Abstract
The early detection of cancers through analysis of circulating DNA could have a substantial impact on morbidity and mortality. To achieve this goal, it is essential to determine the number of mutant molecules present in the circulation of cancer patients and to develop methods that are sufficiently sensitive to detect these mutations. Using a modified version of a recently developed assay for this purpose, we found that patients with advanced colorectal cancers consistently contained mutant adenomatous polyposis coli (APC) DNA molecules in their plasma. The median number of APC DNA fragments in such patients was 47,800 per ml of plasma, of which 8% were mutant. Mutant APC molecules were also detected in >60% of patients with early, presumably curable colorectal cancers, at levels ranging from 0.01% to 1.7% of the total APC molecules. These results have implications for the mechanisms through which tumor DNA is released into the circulation and for diagnostic tests based on this phenomenon.
Keywords: colorectal cancer, plasma DNA, tumor suppressor gene, circulating DNA, diagnosis
The probability of curing cancers through surgery alone is high in individuals whose primary tumors are detected at a relatively early stage. Such early detection is therefore one of the most promising approaches for limiting cancer morbidity and mortality in the future (1). At present, Pap smears can be used to detect cervical cancers, mammography can detect breast cancers, serum PSA (prostate-specific antigen) levels can signify the presence of prostate cancer, and colonoscopy and fecal occult blood tests can detect colon cancers (2). However, problems with sensitivity, specificity, cost, or compliance have complicated widespread implementation of many of these tests (3–5). Moreover, methods for the early detection of most other cancer types are not yet available.
The discovery of the genetic bases of neoplasia has led to new approaches to detect tumors noninvasively (6–8). Several of these approaches rely on the ex vivo detection of mutant forms of the oncogenes and tumor suppressor genes that are responsible for the initiation and progression of tumors. This approach was first used to detect bladder and colon tumors through examination of urine and stool, respectively (9, 10), and has since been used to detect several other tumor types (11–14). Because the mutant genes are not only “markers” for cancer but also the proximate causes of tumor growth (1), they have major conceptual advantages over conventional markers such as fecal occult blood or serum PSA. In particular, conventional markers are not pathogenically involved in the tumorigenic process and are much less specific for neoplasia than are mutations.
The evaluation of patient blood samples for mutant DNA molecules is a particularly attractive approach because such tests could detect many different forms of cancers. Additionally, blood can be easily obtained from patients during routine outpatient visits, and methods for preparing and storing plasma and serum are well known and reliable. Accordingly, numerous studies have attempted to identify abnormal forms or quantities of DNA in plasma or serum (6, 11–15). Unfortunately, the results of many of these studies are contradictory. Some report high detection rates of cancers, and others report very low detection rates, despite the use of similar techniques and patient cohorts. Moreover, several studies have shown that loss of heterozygosity is routinely detectable in circulating DNA, even in patients with relatively nonaggressive tumors. To detect loss of heterozygosity in such samples, the neoplastic cells within a tumor must contribute >50% of the total circulating DNA.
The above studies, although promising, lead to several questions that must be answered to engender confidence in the use of circulating, abnormal DNA as a biomarker of malignancy. First, how many copies of a given gene fragment are present in the circulation in cancer patients? Second, what is the nature of this DNA (e.g., intact vs. degraded)? Third, what fraction of these gene fragments have an abnormal (e.g., mutant) DNA sequence? And, fourth, how does this fraction vary with stage of disease? To answer these questions, it was necessary to develop technologies that could simultaneously quantify the number of normal and mutant DNA molecules in a given sample, even when the fraction of mutant molecules was very small. In the current study, we employ such a technology to investigate circulating DNA in patients with colorectal tumors.
Materials and Methods
Sample Collection, DNA Extraction, and Sequencing. Detailed methods for these procedures are provided in the supporting information, which is published on the PNAS web site.
Real-Time PCR. Primers were designed to generate ≈100-bp amplicons that included one or more mutation sites. A universal tag (5′-TCCCGCGAAATTAATACGAC-3′) was added to the 5′ end of either the forward or reverse primer used to generate each amplicon. The sequences of these primers are listed in the supporting information. PCR was performed in 50-μl reactions containing 10 μl of 5× Phusion HF buffer, a 0.2 mM concentration of each dNTP, a 1 μM concentration of each primer, 1:50,000 dilution of SYBR green I (Invitrogen), 1.5 units of Phusion DNA polymerase (NEB, Beverly, MA), and 15 μl of purified plasma DNA (equivalent to 100 μl of plasma) or genomic DNA purified from normal mononuclear cells of the blood of healthy volunteers. The amplifications were carried out with an iCycler (Bio-Rad) under the following conditions: 98°C for 1 min; 98°C for 10 s, 70°C for 10 s, and 72°C for 10 s 3 times; 98°C for 10 s, 67°C for 10 s, and 72°C for 10 s 3 times; 98°C for 10 s, 64°C for 10 s, and72°C for10 s 3 times; and 98°C for 10 s, 61°C for 10 s, and 72°C for 10 s 30 times. Each reaction was performed in duplicate, and a calibration curve was generated in each 96-well plate by using various amounts of normal human genomic DNA. The concentration of PCR products was determined by using a PicoGreen dsDNA quantification assay (Invitrogen).
BEAMing. A common oligonucleotide (5′-TCCCGCGAAATTAATACGAC-3′) was synthesized with a dual biotin group at the 5′ end and with a six-carbon linker (C6) between the biotin and the other nucleotides (Integrated DNA Technologies, Coralville, IA). This oligonucleotide was coupled to streptavidin-coated magnetic beads (MyOne, Dynal, Oslo) according to the protocol described in ref. 16. The water-in-oil emulsions were prepared by modifications of the methods described by Ghadessy and Holliger (17) and Bernath et al. (18). For each emulsion PCR, a 240-μl aliquot of an aqueous PCR mix was added to 960 μl of 7% (wt/vol) Abil EM90 (Degussa Goldschmidt Chemical, Hopewell, VA) in mineral oil (Sigma). The aqueous phase contained 67 mM Tris·HCl (pH 8.8), 16.6 mM (NH4)2SO4, 6.7 mM MgCl2, 10 mM 2-mercaptoethanol, a 0.2 mM concentration of each dNTP, 0.05 μM forward primer (5′-TCCCGCGAAATTAATACGAC-3′), 8 μM reverse primer, 0.2 units/μl Platinum Taq polymerase (Invitrogen), 3 × 105 per μl oligonucleotide-coupled beads, and 0.1 pg/μl template DNA. The reverse primers are listed in the supporting information. The water–oil mix was vortexed for 10 s and then emulsified for 50 s by using an Ultra-Turrax homogenizer (T25 basic, IKA, Wilmington, NC) with a disposable OmniTip (Omni International, Waterbury, CT) at the minimum speed. The emulsions were aliquoted into 8 wells of a 96-well PCR plate and cycled under the following conditions: 94°C for 2 min; 94°C for 10 s, 58°C for 15 s, and 70°C for 15 s 50 times. After PCR, the emulsions were pooled into a 15-ml tube and demulsified through the addition of 10 ml of NX buffer (100 mM NaCl/1% Triton X-100/10 mM Tris·HCl, pH 7.5/1 mM EDTA/1% SDS). After vortexing for 10 s, the beads were pelleted by centrifugation for 5 min at 4,100 × g. The top phase was removed, and the beads were resuspended in 800 μl of NX buffer and transferred to a 1.5-ml tube. The beads were collected by using a magnet (MPC-S, Dynal) and washed with 800 μl of wash buffer (20 mM Tris·HCl,pH8.4/50 mM KCl). The double-stranded DNA on the beads was converted to single-stranded DNA by incubation in 800 μl of 0.1 M NaOH for 2 min at room temperature. The beads were washed twice with 800 μl of wash buffer, using the magnet, and finally resuspended in 200 μl of wash buffer. Single base extension and flow cytometry were performed as described in the supporting information.
Results
Circulating Mutant DNA Is Degraded. We used real-time PCR or digital PCR to determine the number of total circulating APC (adenomatous polyposis coli) genes in 33 patients with colorectal tumors and 10 age-matched donors without any tumors. The number of APC gene copies was significantly higher in advanced stage patients (Dukes' D) than in patients with early stage cancers (P < 0.0001, Student's t test), consistent with previous studies (19, 20). In advanced stage patients, the median number of APC gene fragments per ml of plasma was 47,800, whereas the median number was 3,500 and 4,000 for patients with Dukes' A and Dukes' B cancers, respectively (Table 1). There was no significant difference between the number of circulating copies in early stage cancer patients (Duke's A or B), patients with adenomas (4,300 APC fragments per ml of plasma), and normal individuals (3,460 APC fragments per ml of plasma; range of 1,150–8,280 fragments per ml).
Table 1. Quantification of APC mutations in plasma.
| Patient no. | Sex/age, yr | Site | Dukes' stage (tumor node metastasis stage) | Diameter of lesion, cm | Mutation identified in primary tumor (codon) | Fragments per ml of plasma | No. of fragments analyzed | Percentage of mutant fragments, % |
|---|---|---|---|---|---|---|---|---|
| 1 | M/50 | Ascending colon | Adenoma | 3.0 | C4348T (1450) | 2,600 | 2,350 | 0.002 |
| 2 | M/67 | Descending colon | Adenoma | 2.5 | C4285T (1429) | 5,080 | 5,080 | 0.001 |
| 3 | M/54 | Rectum | Adenoma | 4.0 | G3856T (1286) | 4,150 | 4,150 | 0.002 |
| 4 | F/82 | Rectum | Adenoma | 3.0 | 4147–4148insA (1383) | 1,350 | 1,350 | 0.001 |
| 5 | F/65 | Rectum | Adenoma | 1.0 | C4067G (1356) | 4,260 | 4,260 | 0.001 |
| 6 | F/71 | Ascending colon | Adenoma | 4.0 | G3856T (1286) | 4,150 | 4,150 | 0.001 |
| 7 | M/68 | Cecum | Adenoma | 6.5 | C4285T (1429) | 4,760 | 4,760 | 0.003 |
| 8 | M/93 | Ascending colon | Adenoma | 0.8 | A4345T (1449) | 4,320 | 4,320 | 0.001 |
| 9 | F/78 | Ascending colon | Adenoma | 3.0 | C4216T (1406) | 28,570 | 28,570 | 0.001 |
| 10 | F/59 | Sigmoid colon | Adenoma | 5.0 | 4661–4662insA (1554) | 2,160 | 2,160 | 0.002 |
| 11 | F/73 | Ascending colon | Adenoma | 5.0 | C4348T (1450) | 8,000 | 8,000 | 0.02 |
| Median/mean | 4,300/6,300 | 0.02* | ||||||
| Mutant plasma samples per samples analyzed | 1/11 (9) | |||||||
| 12 | F/81 | Sigmoid colon | A (T2N0M0) | 4.0 | G4189T (1397) | 7,900 | 12,000 | 0.01 |
| 13 | F/75 | Sigmoid colon | A (T2N0M0) | 2.5 | 3927–3931del AAAGA (1309) | 2,160 | 2,160 | 0.001 |
| 14 | M/60 | Sigmoid colon | A (T2N0M0) | 3.0 | 3927–3931del AAAGA (1309) | 4,600 | 6,900 | 0.04 |
| 15 | M/79 | Right colic flexure | A (T2N0M0) | 3.0 | 4470delT (1490) | 4,600 | 3,696 | 0.03 |
| 16 | M/70 | Ileocecal | A (T2N0M0) | 2.5 | 4481delA (1494) | 6,200 | 3,105 | 0.07 |
| 17 | F/68 | Ascending colon | A (T2N0M0) | 3.5 | C4348T (1450) | 2,170 | 2,170 | 0.001 |
| 18 | F/66 | Sigmoid colon | A (T1N0M0) | 2.5 | 3927–3931del AAAGA (1309) | 1,920 | 1,920 | 0.001 |
| 19 | M/68 | Rectum | A (T2N0M0) | 5.5 | C3907T (1303) | 2,300 | 1,170 | 0.12 |
| Median/mean | 3,500/4,000 | 0.04/0.04* | ||||||
| Mutant plasma samples per samples analyzed | 5/8 (63) | |||||||
| 20 | F/65 | Cecum | B (T3N0M0) | 3.5 | G4396T (1466) | 5,300 | 5,300 | 0.002 |
| 21 | M/71 | Sigmoid colon | B (T3N0M0) | 3.0 | C4348T (1450) | 2,100 | 1,863 | 0.19 |
| 22 | M/37 | Descending colon | B (T4N0M0) | 10.0 | C4330T (1444) | 5,400 | 4,887 | 1.28 |
| 23 | M/64 | Sigmoid colon | B (T3N0M0) | 6.5 | C4099T (1367) | 3,810 | 3,810 | 0.001 |
| 24 | M/72 | Sigmoid colon | B (T3N0M0) | 3.0 | C4012T (1338) | 4,800 | 4,800 | 0.03 |
| 25 | F/82 | Hepatic flexure | B (T3N0M0) | 4.0 | C4099T (1367) | 3,840 | 3,840 | 1.46 |
| 26 | M/83 | Ascending colon | B (T3N0M0) | 6.0 | 4470delT (1490) | 1,600 | 1,404 | 1.75 |
| 27 | M/61 | Sigmoid colon | B (T3N0M0) | 4.0 | 4260–4261delCA (1420) | 4,200 | 4,200 | 0.001 |
| Median/mean | 4,000/3,900 | 1.28/0.94* | ||||||
| Mutant plasma samples per samples analyzed | 5/8 (63) | |||||||
| 28 | F/83 | Ascending colon | D (T3N2M1) | 5.0 | 4661–4662insA (1554) | 230,000 | 24,857 | 5.6 |
| 29 | M/55 | Sigmoid colon | D (T3N0M1) | 3.0 | G3925T (1309) | 69,600 | 1,636 | 27.4 |
| 30 | F/33 | Descending colon | D (T4N1M1) | 5.0 | C4067A (1356) | 18,000 | 491 | 10.5 |
| 31 | M/64 | Sigmoid colon | D (T4N2M1) | 6.0 | T4161A (1387) | 26,000 | 975 | 1.9 |
| 32 | M/56 | Rectum | D (T3N2M1) | 3.0 | 4468–4469delCA (1490) | 103,200 | 1,187 | 18.9 |
| 33 | F/60 | Rectum | D (T3N2M1) | 4.0 | 4059–4060insT (1354) | 8,400 | 850 | 2.0 |
| Median/mean | 47,800/75,900 | 8.05/11.05* | ||||||
| Mutant plasma samples per samples analyzed | 6/6 (100) | |||||||
Calculated only for samples in which the percentage of mutant fragments was significantly higher than in control samples (i.e., >0.003%; printed in boldface). M, male; F, female
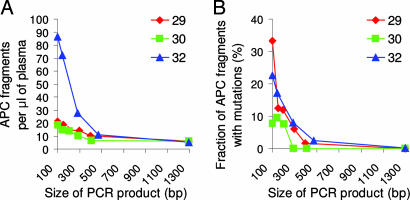
To determine the size of mutant gene fragments in circulating DNA, we analyzed plasma DNA from three patients with advanced colorectal cancers (Dukes' D, metastatic to liver) who were shown to contain APC gene mutations in their tumors. By varying the size of the amplicons, it was possible to determine the number of normal and mutant gene fragments by sequencing the PCR products derived from one or a few template molecules (detailed in the supporting information). The size of the amplicons varied from 100 to 1,296 bp and encompassed the mutation present in each patient. The number of total APC fragments (WT plus mutant) increased by 5- to 20-fold as the size of the amplicons decreased from 1,296 to 100 bp (Fig. 1A). The fraction of mutant molecules was strikingly dependent on size of the amplicon, increasing by >100-fold over the size range tested (Fig. 1B).
Fig. 1.
Effect of the PCR amplicon size on plasma DNA concentration and mutation frequency. (A) The concentration of total APC fragments (WT plus mutant) of various sizes was determined by using digital PCR of plasma DNA from three different patients (patients 29, 30, and 32). (B) The fraction of mutant APC fragments was determined by digital sequencing of PCR products.
We conclude that the mutant DNA fragments present in the circulation of cancer patients are degraded compared with the circulating DNA derived from nonneoplastic cells. This conclusion is consistent with previous studies of other tumor types (21, 22) and has important implications for the detection of such mutant molecules.
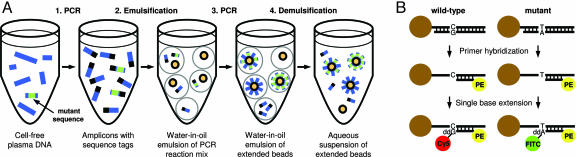
Development of a Quantitative Assay for Detection of Rare Mutations. The results described above were obtained by sequencing hundreds of PCR products, each derived from one or a few DNA template molecules. In preliminary studies, we found that such digital PCR-based techniques were sufficiently sensitive to detect circulating mutant DNA molecules in patients with advanced cancers but not in patients with early stage cancers. To increase the sensitivity and reliability of these assays, we developed an extension of BEAMing (which derives its name from its principal components: beads, emulsion, amplification, and magnetics) that allowed us to examine many more template molecules in a convenient fashion. The approach consists of four steps. (i) Real-time PCR was used to determine the number of total APC gene fragments in the plasma sample (Fig. 2A, step 1). (ii) BEAMing was used to convert the amplified plasma DNA into a population of beads (Fig. 2A, steps 2–4). (iii) The mutational status of the extended beads was determined by single base extension (Fig. 2B). (iv) Flow cytometry was used to simultaneously measure the FITC, Cy5, and phycoerythrin (PE) signals of individual beads.
Fig. 2.
Schematic of the BEAMing-based assay. (A) Extended beads were prepared by modifications of the BEAMing procedure described by Dressman et al. (16). (B) Single base extensions were performed on the extended beads. Normal DNA sequences contained a G at the queried position; mutant sequences contained an A.
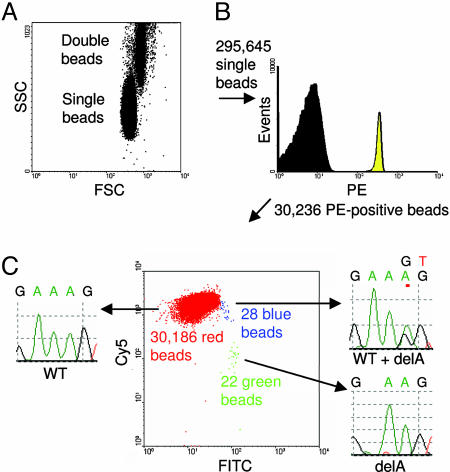
Fig. 3 shows a representative flow cytometry result wherein the interpretation of the profiles was confirmed experimentally. In the example shown, 342,573 beads were analyzed by flow cytometry. The single bead population (295,645) was used for the fluorescence analysis (Fig. 3A). Of these, 30,236 exhibited a PE signal (Fig. 3B), indicating that they had been extended during the emulsion PCR. The FITC and Cy5 signals reflected the number of beads containing mutant or WT sequences, respectively. Beads containing the WT DNA sequences (30,186) had high Cy5 but background FITC signal (“red beads” in Fig. 3C). Beads extended only with mutant DNA sequences (22) had high FITC signals but background Cy5 signals (“green beads”). Twenty-eight had both FITC and Cy5 signals (“blue beads”). Such dual-labeled beads resulted from either the presence of both a WT and mutant template in the droplet containing the bead or an error in the early cycles of the emulsion PCR (see below). These dual-labeled beads were eliminated from analysis, and only homogeneously labeled beads were considered for the enumeration of mutations. Note that this conservative analysis strategy results in a slight underestimation of the fraction of mutations, because it excludes mutants that were present in droplets that also contained one or more WT fragments. Beads in each of these three populations were collected by flow sorting, and single beads from the sort were used as templates in conventional DNA sequencing. All 131 beads subjected to sequencing analysis showed the expected patterns, with examples illustrated in Fig. 3C.
Fig. 3.
Processing of flow cytometry data obtained by BEAMing. (A) Dot plot of forward-scatter (FSC) and side-scatter (SSC) signals of beads. (B) Histogram of single beads with regard to PE signal. (C) Dot plot showing the Cy5 and FITC fluorescence intensity profiles of PE-positive beads. The beads clustered in three distinct populations colored red, green, and blue. Sequencing of individual beads sorted from each population showed that the red and green beads contained homogeneous WT and mutant sequences, respectively; the blue beads contained a mixture of WT and mutant sequences.
Limits to the Sensitivity of Assays for Plasma DNA Mutations. The results described above show that the BEAMing approach can, in principle, detect a very small fraction of fragments containing mutant sequences within a much larger pool of fragments containing WT sequence. Because >50 million beads are used in a single emulsion PCR and flow cytometry can be performed at speeds of >50,000 beads per s, the capacity to enumerate such mutations is not limited by the beads themselves. Instead, two other features limit the sensitivity. First, there is a finite number of DNA fragments present in clinical samples. As noted above, this number ranged from 1,350 to 230,000 fragments per ml in the patients with tumors (Table 1) and from 1,150 to 8,280 fragments per ml in control patients, which gives an upper bound to the sensitivity of the assays. For example, a calculation using the Poisson distribution shows that if 4,000 fragments were analyzed, the mutation fraction in circulating DNA would have to be >1 in 1,333 fragments (i.e., 3 divided by the number of total fragments analyzed) for the assay to achieve 95% sensitivity. A second limiting feature is the error rates of the polymerases used for PCR. In our approach, two PCR steps are used: The first is a conventional PCR that employs plasma DNA fragments as templates, and the second is an oil-in-water emulsion PCR that uses the initial PCR products as templates. In the emulsion PCR, errors occurring during the early rounds of PCR can result in heterogeneous beads containing both WT and mutant sequences. These beads are easily eliminated from consideration, as described in Fig. 3C. However, the errors introduced in the first PCR cannot be eliminated, because they give rise to beads with homogeneous mutant sequences, indistinguishable from those resulting from genuine mutations in the original plasma DNA templates.
The fraction of mutant molecules present after the first PCR equals the product of the mutation rate of the polymerase and the number of cycles carried out. BEAMing provides a quantitative way to determine the error rate of any polymerase used in PCR without requiring cloning in bacterial vectors (M.L., F.D., S.N.G., K.W.K., and B.V., unpublished data). Of 19 different base changes evaluated in normal DNA, the error rates with the polymerase used in the current study averaged 3.0 × 10-7 mutations per bp per PCR cycle and ranged from 1.7 × 10-7 to 6.5 × 10-7 mutations per bp per PCR cycle, depending on the mutation site assessed. As a result, we only scored plasma samples as positive for mutations if their frequency in the sample was significantly higher than the maximum error rate of polymerase found experimentally (i.e., 1.95 × 10-5 after 30 cycles). As a result of the relatively low error rate with the polymerase used, it was the number of molecules present in the original plasma sample, rather than the polymerase error rate per se, that limited sensitivity.
These issues suggest that the sensitivity of assays for circulating mutant DNA could be increased in the future by (i) the development of new or modified polymerases with reduced error rates and (ii) the use of more plasma per assay (i.e., more template molecules).
Quantification of Mutant APC Fragments in Plasma from Patients with Colorectal Tumors. Based on the principles derived from the experiments described above, we determined whether fragments of tumor DNA could be detected in patients with colorectal tumors of various types. We selected APC gene mutations for this assessment, because >85% of colorectal tumors contain mutations of this gene, irrespective of tumor stage (23). Mutations within codon 1209–1581 of APC, containing most previously identified mutations, were evaluated by sequencing of DNA purified from the tumors of 56 patients. Mutations were observed in 33 of these patients (59%), and, as expected, the proportion of tumors with these mutations did not differ significantly among tumors of various stages (see the supporting information).
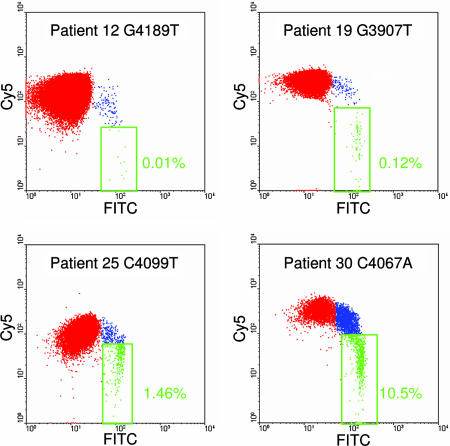
A BEAMing assay was then designed for each of the mutations identified in the 33 tumors and applied to the DNA purified from the plasma of the corresponding patients (Table 1). In each case, DNA from normal lymphocytes or plasma from patients without cancer was used as a negative control. DNA from the tumors of the 33 patients was used as a positive control. All six patients with advanced lesions (Dukes' D, defined as having at least one distant metastatic lesion) were found to contain mutant DNA fragments in their plasma. Among 16 patients harboring cancers with a favorable prognosis (Dukes' A or B, defined as having no lymph node involvement and no distant metastases), 10 (63%) were found to contain mutant DNA fragments in their plasma. In contrast, among 11 patients with large, benign tumors (adenomas), only 1 patient's plasma was found to contain mutant DNA fragments. Representative flow cytometric results are shown in Fig. 4 and summarized in Table 1.
Fig. 4.
Examples of flow cytometric profiles of beads generated from plasma DNA (patient 16). Cy5 and FITC fluorescence intensity profiles of PE-positive beads from four patients are shown. The patients, mutations, and fraction of mutant APC fragments are indicated.
The fraction of mutant molecules found in the plasma of the 17 cases with detectable mutations also varied according to tumor stage (P < 0.0001, Fisher exact test). In the advanced cases (Dukes' D), an average of 11.1% (range of 1.9–27%) of the total APC gene fragments were mutant. In patients without metastases (Dukes' B), an average of 0.9% (range of 0.03–1.75%) of the plasma APC gene fragments were mutant. In patients with lower stage tumors (Dukes' A), the fraction was even lower, averaging 0.04% (range of 0.01–0.12%). And in the one patient with a benign tumor, only 0.02% of the plasma DNA fragments were mutant. The median fraction of positive beads found in the control DNA samples from patients without cancer was 0.0009% (range of 0.003–0.0005%). The mutations in the control samples likely resulted from PCR errors, as noted above.
Table 1 also lists the concentration of total APC fragments (WT plus mutant) in these patients' plasma. There was no direct relationship between the concentration of total APC fragments and the mutational load. Although patients with advanced cancers tended to have higher concentrations of total APC fragments than the other patients, this increase was not due to DNA from neoplastic cells. Furthermore, no correlation was found between tumor burden (volume of primary tumor plus metastatic sites) and either the concentration of APC fragments or percentage of mutant APC fragments in the circulation.
Discussion
The data described above conclusively demonstrate that APC gene fragments from the neoplastic cells of colorectal tumors can be found in the circulation and that the number of such fragments depends on tumor stage. These results have implications for both colorectal tumor biology and for practical diagnostic tests, as discussed below.
Source of Plasma DNA. Previous studies have shown that the total DNA concentration in the plasma of cancer patients is often elevated (19, 20). Our results support this conclusion only in advanced stage patients, in that more total APC gene fragments (WT plus mutant) were present in the plasma of patients with Dukes' D cancers than in those with earlier stage tumors. Our results additionally show that this “extra” DNA in advanced stage patients is not derived from the neoplastic cells themselves, because only a minor fraction of the circulating APC fragments are mutant, whereas all of the neoplastic cell's APC fragments are mutant.
But there are still a large number of mutant DNA fragments circulating in cancer patients. Assuming that the volume of distribution of DNA at steady state is similar to that of oligonucleotides in primates (60–70 ml/kg), an 8% fraction of mutant molecules among 47,800 fragments per ml of plasma (as in Dukes' D patients) would correspond to 1.6 × 107 mutant fragments present in a 70-kg person at any given time (24). The half-life of this tumor DNA is estimated at 16 min, based on the data obtained from clearance of fetal DNA in maternal plasma (25), which translates to ≈6 × 108 mutant fragments released from the tumor each day. For patients with a tumor load 100 g in size (≈3 × 1010 neoplastic cells), we thereby estimate that 3.3% of the tumor DNA is fed into the circulation on a daily basis. For a Dukes' B cancer of 30 g in which 1.3% of the 4,000 circulating APC fragments per ml of plasma are mutant, the corresponding estimate is that 0.15% of the tumor DNA is fed into the circulation each day.
So how do mutant APC gene fragments get into the plasma? Several clues are provided by our data. The ability to get into the circulation was clearly not related to tumor size, because the benign tumors we studied were as large as the cancers (Table 1), yet the former rarely gave rise to detectable mutant DNA fragments. Similarly, there was no significant correlation between the tumor load (including metastatic deposits) and the amount of mutant DNA in the circulation. In contrast, the degree of invasion was indeed correlated with the number of circulating DNA fragments. Those lesions that weren't invasive (benign tumors) did not commonly feed mutant DNA molecules into the plasma. As tumors invaded through more layers of the intestinal wall in Dukes' B vs. Dukes' A tumors, and through the intestine to distant sites in Dukes' D vs. Dukes' B tumors, the number of circulating mutant DNA molecules progressively increased (Fig. 5).
Fig. 5.
Fraction of mutant APC gene fragments in the plasma of patients with various colorectal tumors [adenomas (Ad) and Dukes' stage A, B, and D carcinomas]. In each mutation analyzed, DNA from normal lymphoid cells or plasma DNA from healthy donors was used as a control (Normal). The “mutants” observed in assays with normal cellular DNA represent errors generated during the PCR process rather than mutations present in the template DNA (see text). The red lines represent the mean, minimum, and maximum values of the normal controls.
Another clue is provided by the size of the mutant DNA molecules. The data in Fig. 1 show that mutant sequences are enriched in small DNA fragments and could not be identified at all in fragments of 1,296 bp.
Based on these observations, we propose that the mutant DNA fragments found in the circulation are derived from necrotic neoplastic cells that had been engulfed by macrophages. As tumors enlarge and invade, they are more likely to outgrow their blood supply. Thus, invasive tumors generally contain large regions of necrosis, whereas benign tumors rarely do (26–29). Necrotic cells are not thought to release DNA into the extracellular milieu (30). However, cells that die from necrosis or apoptosis are routinely phagocytosed by macrophages or other scavenger cells. Interestingly, it has been shown that macrophages that engulf necrotic cells release digested DNA into the medium, whereas macrophages that engulf apoptotic cells do not (30). Moreover, the size of the DNA released from macrophages is small (30). All of these observations are consistent with a model wherein hypoxia induces necrosis of tumors, leading to the phagocytosis of tumor cells and the subsequent release of the digested DNA into the circulation. As tumors become more aggressive, the degree of this necrosis increases and the absolute amount of circulating mutant DNA correspondingly rises. Because necrosis involves the killing of neoplastic cells and surrounding stromal and inflammatory cells within the tumor, the DNA released from necrotic regions is likely to contain WT DNA sequences as well as mutant sequences. This phenomenon may explain the increase in total (nonmutant) circulating DNA observed in the plasma of patients with advanced cancers.
Clinical Implications. The ability to detect and quantify mutant DNA molecules in the circulation has obvious clinical importance, and this line of research has been pursued by several investigators. Our results inform the field in several ways. First, it is unlikely that circulating mutant DNA could be used to detect premalignant tumors, based on the fact that we were unable to detect such DNA even in very large adenomas. Second, it is unlikely that loss of heterozygosity detection or other techniques that require a majority of the circulating DNA to be derived from neoplastic cells will allow such detection, at least in colorectal cancers, because the proportion of mutant DNA fragments in plasma was small, averaging only 11% of the total DNA fragments even in large, metastatic cancers.
On the positive side, our data show that even relatively early cancers give rise to circulating mutant DNA fragments that can be detected with sufficiently sensitive and specific assays. In fact, >60% of cancers that had not yet metastasized gave rise to detectable mutant fragments in plasma. Even Dukes' A tumors, which are by definition barely invasive, were detectable with BEAMing-based assays. Virtually all Dukes' A tumors and most Dukes' B tumors can be cured with conventional surgery alone, without the need for adjuvant therapies (31).
In practical terms, plasma-based assays for mutant DNA fragments are inferior in several ways to more conventional techniques for early colorectal cancer detection. Colonoscopy is the gold standard, with sensitivity rates of >80% for adenomas and >90% for cancers (32). In particular, adenomas detected by colonoscopy can often be removed through the colonoscope, alleviating the need for surgery. Unfortunately, a variety of issues limits the widespread applicability of colonoscopy (either conventional or virtual) to the screening of asymptomatic patients (3, 5, 33), a fact that has stimulated the development of noninvasive technologies. One of the most promising of these noninvasive technologies is the analysis of fecal DNA for mutations (34). Because of the frequent presence of mutant DNA molecules in feces from both adenomas and early cancers, fecal DNA analysis is superior to plasma with regard to sensitivity. However, plasma-based assays have potential advantages with regard to ease of implementation and compliance.
For many tumor types, there are currently no alternative methods for presymptomatic diagnosis, unlike the case with colorectal cancers. In these other tumor types, the evaluation of circulating DNA could be particularly useful. Even if such assays could detect only a fraction of patients with treatable cancers, much morbidity and mortality could be averted.
Supplementary Material
Acknowledgments
We thank Natalie Silliman and Janine Ptak for help with DNA sequencing and Dr. Zornig (head of surgery at Israelitic Hospital, Hamburg) and Dr. Doerner (head of surgery at Clinic Alten Eichen, Hamburg) for valuable collaboration. This work was supported by the National Colorectal Cancer Research Alliance, The Clayton Fund, and National Institutes of Health Grants CA43460, CA57345, and CA62924.
Author contributions: F.D., K.A.D., H.J., K.W.K., and B.V. designed research; F.D., M.L., and S.S. performed research; D.D., Y.H., D.S., K.A.D., and H.J. contributed new reagents/analytic tools; F.D., L.A.D., S.N.G., K.W.K., and B.V. analyzed data; and F.D., S.N.G., H.J., K.W.K., and B.V. wrote the paper.
Conflict of interest statement: Under a licensing agreement between EXACT Sciences and The Johns Hopkins University, K.W.K. and B.V. are entitled to a share of royalties received by the university on sales of products related to digital PCR. Under a licensing agreement between Agencourt Biosciences Corporation and The Johns Hopkins University, D.D., K.W.K., and B.V. are entitled to a share of royalties received by the university on sales of products related to the use of BEAMing for preparing templates for DNA sequencing. The terms of these arrangements are being managed by The Johns Hopkins University in accordance with its conflict of interest policies.
Abbreviations: APC, adenomatous polyposis coli; PE, phycoerythrin.
References
- 1.Vogelstein, B. & Kinzler, K. W. (2004) Nat. Med. 10, 789-799. [DOI] [PubMed] [Google Scholar]
- 2.Smith, R. A., Cokkinides, V. & Eyre, H. J. (2005) CA Cancer J. Clin. 55, 31-44, and quiz, 55-56. [DOI] [PubMed] [Google Scholar]
- 3.Breen, N. & Meissner, H. I. (2005) Annu. Rev. Public Health 26, 561-582. [DOI] [PubMed] [Google Scholar]
- 4.Ransohoff, D. F. (2005) Nat. Rev. Cancer 5, 142-149. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan, R. M. (2005) Recent Results Cancer Res. 166, 315-334. [DOI] [PubMed] [Google Scholar]
- 6.Sidransky, D. (2002) Nat. Rev. Cancer 2, 210-219. [DOI] [PubMed] [Google Scholar]
- 7.Verma, M. & Srivastava, S. (2003) Recent Results Cancer Res. 163, 72-84, and discussion, 264-266. [DOI] [PubMed] [Google Scholar]
- 8.Jaffer, F. A. & Weissleder, R. (2005) J. Am. Med. Assoc. 293, 855-862. [DOI] [PubMed] [Google Scholar]
- 9.Sidransky, D., Von Eschenbach, A., Tsai, Y. C., Jones, P., Summerhayes, I., Marshall, F., Paul, M., Green, P., Hamilton, S. R., Frost, P., et al. (1991) Science 252, 706-709. [DOI] [PubMed] [Google Scholar]
- 10.Sidransky, D., Tokino, T., Hamilton, S. R., Kinzler, K. W., Levin, B., Frost, P. & Vogelstein, B. (1992) Science 256, 102-105. [DOI] [PubMed] [Google Scholar]
- 11.Burchill, S. A. & Selby, P. J. (2000) J. Pathol. 190, 6-14. [DOI] [PubMed] [Google Scholar]
- 12.Goessl, C. (2003) Expert Rev. Mol. Diagn. 3, 431-442. [DOI] [PubMed] [Google Scholar]
- 13.Lotze, M. T., Wang, E., Marincola, F. M., Hanna, N., Bugelski, P. J., Burns, C. A., Coukos, G., Damle, N., Godfrey, T. E., Howell, W. M., et al. (2005) J. Immunother. 28, 79-119. [DOI] [PubMed] [Google Scholar]
- 14.Bremnes, R. M., Sirera, R. & Camps, C. (2005) Lung Cancer 49, 1-12. [DOI] [PubMed] [Google Scholar]
- 15.Muller, H. M. & Widschwendter, M. (2003) Expert Rev. Mol. Diagn. 3, 443-458. [DOI] [PubMed] [Google Scholar]
- 16.Dressman, D., Yan, H., Traverso, G., Kinzler, K. W. & Vogelstein, B. (2003) Proc. Natl. Acad. Sci. USA 100, 8817-8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghadessy, F. J. & Holliger, P. (2004) Protein Eng. Des. Sel. 17, 201-204. [DOI] [PubMed] [Google Scholar]
- 18.Bernath, K., Hai, M., Mastrobattista, E., Griffiths, A. D., Magdassi, S. & Tawfik, D. S. (2004) Anal. Biochem. 325, 151-157. [DOI] [PubMed] [Google Scholar]
- 19.Leon, S. A., Shapiro, B., Sklaroff, D. M. & Yaros, M. J. (1977) Cancer Res. 37, 646-650. [PubMed] [Google Scholar]
- 20.Sozzi, G., Conte, D., Mariani, L., Lo Vullo, S., Roz, L., Lombardo, C., Pierotti, M. A. & Tavecchio, L. (2001) Cancer Res. 61, 4675-4678. [PubMed] [Google Scholar]
- 21.Giacona, M. B., Ruben, G. C., Iczkowski, K. A., Roos, T. B., Porter, D. M. & Sorenson, G. D. (1998) Pancreas 17, 89-97. [DOI] [PubMed] [Google Scholar]
- 22.Jahr, S., Hentze, H., Englisch, S., Hardt, D., Fackelmayer, F. O., Hesch, R. D. & Knippers, R. (2001) Cancer Res. 61, 1659-1665. [PubMed] [Google Scholar]
- 23.Kinzler, K. W. & Vogelstein, B. (1996) Cell 87, 159-170. [DOI] [PubMed] [Google Scholar]
- 24.Yu, R. Z., Geary, R. S., Monteith, D. K., Matson, J., Truong, L., Fitchett, J. & Levin, A. A. (2004) J. Pharm. Sci. 93, 48-59. [DOI] [PubMed] [Google Scholar]
- 25.Lo, Y. M., Zhang, J., Leung, T. N., Lau, T. K., Chang, A. M. & Hjelm, N. M. (1999) Am. J. Hum. Genet. 64, 218-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomlinson, R. H. & Gray, L. H. (1955) Br. J. Cancer 9, 539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerar, A., Zidar, N. & Vodopivec, B. (2004) Pathol. Res. Pract. 200, 657-662. [DOI] [PubMed] [Google Scholar]
- 28.Chen, S., Yu, L., Jiang, C., Zhao, Y., Sun, D., Li, S., Liao, G., Chen, Y., Fu, Q., Tao, Q., et al. (2005) J. Clin. Oncol. 23, 1538-1547. [DOI] [PubMed] [Google Scholar]
- 29.Leek, R. D., Landers, R. J., Harris, A. L. & Lewis, C. E. (1999) Br. J. Cancer 79, 991-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi, J. J., Reich, C. F., III, & Pisetsky, D. S. (2005) Immunology 115, 55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyerhardt, J. A. & Mayer, R. J. (2005) N. Engl. J. Med. 352, 476-487. [DOI] [PubMed] [Google Scholar]
- 32.Winawer, S., Faivre, J., Selby, J., Bertaro, L., Chen, T. H., Kroborg, O., Levin, B., Mandel, J., O'Morain, C., Richards, M., et al. (2005) Ann. Oncol. 16, 31-33. [DOI] [PubMed] [Google Scholar]
- 33.Lieberman, D. A. & Atkin, W. (2004) Aliment. Pharmacol. Ther. 19, Suppl. 1, 71-76. [DOI] [PubMed] [Google Scholar]
- 34.Ahlquist, D. A. & Shuber, A. P. (2002) Clin. Chim. Acta 315, 157-168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.