixed and probed with an anti-LtrA antibody preparation, followed by IgG-FITC secondary antibody. As shown in Fig. 3, the native LtrA protein detected in this assay had essentially the same polar localization pattern seen for GFP/LtrA fusions, with two foci at the poles in smaller cells and an extra focus in the middle in larger cells. By contrast, untransformed control cells showed only low background fluorescence (data not shown).
Fig. 3.
Immunofluorescence microscopy. E. coli HMS174(DE3) containing pACD2X was induced with 250 μM IPTG for 2 h at 37°C. After fixation, LtrA protein was detected with anti-LtrA antibody, followed by goat anti-rabbit IgG-FITC secondary antibody. (Scale bar ≈ 2 μm.)
Polar Localization Occurs at Different Levels of LtrA Expression. We next tested whether the localization pattern of GFP/LtrA fusions might be affected by their expression level. For these experiments, we used both the RNP expression construct pACD2X–GFP/LtrA and the protein-only expression construct pAC–GFP/LtrA at different temperatures (25°C, 30°C, and 37°C), IPTG concentrations (50 or 250 μM), and induction times (60 to 180 min). Similar results were obtained for both constructs (Table 1; Fig. 8, which is published as supporting information on the PNAS web site; and data not shown). Under all conditions tested, polar localization was seen in >95% of the cells. Furthermore, the same polar localization was seen when LtrA was expressed at a lower level from the lac promoter (plac–GFP/LtrA) with IPTG induction or by “leaky” expression without IPTG induction. We saw no inclusion bodies by phase contrast microscopy, and inclusion bodies are generally distributed throughout the cell rather than pole-localized as for LtrA (26).
Polar Localization of LtrA Occurs Over a Wide Range of Growth Rates. Coros et al. (9) studying the retrotransposition of Ll.LtrB to ectopic sites in E. coli found that the clustering of insertion sites in the Ori and Ter regions was most pronounced (93% of insertions) in slowly growing cells induced with high IPTG concentrations (1 mM; doubling time, 60 min at 37°C), and somewhat less pronounced (80% of insertions) in more rapidly growing cells induced with lower IPTG concentrations (100 μM; doubling time, 23 min at 37°C). We found that LtrA remained predominantly pole localized at IPTG concentrations ranging from 50 μM to 1 mM (induction time, 60 min at 37°C), where doubling times ranged from 30 to 85 min (Table 1).
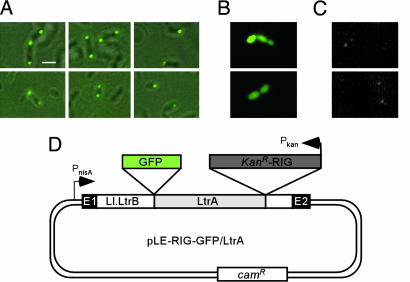
Localization of LtrA in L. lactis. To investigate the intracellular localization of LtrA in L. lactis, we modified an existing construct, denoted pLE-RIG (8), to express an N-terminal LtrA–GFP fusion. The modified construct (pLE-RIG–GFP/LtrA) (Fig. 4D) contains the full-length Ll.LtrB intron and short flanking exons cloned behind an inducible nisA promoter, with the GFP/LtrA fusion expressed from the normal location within intron. After transformation into L. lactis strain NZ9800 and induction with nisin, 90% of the cells showed one (35%) or two foci (55%) apposed to or near the membrane on opposite sides of the cell, with an additional 5% showing two foci on opposite sides plus a third focus in the middle; the remaining 5% of cells showed one focus elsewhere in the cell (Fig. 4A and Table 1). Although many of the cells were roughly spherical, pole localization could be clearly discerned in those cells that were somewhat elongated or growing in chains. Thus, the intracellular localization of the GFP/LtrA fusion in L. lactis appears similar to that in E. coli. By contrast, GFP expressed by itself gave uniform fluorescence throughout the cell (Fig. 4B).
Fig. 4.
LtrA is pole-localized in L. lactis.(A and B) Fluorescence microscopy. L. lactis NZ9800 containing pLE-RIG–GFP/LtrA or pLE-RIG–GFP, respectively, was grown and induced with nisin. (A) GFP fluorescence is superimposed over phase contrast images. (C) Detection of LtrA expressed from the endogenous Ll.LtrB intron in L. lactis NZ9800 by immunofluorescence microscopy. (D) pLE-RIG–GFP/LtrA contains the Ll.LtrB intron and short flanking exons (E1 and E2) cloned downstream of an inducible nisA promoter (PnisA). The ORF encoding the GFP/LtrA fusion is located in intron domain IV just upstream of a kanR–RIG marker (8). (Scale bar ≈ 2 μm.)
We also attempted to localize LtrA synthesized from the endogenous chromosomal copy of Ll.LtrB in L. lactis strain NZ9800 by immunofluorescence microscopy with anti-LtrA antibody. Immunoblots showed that the level of LtrA expressed from the endogenous element was <2% that from nisin-induced pLE-RIG–GFP/LtrA (data not shown). The immunofluorescence microscopy showed correspondingly very light foci with the same localization pattern found for the GFP/LtrA fusion (Fig. 4C). These foci were quickly bleached but not observed when the primary anti-LtrA antibody was omitted, nor in NZ9800ΔltrB, which is deleted for the endogenous Ll.LtrB intron (ref. 8 and data not shown). Thus, despite qualifications required by the low expression level, LtrA synthesized from the endogenous integrated element also appears to be pole-localized.
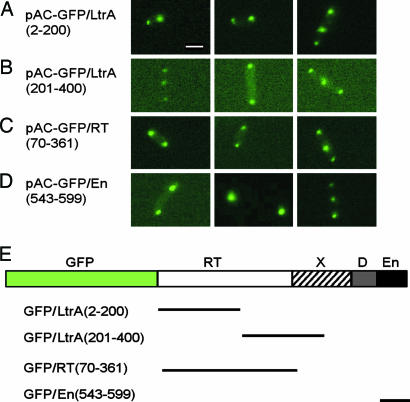
Localization of GFP Fusions with Different LtrA Subsegments. To determine whether a specific region of LtrA is responsible for its polar localization, we tested GFP fusions with different subsegments of the protein. All LtrA subsegments tested, including three nonoverlapping regions (amino acid residues 2–200 and 201–400 and the En region), showed pole localization patterns similar to that of full-length LtrA (Fig. 5 and Table 1). In other experiments, we also found polar localization from the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein) (27), which is unrelated to LtrA but has similar size (637 aa) and basicity (not shown). We note, however, that CYT-18 is closely related to E. coli TyrRS, whose intracellular localization is unknown. Together, these results suggest either that signals responsible for the polar localization are common and redundant or that some physical property of the protein (e.g., positively charged regions) is responsible for the polar localization.
Fig. 5.
Localization of GFP fusions with different subsegments of LtrA. (A–D) Fluorescence microscopy. E. coli HMS174(DE3) containing derivatives of pAC–GFP/LtrA with GFP fused to different subsegments of LtrA was induced with 250 μM IPTG at 37°C for 1 h. Synthesis of the correct-sized protein was confirmed by SDS/PAGE and Coomassie blue staining (A–C) or by SDS/PAGE and immunoblotting with anti-GFP antibody (D). (E) Schematic of the LtrA protein with segments used in the GFP fusions delineated below. (Scale bar ≈ 2 μm.)
Localization of LtrA in an oriC– Strain. The polar localization of LtrA could account for the clustering of Ll.LtrB-insertion sites in the Ori and Ter regions, which are similarly localized for much of the cell cycle (14–16). In principle, the polar localization of LtrA could reflect its interaction with components that are bound to the origin and/or terminus of DNA replication, with exposed DNA sites in these regions, or with pole-localized cellular components, possibly the same ones that dictate the polar localization of oriC (see Discussion). Although the cellular components responsible for oriC localization are not known, the pole localization of oriC-linked sequences remains in oriC– strains, which use alternative Hfr replication origins, and is thought to reflect the recognition of “centromere-like” sequences linked to oriC (28).
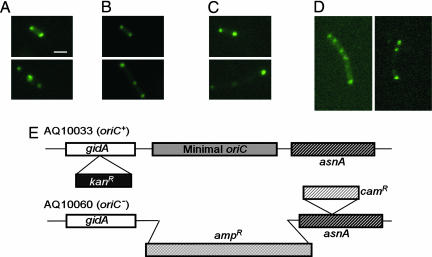
To test whether or not the polar localization of LtrA depends on the functioning of oriC, we examined the localization of the GFP/LtrA fusion in the oriC– strain AQ10060(DE3) and its oriC+ parent AQ10033(DE3) (Fig. 6). Because the minimal oriC in AQ10066 was replaced by an ampR gene, we used an alternative intron expression plasmid pACSD2–GFP/LtrA carrying a spcR marker. In the oriC+ strain AQ10033(DE3), the GFP/LtrA fusion showed the same polar localization pattern as in HMS174(DE3), with the majority of cells having two foci near the poles (Fig. 6 A and B and Table 1). Most (72.1%) of the oriC– AQ10060(DE3) cells were of normal size, and in these cells GFP/LtrA showed polar localization (Fig. 6C and Table 1), which is therefore not dependent on the function of oriC. Additionally, oriC– cells have an increased tendency to form filaments in which oriC-linked sequences are found at multiple foci throughout the cell (28). In our experiments, 27.9% of the oriC– cells formed filaments, and these likewise had multiple GFP/LtrA fluorescence foci distributed throughout the cell (two examples are shown in Fig. 6D). Thus, the disrupted processes that lead to filament formation and mislocalization of oriC-linked sequences in some oriC– cells also lead to mislocalization of LtrA.
Fig. 6.
Polar localization of LtrA is not dependent on oriC function. (A–D) Fluorescence microscopy. E. coli HMS174(DE3) containing pACD2X–GFP/LtrA (oriC+) (A) or AQ10033(DE3) oriC+ (B) and AQ10060(DE3) oriC– (C and D) containing pACSD2–GFP/LtrA were induced with 250 μM IPTG for 1 h at 30°C. (C and D) Examples of AQ10060(DE3) oriC– cells with normal and filamentous morphology. (E) Diagram of the oriC region in E. coli AQ10033 and AQ10060 in which the minimal oriC is replaced by an ampR gene. (Scale bar ≈ 2 μm.)
LtrA Interferes with Polar Localization of the Shigella IcsA Protein. The Shigella outer-membrane protein IcsA (VirG) localizes to the old pole of the bacterium to mediate polarized assembly of an actin tail that pushes the bacterium through the cytoplasm of infected mammalian cells and into adjacent cells (22, 29). In E. coli, GFP fusions of two nonoverlapping IcsA fragments (IcsA1–104 and IcsA507–620) both showed polar localization patterns (22, 30). Among proteins reported in the literature to be pole-localized in E. coli, the localization patterns of IcsA1–104 and IcsA507–620 appeared to us particularly similar to that of LtrA.
To test whether IcsA507–620 and LtrA compete for pole localization determinants, the two proteins were coexpressed in E. coli HMS174(DE3). The IcsA507–620–GFP fusion protein expressed by itself showed the expected polar localization pattern but with the proportion of cells containing only a single focus at one pole higher than that for LtrA (Fig. 7A and Table 1). Significantly, the coexpression of LtrA interfered with the pole localization of the IcsA507–620–GFP fusion, resulting in a high proportion of cells (70.5%) showing diffused fluorescence (Fig. 7 A and B and Table 1). The remaining cells showed pole localization (22.8%) or one fluorescent focus elsewhere (6.7%). We note that about a third of 22.8% of cells tabulated as showing pole localization in this experiment also had high dispersed background fluorescence, suggesting incomplete interference. Immunoblots with anti-GFP antibody showed that the coexpression of LtrA reduced IcsA507–620–GFP expression by only about one-third (data not shown). In reciprocal experiments, neither full-length IcsA with its signal peptide deleted (IcsAΔSP) (Fig. 7 C and D) nor IcsA507–620 fused to truncated GFP (pBAD24-icsA507–620::gfpt) (data not shown) displaced LtrA from the poles. High-level expression of IcsA507–620 fused to truncated GFP was confirmed by immunoblotting (data not shown). Together, these findings suggest that LtrA has a higher affinity for a required positional determinant than does IcsA.
Fig. 7.
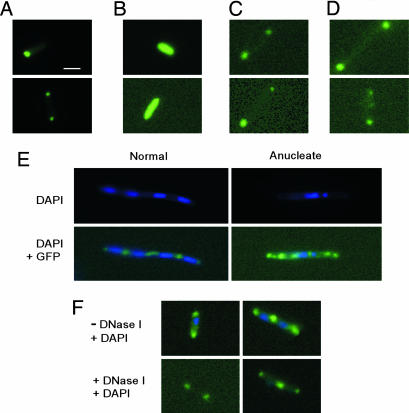
LtrA and IcsA may use related localization mechanisms. (A–D) Competition experiments with E. coli HMS174(DE3) expressing IcsA507–620–GFP (pBAD24-icsA507–620::gfp) (A), IcsA507–620–GFP (pBAD24-icsA507–620::gfp) plus LtrA (pACD2X) (B), GFP/LtrA (placGFP/LtrA) (C), and GFP/LtrA (placGFP/LtrA) plus IcsA with the signal peptide deleted (pACD2X-IcsAΔSP) (D). Cells were induced with 0.2% l-arabinose (pBAD promoter) and/or 250 μM IPTG (T7lac promoter) for 4 h at 37°C. (E) Effect of aztreonam. E. coli HMS174(DE3) containing pAC–GFP/LtrA was grown in LB medium containing chloramphenicol and induced with 250 μM IPTG in the presence of 1 μg/ml aztreonam for 2 h at 37°C. DAPI was added to stain DNA. (F) Effect of DNase I on GFP/LtrA localization. HMS174(DE3) containing pACD2X–GFP/LtrA was induced with 250 μM IPTG at 37°C for 2 h and stained with DAPI. A portion of the cells was incubated with 100 μg/ml lysozyme (Sigma) and 100 units/ml DNase I (Invitrogen) for 1 h at room temperature before fluorescence microscopy. (Scale bar ≈ 2 μm.)
Finally, Janakiraman and Goldberg (22) obtained further insight into the mechanism of IcsA localization by treating E. coli with aztreonam to inhibit the cell division protein FtsI. In the resulting filamentous cells, IcsA507–620 foci were no longer confined to the poles but also appeared at regularly spaced intervals between nucleoids and in anucleate segments, with the spacing between foci suggesting localization to potential cell division sites. We found that GFP–LtrA foci behaved similarly in filamentous aztreonam-treated HMS174(DE3), with a spacing between foci of 3.4 ± 1.0 μm compared with 3.0 ± 0.8 μm for pole-localized foci in untreated cells (Fig. 7E and data not shown). Notably, as for IcsA, this spacing was maintained in anucleate segments that appeared in ≈5% of the filamentous cells (Fig. 7E Right). Additionally, the polar localization of GFP/LtrA remained in untreated HMS174(DE3) after intracellular DNA was largely digested by DNase I in the presence of lysozyme (Fig. 7F). Together, these findings indicate that the localization of GFP/LtrA is not the result of nucleoid occlusion and are consistent with localization to potential cell division sites.
Discussion
We find that a group II intron-encoded RT, the LtrA protein encoded by the Ll.LtrB intron, is localized to cellular poles in E. coli and L. lactis. The pole localization in E. coli is seen over a wide range of cellular growth rates and LtrA expression levels and occurs with or without coexpression of the Ll.LtrB intron RNA, which assembles with LtrA into RNPs that mediate intron mobility. Furthermore, we show that LtrA interferes with the pole localization of the Shigella IcsA protein, a mediator of polarized actin filament assembly, and that LtrA and IcsA507–620 localize similarly in filamentous E. coli cells induced by treatment with aztreonam. These findings suggest that LtrA and IcsA may use related localization mechanisms, possibly those that localize proteins to potential cell division sites, as suggested for IcsA (22). The polar localization of LtrA likely contributes to the intron's propensity to integrate in the Ori and Ter regions of the E. coli chromosome (see the Introduction).
The ability of LtrA to compete with IcsA for pole localization suggests that LtrA localization is dictated by a physiologically relevant mechanism involving interaction with a limited number of localized cellular binding sites that can be occupied by LtrA or IcsA. Our results and those for IcsA are compatible with these binding sites being protein or other types of receptor molecules located in the cytoplasm or on the cytoplasmic face of the inner membrane (31). It may be pertinent that in mitochondrial systems there have been persistent speculative indications that group II intron splicing factors are associated with the inner membrane (32).
Charles et al. (30) found that two nonoverlapping segments of IcsA pole-localize independently and suggested that these contain redundant localization signals. Here, we find pole localization for three nonoverlapping segments of LtrA and for the unrelated N. crassa mitochondrial tyrosyl tRNA synthetase (CYT-18 protein). These findings could reflect that pole localization of LtrA and IcsA is dictated by common, redundant signals or some physical property of the proteins, such as regions of high basicity. The amino acid sequences responsible for pole localization of IcsA are not known. Computer analysis (meme/mast) reveals a number of short sequence motifs of varying stringencies that are common to IcsA507–620 and the three localized LtrA subsegments, but their significance, if any, remains to be evaluated.
We find that the pole localization of LtrA is independent of oriC function, suggesting that it is not dictated by interaction with DNA structural features or protein components associated with active origins (e.g., hemimethylated DNA or SeqA) (33). It is possible that the pole localization of LtrA and perhaps other proteins found at the cellular poles is dictated by interaction with the same cellular machinery responsible for the pole localization of the Ori region. This possibility is consistent with the appearance of a third LtrA focus at midcell in larger cells, which may be about to undergo cell division, as well as the altered localization of both LtrA and oriC-linked sequences in filamentous cells derived from oriC– mutants (Fig. 6D) (28). It is also possible that LtrA localization is dictated by interaction with membrane components that pole-localize independently of the Ori region.
Although the polar localization of LtrA can account for the preferential insertion of Ll.LtrB in the Ori and Ter regions of the E. coli chromosome, it is clearly not the only factor that contributes to dictating integration sites. Thus, Coros et al. (9) found that the very strong clustering of Ll.LtrB retrotransposition sites in the Ori and Ter regions was modulated somewhat in more rapidly growing cells, whereas we find LtrA remains largely pole-localized in slowly and rapidly growing cells. Furthermore, Coros et al. (9) characterized retrotransposition events as occurring by DNA–endonuclease-dependent or -independent pathways, depending on whether the target site could support second-strand cleavage by LtrA's En domain. The predicted En-dependent events showed a bias for the Ori and Ter regions, whereas the predicted En-independent events, which may require nascent strands at DNA replication forks to prime reverse transcription, favored the Ori region, with a gradient toward the Ter region. Additionally, Ll.LtrB retrotransposition sites were found to be uniformly distributed throughout the chromosome in L. lactis (8), where we find LtrA is localized to discrete foci at opposite ends of the cell. These findings suggest that chromosome packaging and access to DNA replication forks also play a role in dictating group II intron insertion sites and that the relative contribution of these factors may differ significantly between E. coli and L. lactis (see also ref. 9).
Ll.LtrB RNPs bind DNA nonspecifically and search for target sites by facilitated diffusion along DNA similar to mechanisms used by site-specific DNA-binding proteins (10). The pole localization of LtrA in E. coli may concentrate group II intron RNPs in proximity to exposed DNA segments in the Ori and Ter regions, thereby facilitating their initial nonspecific DNA binding, after which the RNPs can search for target sites along the DNA. This scenario readily accounts for the preferential insertion of Ll.LtrB insertions sites in the Ori and Ter regions, whereas the distal regions may be more or less accessible to the RNPs depending on growth conditions. The polar regions may also be favorable sites for interaction with components of the DNA replication machinery, such as Pol III, which is pole-localized in nonreplicating cells (34). Such interactions may facilitate access of group II intron RNPs to DNA replication forks and/or minimize the interval between the initial steps of intron mobility and downstream steps, such as second-strand synthesis, which may depend on host DNA replication (17). Finally, the pole localization of LtrA could potentially link group II intron mobility to the cell division machinery and/or facilitate the segregation of group II intron RNPs to daughter cells. The latter could be particularly beneficial for many group II introns found in plasmids, which may themselves have unreliable segregation mechanisms (35). It remains to be seen whether other transposable elements use analogous intracellular localization strategies.
Supplementary Material
Acknowledgments
We thank Marlene Belfort, Marcia Goldberg, Georg Mohr, Roland Saldanha, and Jim Walker for comments on the manuscript; Gary Dunny for anti-LtrA antibody; and Marlene Belfort (Wadsworth Center, Albany, NY), George Georgiou (University of Texas), Marcia Goldberg, Shelly Payne (University of Texas), and Jim Walker for strains and plasmids. This work was supported by National Institutes of Health Grant GM37949.
Author contributions: J.Z. and A.M.L. designed research; J.Z. performed research; J.Z. and A.M.L. analyzed data; and J.Z. and A.M.L. wrote the paper.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 20, 2004.
Abbreviations: D domain, DNA-binding domain; En domain, DNA–endonuclease domain; IPTG, isopropyl β-d-1-thiogalactopyranoside; RNP, ribonucleoprotein; RT, reverse transcriptase; Ter region, terminus region; Ori region, origin region; RIG, retrotransposition indicator gene; CamR, chloramphenicol resistance; TetR, tetracycline resistance; AmpR, ampicillin resistance.
References
- 1.Belfort, M., Derbyshire, V., Parker, M. M., Cousineau, B. & Lambowitz, A. M. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol., Washington, DC), pp. 761–783.
- 2.Lambowitz, A. M. & Zimmerly, S. (2004) Annu. Rev. Genet. 38, 1–35. [DOI] [PubMed] [Google Scholar]
- 3.Cui, X., Matsuura, M., Wang, Q., Ma, H. & Lambowitz, A. M. (2004) J. Mol. Biol. 340, 211–231. [DOI] [PubMed] [Google Scholar]
- 4.Blocker, F. J., Mohr, G., Conlan, L. H., Qi, L., Belfort, M. & Lambowitz, A. M. (2005) RNA 11, 14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.San Filippo, J. & Lambowitz, A. M. (2002) J. Mol. Biol. 324, 933–951. [DOI] [PubMed] [Google Scholar]
- 6.Zhong, J. & Lambowitz, A. M. (2003) EMBO J. 22, 4555–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao, J., Zhong, J. & Lambowitz, A. M. (2005) Nucl. Acids. Res. 33, 3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichiyanagi, K., Beauregard, A., Lawrence, S., Smith, D., Cousineau, B. & Belfort, M. (2002) Mol. Microbiol. 46, 1259–1272. [DOI] [PubMed] [Google Scholar]
- 9.Coros, C. J., Landthaler, M., Piazza, C. L., Beauregard, A., Esposito, D., Perutka, J., Lambowitz, A. M. & Belfort, M. (2005) Mol. Microbiol. 56, 509–524. [DOI] [PubMed] [Google Scholar]
- 10.Aizawa, Y., Xiang, Q., Lambowitz, A. M. & Pyle, A. M. (2003) Mol. Cell 11, 795–805. [DOI] [PubMed] [Google Scholar]
- 11.Singh, N. N. & Lambowitz, A. M. (2001) J. Mol. Biol. 309, 361–386. [DOI] [PubMed] [Google Scholar]
- 12.Lambowitz, A. M., Mohr, G. & Zimmerly, S. (2005) in Homing Endonucleases and Inteins, Nucleic Acids and Molecular Biology, eds. Belfort, M., Derbyshire, V., Stoddard, B.L. & Wood, D.W. (Springer, Heidelberg, Germany), Vol. 16, pp. 121–145. [Google Scholar]
- 13.Zhong, J., Karberg, M. & Lambowitz, A. M. (2003) Nucl. Acids Res. 31, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niki, H. & Hiraga, S. (1998) Genes Dev. 12, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niki, H., Yamaichi, Y. & Hiraga, S. (2000) Genes Dev. 14, 212–223. [PMC free article] [PubMed] [Google Scholar]
- 16.Draper, G. C. & Gober, J. W. (2002) Annu. Rev. Microbiol. 56, 567–597. [DOI] [PubMed] [Google Scholar]
- 17.Smith, D., Zhong, J., Matsuura, M., Lambowitz, A. M. & Belfort, M. (2005) Genes Dev., in press. [DOI] [PMC free article] [PubMed]
- 18.Bates, D. B., Asai, T., Cao, Y., Chambers, M. W., Cadwell, G. W., Boye, E. & Kogoma, T. (1995) Nucl. Acids Res. 23, 3119–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard-Flanders, P., Simson, E. & Theriot, L. (1964) Genetics 49, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karberg, M., Guo, H., Zhong, J., Coon, R., Perutka, J. & Lambowitz, A. M. (2001) Nat. Biotechnol. 19, 1162–1167. [DOI] [PubMed] [Google Scholar]
- 21.Crameri, A., Whitehorn, E. A., Tate, E. & Stemmer, W. P. (1996) Nat. Biotechnol. 14, 315–319. [DOI] [PubMed] [Google Scholar]
- 22.Janakiraman, A. & Goldberg, M. B. (2004) Proc. Natl. Acad. Sci. USA 101, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo, H., Karberg, M., Long, M., Jones, J. P., III, Sullenger, B. & Lambowitz, A. M. (2000) Science 289, 452–457. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez de Palencia, P., Nieto, C., Acebo, P., Espinosa, M. & Lopez, P. (2000) FEMS Microbiol. Lett. 183, 229–234. [DOI] [PubMed] [Google Scholar]
- 25.Singh, R. N., Saldanha, R. J., D'Souza, L. M. & Lambowitz, A. M. (2002) J. Mol. Biol. 318, 287–303. [DOI] [PubMed] [Google Scholar]
- 26.Carrió, M. M. & Villaverde, A. (2005) J. Bacteriol. 187, 3599–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kittle, J. D., Jr., Mohr, G., Gianelos, J. A., Wang, H. & Lambowitz, A. M. (1991) Genes Dev. 5, 1009–1021. [DOI] [PubMed] [Google Scholar]
- 28.Gordon, G. S., Shivers, R. P. & Wright, A. (2002) Mol. Microbiol. 44, 501–507. [DOI] [PubMed] [Google Scholar]
- 29.Goldberg, M. B., Barzu, O., Parsot, C. & Sansonetti, P. J. (1993) J. Bacteriol. 175, 2189–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charles, M., Perez, M., Kobil, J. H. & Goldberg, M. B. (2001) Proc. Natl. Acad. Sci. USA 98, 9871–9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janakiraman, A. & Goldberg, M. B. (2004) Trends Microbiol. 12, 518–525. [DOI] [PubMed] [Google Scholar]
- 32.Luban, C., Beutel, M., Stahl, U. & Schmidt, U. (2005) Gene 385, 72–79. [DOI] [PubMed] [Google Scholar]
- 33.Slater, S., Wold, S., Lu, M., Boye, E., Skarstad, K. & Kleckner, N. (1995) Cell 82, 927–936. [DOI] [PubMed] [Google Scholar]
- 34.Onogi, T., Ohsumi, K., Katayama, T. & Hiraga, S. (2002) J. Bacteriol. 184, 867–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichiyanagi, K., Beauregard, A. & Belfort, M. (2003) Proc. Natl. Acad. Sci. USA 100, 15742–15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.