Abstract
Bacterial nucleoid organization is believed to have minimal influence on the global transcription program. Using an altered bacterial histone-like protein, HUα, we show that reorganization of the nucleoid configuration can dynamically modulate the cellular transcription pattern. The mutant protein transformed the loosely packed nucleoid into a densely condensed structure. The nucleoid compaction, coupled with increased global DNA supercoiling, generated radical changes in the morphology, physiology, and metabolism of wild-type K-12 Escherichia coli. Many constitutive housekeeping genes involved in nutrient utilization were repressed, whereas many quiescent genes associated with virulence were activated in the mutant. We propose that, as in eukaryotes, the nucleoid architecture dictates the global transcription profile and, consequently, the behavior pattern in bacteria.
Keywords: bacterial HU, nucleoid condensation, virulence
The highly organized eukaryotic chromosome requires elaborate remodeling for coordination of transcription processes. The bacterial chromosome, however, constitutes a comparatively open and expanded structure, accessible throughout the cell cycle to DNA-binding proteins, polymerases, and ribosomes (1, 2). The apparent lack of a systemic hierarchy of nucleoid organization in bacteria was attributed to a low stability of histone-like protein–DNA complexes and the dynamic nature of the bacterial chromosome (3). In the absence of any perceptible higher-order chromosome organization imposing a general level of restriction on the accessibility of bacterial promoters, gene expression is believed to be regulated by operon-specific factors, adjacent DNA control elements and local DNA architecture; global nuclear organization is thought to contribute minimally to overall control of cellular processes involving DNA as substrate (4). Considering the exquisite precision with which bacteria can modulate their gene-expression profile to various environmental challenges, the ostensible lack of influence of chromosome organization over global gene regulation is surprising.
Based on its small size, basic nature, cellular abundance, and sequence-independent DNA-binding capacity, the nucleoid-associated protein HU has long been characterized as the bacterial counterpart of eukaryotic histones (5). HU was initially attributed with the capability to form nucleosome-like structures in bacterial chromosomes (6), but subsequent studies have resulted in conflicting reports about the exact role of HU in chromosome compaction (7, 8). In almost all bacteria except enterobacteriaciae, including Eschericiha coli, HU exists as an 18-kDa homodimer. In E. coli, HU is a heterodimer of two subunits, HUα and HUβ.
Using a gain-of-function HUα mutant, we demonstrate that nucleoid structural reorganization in bacteria can directly induce a radical change in the gene-expression profile, resulting in dramatic changes in cellular morphology and physiology.
Materials and Methods
Bacterial Strains, Media, and Growth Conditions. Mutagenesis of HUα and integration of the mutant hupA gene into the chromosome have been described in ref. 9. A spectinomycin-resistance cassette was used as a marker in both the plasmid and chromosomal constructs of the hupA mutant. Although the spectinomycin-resistance marker, on its own, did not show any of the phenotypes displayed by HUαE38K,V42L, there was a sharp reduction in the mutant HUα expression when some other selective markers were used as substitutes, the basis of which is not known. For creating pHU-GFP6, hupA was amplified with the primer set CGAAGCTTATGAACAAGACTAACTGATTG (HindIII-HU forward) and CCACCGGTTTAACTGCGTCTTTCAGTGC (AgeI-HU reverse). The amplified DNA product, which lacked the last four nucleotides of the hupA gene, was cleaved and cloned between the HindIII and AgeI sites of plasmid pGFPuv (Clontech), creating a HUα–GFPuv translational fusion. For plasmid pPROU37, the proU promoter was amplified as a 612-bp fragment (-240 to +372) by using PCR primers with 5′ EcoRI and 3′ PstI restriction sites and cloned between the EcoRI and PstI sites of plasmid pSA850, which contains the ρ-independent transcription terminator immediately downstream of the PstI site. LB containing 0.4% glucose was used for growth experiments. The metabolic profile of the wild-type and mutant strains was assayed by using GN2 plates (BioLog Chemical, San Diego).
Nucleoid Isolation and Reassembly with HU. Intact nucleoids were extracted from strain DM0100 (ΔhupAhupB), following the procedure of Zimmerman and Murphy (10). Isolated nucleoids were incubated with wild-type HUα and HUα mutant at a (HUα)2:nucleoid DNA molar ratio of 4 nM:0.3 fM in a buffer containing 10 mM Tris (pH 8.0) and 50 mM KCl for 10 min on ice. DAPI was added to a final concentration of 5 μg/ml, and the resultant nucleoids were visualized by fluorescence microscopy.
Fluorescence Microscopy. Mutant E. coli cells with pHU–GFPuv were grown at 37°C until OD600 nm 0.5 and induced with 1 mM isopropyl β-d-thiogalactoside (IPTG). Samples were taken out at various times and stained with N-(3-triethylammoniumpropyl)-4(6-(4(diethylamino)phenyl)hexatrienyl)pyridinium dibromide (FM 4–64) for the visualization of the lipid membrane. Microscopy was performed on an Eclipse E1000 microscope (Nikon) with a Sensicam QE charge-coupled-device camera (Cooke, Romulus, MI) controlled by IP Labs software (Scanalytics, Rockville, MD) with filter sets for DAPI or differential interference contrast.
Other experimental methods are described in Supporting Text, which is published as supporting information on the PNAS web site.
Results
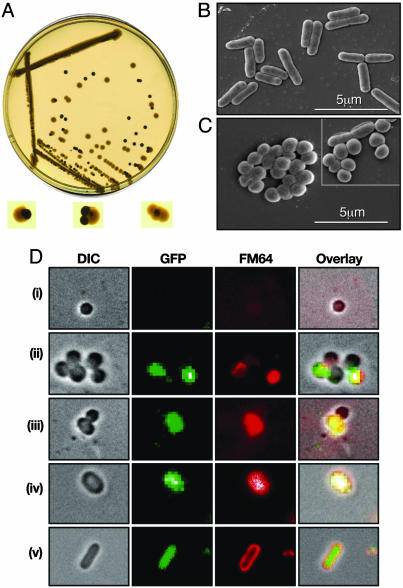
Changes in Colony Architecture and Cell Morphology by an HUα Mutant. While performing mutagenesis of the hupA gene in E. coli K-12 strain MG1655 to identify HUα mutants specifically defective in DNA looping involved in the transcription repression of the gal operon (9), we encountered a chromosomal mutant which superrepressed the gal transcription in vivo. The mutant HUα had two amino acid substitutions, E38K and V42L. The HUαE38K,V42L mutant displayed changes in its phenotype, as described below. We cloned the hupA gene encoding HUαE38K,V42L with its own promoter, along with a spectinomycin-resistance cassette engineered as an adjacent selective marker into a plasmid and introduced it into a ΔrecA E. coli strain. Based on colony morphology, two kinds of transformants were obtained. Some were flat and translucent with irregular edges, resembling the parental wild-type characteristic. Others appeared to be opaque, glossy, and round, with smooth edges. When the transformants belonging to the second class were grown on nonselective LB plates, the smooth, dense colonies segregated into the rough, translucent form (Fig. 1A). Restreaking on selective and nonselective plates and plasmid extraction from the two kinds of colony forms showed that the round, smooth colonies emerged as a result of the plasmid-borne mutant hupA gene and, upon the loss of the plasmid, the cells reverted back to their normal colony morphology.
Fig. 1.
Effect of HUαE38K,V42L substitution on colony morphology and cell architecture. (A) Conversion of colony morphology of wild-type E. coli cells carrying the mutant hupA plasmid, upon losing the mutant clone, on nonselective plates. (B and C) Scanning electron micrographs of wild-type E. coli (B) and hupA mutant (C) cells. (Inset) Revertant cells from regions of papillae around the hupA mutant colonies. (D) Conversion of cellular morphology of hupA mutant upon induction of plasmid-borne wild-type HUα–GFP. Cell shape, GFP fluorescence, membrane stain, and overlay of all three fields at 0 (i), 45 (ii), 90 (iii), 135 (iv), and 180 min (v) after the addition of IPTG. DIC, differential interference contrast microscopy.
The mutant hupA gene was transferred to the chromosome of the wild-type E. coli strain MG1655 at the hupA locus (strain SK3842). The chromosomal hupA mutant also had dense, smooth, round colonies. The mutant hupA phenotype was epistatic to the wild-type hupB allele. Transmission electron microscopy of SK3842 showed that the mutant cells had lost the elongated, rod-shaped appearance that is characteristic of wild-type E. coli (average length 2.45 ± 0.53 μm) and assumed a diminutive coccoid morphology (average length 0.78 ± 0.37 μm) (Fig. 1 B and C). Cells from papillae generated around the mutant colonies on plates frequently included phenotypic revertants that resembled the wild-type cell (Fig. 1C Inset). The E38K, V42L change appeared to be gain-of-function mutations; deletion of the mutant hupA allele or replacement of it with the wild-type allele restored normal morphology (data not shown).
To show that the morphological changes exhibited by SK3842 were a direct consequence of HUαE38K,V42L expression, and the effects could be reversed by complementation with a large excess of wild-type HUα, we introduced a multicopy plasmid containing an inducible wild-type HUα–GFP fusion into the mutant strain. In the absence of HUα–GFP expression, the mutant cells remained coccoid and were poorly stained by the membrane stain FM-64 (Fig. 1Di). Upon induction with IPTG, some of the coccoid cells began to express the HUα–GFP fluorescence and, concomitantly, incorporate the membrane stain (Fig. 1Dii), indicating a shift toward normalcy in the membrane architecture. With continued induction of the wild-type HUα fusion protein, some of the spherical mutant cells started to adopt a slightly elongated ovoid shape, both in dividing (Fig. 1Diii) and resting (Fig. 1Div) cells. The GFP fluorescence in these transitional cells confirmed that the morphological transformations were not random occurrences but effected by the overexpression of the wild-type HUα fusion. Finally, these ovoid cells changed into regular elongated rods, characteristic of wild-type E. coli (Fig. 1Dv).
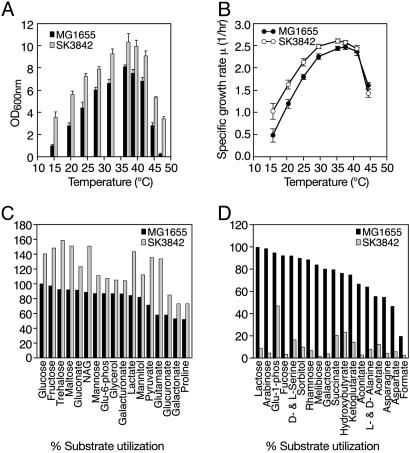
Improved Growth Characteristics and a Wider Temperature-Adaptability Profile of the hupA Mutant Cells. In rich media, the final cell yield of SK3842 at 37°C was significantly higher (≈30%) than the wild type (Fig. 2A). This difference in the final cell density was even more striking at nonoptimal temperatures. At both 15°C and 47°C, the mutant reached high cell concentrations, whereas the wild type was severely crippled in its ability to achieve healthy cell growth. The growth rate of the mutant (2.6 generations per h) was also notably higher than that of the wild type (2.43 generations per h) at 37°C (Fig. 2B). The difference in the growth rate was also more pronounced at lower temperatures. But, at temperatures above 42°C, the growth rate of the mutant was actually lower than that of the wild type, in spite of the mutant's higher final cell density.
Fig. 2.
Effect of HUαE38KV42L substitution on growth and metabolism. (A) Saturation cell densities of the wild type and hupA mutant at different temperatures. (B) Specific growth rates of the wild type and hupA mutant at different temperatures. (C and D) Differences between the wild type and hupA mutant in utilization of individual substrates as sole carbon source. (C) Substrates that were used better by the hupA mutant. Growth (A600 nm)ofthe wild type on glucose was arbitrarily set at 100%, and all other values for growth on different substrates were expressed as percentages relative to growth of the wild type on glucose. (D) Substrates that were used poorly by the hupA mutant. Growth (A600 nm) of the wild type on lactose was arbitrarily set at 100%, and all other values were expressed as relative percentages.
The Mutant Strain Exhibited Altered Anabolic and Catabolic Potential. Investigation of the nutritional requirements showed that the mutant was prototrophic for only the amino acids aspartate, glutamate, asparagine, glutamine, cysteine, lysine, alanine, and tryptophan. The mutant also had a requirement for nicotinic acid for growth in minimal media. The mutant hupA allele also had pronounced pleiotropic effects on carbon utilization, as determined by its ability to grow on different substrates in BioLog plates (Fig. 2 C and D). SK3842 showed little or no growth on many sugars (galactose, lactose, arabinose, melibiose, fucose, and rhamnose), TCA cycle intermediates (succinate, cis-aconate and α-ketoglutarate), amino acids (d- and l-alanine, asparagines, and aspartate), fermentation acids (formate and acetate), and short-chain fatty acids (α-hydroxybutyric acid). However, the mutant showed enhanced growth on glucose, fructose, and mannose; polyols (glycerol and mannitol); hexonates and hexuronates (gluconate, glucuronate, and galacturonate); fermentation acids (pyruvate and lactate); and proline, glutamate, and trehalose.
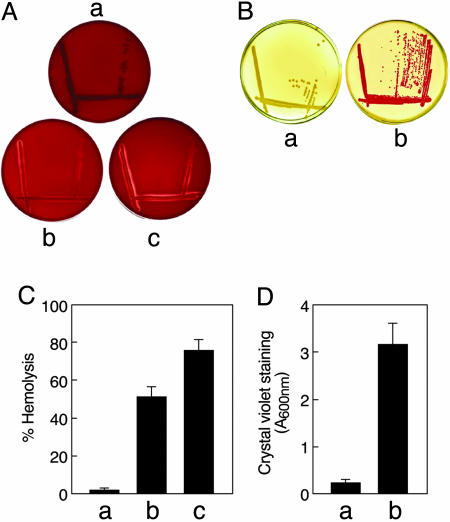
Quiescent Virulence Genes Were Activated in the Mutant. A number of genes that are quiescent under normal laboratory growth conditions were expressed in the mutant. These include genes coding for dormant virulence factors like hemolysin and curli fibers. The hemolysin encoded by the hlyE gene of E. coli K-12 is a cytolytic protein that causes lysis of mammalian cells by pore formation (11). Curli fibers are thin, aggregative surface structures expressed during mammalian infection and promote formation of biofilms (12). Unlike the wild type, SK3842, when cultured on blood agar plates, showed a clear zone of hemolysis, indicating the expression of the hemolysin by the mutant strain (Fig. 3A). The zone of hemolysis became wider and more prominent when the blood agar plates were supplemented with 5 μg/ml nicotinamide. Quantitative liquid hemolytic assays confirmed that the hemolysin activity in SK3842 was induced to a significant level, which was further enhanced by nicotinamide supplementation (Fig. 3C). SK3842 also grew as deep red colonies on Congo red plates at 37°C, indicating that the mutant strain was expressing curli fibers whereas the wild-type cells were not (Fig. 3B). Because expression of curli fibers is also a strong indicator of biofilm formation, we assessed the potential for biofilm formation by the mutant. Fig. 3D shows that the biofilm formation was much more extensive in the mutant than in the wild type.
Fig. 3.
Expression of quiescent virulence genes in the hupA mutant. (A) Expression of hemolysin by wild type (a), hupA mutant (b) and hupA mutant supplemented with nicotinamide (c) on blood agar plates. (B) Curli-fiber production by wild type (a) and hupA mutant (b) on Congo red plates. (C) Quantitative hemolytic activity of wild type (a), hupA mutant (b), and hupA mutant supplemented with nicotinamide (c), as quantitated by lysis of sheep erythrocytes by culture supernatants. (D) Biofilm production in the wild type and hupA mutant, as measured by crystal violet staining of adhering cells on polystyrene microtitre plates.
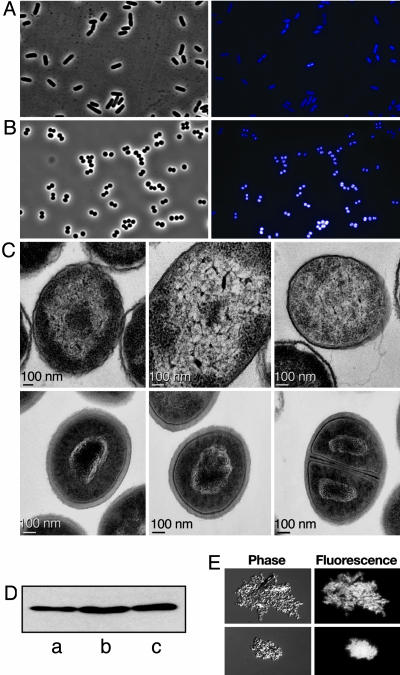
Condensation of the Bacterial Nucleoid by HUαE38K,V42L. E. coli mutants with coccoid morphology (e.g., rodA, pbpA, and mreBCD) are normally defective in growth, cell division, and nucleoid segregation. However, phase-contrast microscopy showed that the mutant cells were remarkably uniform in size and contour, without the appearance of any anucleate, filamentous, or abnormally shaped cells that are usually associated with aberrant cell division and chromosome segregation (Fig. 4B). Most of the cells were engaged in active cell division, reflecting the robust growth rate of the mutant. Consistent with the markedly reduced cell volume of the mutant, the nucleoids appeared to be much more condensed, in comparison with that of the wild type (Fig. 4A).
Fig. 4.
Effect of HUαE38K,V42L substitution on the nucleoid configuration. (A and B) Light (Left) and fluorescence (Right) microscopic observation of wild-type E. coli (A) and hupA mutant (B). (C) Thin-section transmission electron photomicrographs illustrating the nucleoid ultrastructure in wild-type E. coli (Upper) and hupA mutant (Lower). (D) Western blot analysis of HU proteins in cell lysates from wild-type (a), ΔhupB (b), and mutant hupAΔhupB (c) strains. (E) Phase-contrast fluorescence photomicrographs of purified nucleoids from ΔhupAhupB cells complexed with wild-type HUα (Upper) and mutant HUα (Lower) proteins.
Because HU is a major architectural component of the bacterial nucleoid (13), we compared the nucleoid configuration in the wild type and mutant by transmission electron microscopy on thin sections of actively growing cells. The wild-type E. coli nucleoid, as expected, was spread out as a dispersed, lobular structure interspersed with the ribosome-filled, granular cytoplasm (Fig. 4C Upper). In contrast, the nucleoid in SK3842 was organized into a densely condensed unit of tightly coiled DNA packed into a limited space (Fig. 4C, Lower). There was a highly condensed, electron-dense central core with coils and loops of exposed DNA strands arranged in a rosette-shaped structure. Nucleoid condensation was visible during all stages of the cell cycle, from a newly divided cell to a resting cell.
The changes encountered in the mutant could be a result of higher levels of the mutant HUα. Western blot analysis of cellular lysates from log-phase cultures of wild type, ΔhupB, and mutant hupA(ΔhupB) showed that the amount of HUα in all strains was almost identical (Fig. 4D). To investigate whether the nucleoid condensation observed in the mutant was a direct effect of HUαE38K,V42L and not mediated through other nucleoid-condensing proteins, such as H-NS and Dps, we examined the effect of purified HUαE38K,V42L on the morphology of nucleoids isolated from ΔhupAhupB cells (Fig. 4E). When treated with wild-type HUα, the nucleoids looked dispersed and loosely organized, with discrete, nonuniform distribution of DAPI foci. Addition of HUαE38K,V42L, at a similar ratio, produced a highly compact nucleoid structure with a dense, uniformly distributed, and crystalline pattern of DAPI staining. This experiment showed that the HUαE38K,V42L was the primary agent in the nucleoid condensation observed in vivo.
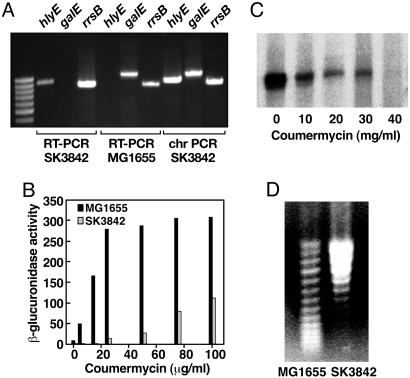
Increased Global DNA Superhelicity in the hupA Mutant. To verify whether the phenotypic changes in the mutant resulted from changes in the transcriptional output of the cell, we performed RT-PCR on RNA from the wild-type and mutant cells, probing three genes: hlyE (expressed only in the mutant), galE (expressed only in the wild type), and rrsB (signature 16S ribosomal RNA gene expressed in both). In the mutant, RT-PCR was positive for hlyE and negative for galE, whereas, in the wild type, the RT-PCR was positive for galE and negative for hlyE (Fig. 5A). The amount of PCR product for the 16S rRNA was slightly higher in the mutant, consistent with the faster growth rate of the mutant compared with the wild type.
Fig. 5.
Effects of HUαE38K,V42L on gene transcription and global DNA supercoiling. (A) RT-PCR of total cellular RNA from wild-type and hupA mutant cells. PCR was performed on reverse-transcribed RNA (RT-PCR) and mutant hupA chromosomal DNA (chromosomal PCR) by using primers for hlyE, galE, and rrsB.(B) β-glucuronidase activity from gal P2--gusA transcription fusion in the presence of different concentrations of coumermycin in wild-type and hupA mutant strains at OD600 nm = 0.5. (C) Northern blot analysis of the effect of different concentrations of coumermycin on chromosomal proU mRNA expression in the hupA mutant. (D) Topoisomer distribution of plasmid pPROU37 isolated from wild-type and hupA mutant cells on agarose gels containing 15 μg/ml chloroquine.
Activation of hlyE and csg, superrepression of gal, and higher abundance of rRNA, may be indicative of a change in global DNA supercoiling in the mutant. We measured the transcription activity of the supercoiling-sensitive promoter galP2 (14) in the presence of increasing concentrations of the gyrase-inhibitor drug coumermycin. In contrast to the wild-type strain, where the β-glucuronidase activity of P2-gus fusion increased sharply to a saturation level (Fig. 5B), the rise in the P2-gus activity in the mutant was delayed and gradual. The alleviation of repression of the galP2 promoter in the mutant, albeit partial, by coumermycin demonstrated that the repressed status of the promoter was caused by excessive negative supercoiling of the chromosomal locus. Northern blot analysis of the mRNA of another supercoiling-sensitive gene proU (15) under varying concentrations of coumermycin showed that proU mRNA was present in a significant amount in the mutant, but its level progressively diminished with DNA relaxation by coumermycin (Fig. 5C) and was ultimately abolished at 60 μg/ml coumermycin concentration.
To confirm the higher degree of negative supercoiling in the mutant, we analyzed the topoisomer distribution of plasmid pPROU37, containing the transcriptionally active PproU promoter from the wild-type and SK3842 strains on chloroquine gels. At a chloroquine concentration of 15 μg/ml, plasmid topoisomers with a higher degree of supercoiling migrate more slowly on the gel. Plasmid from the mutant strain appeared to have significantly higher negative supercoiling in comparison with that from the wild type (Fig. 5D).
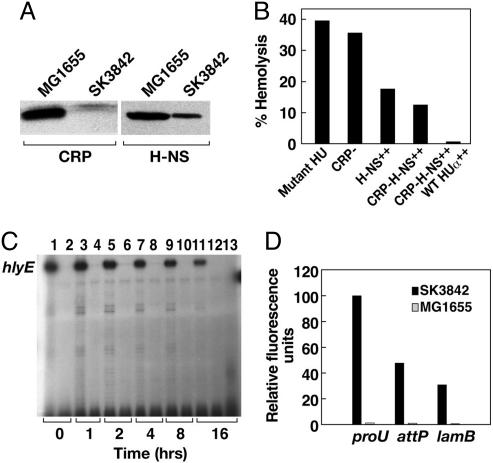
Nucleoid Reorganization Had a Direct Impact on Transcription Pattern. The effect of HUαE38K,V42L on the changes in the transcription profile could be mediated (i) directly, by remodeling the three-dimensional architecture of the nucleoid, thereby changing the susceptibility of individual promoters to the transcription machinery; (ii) indirectly, by modulating the expression of a subset of global regulatory factors that leads to the transcription reorganization; or (iii) through a combination of these two effects. The transcription of hlyE in wild-type E. coli is strictly contingent upon two regulators, the presence of the activator CRP and the absence of the repressor H-NS (16). Western blot analysis showed that, compared with the wild type, the level of CRP in the mutant was drastically diminished (Fig. 6A). The amount of H-NS was also reduced, but not totally eliminated, in the mutant. To corroborate the influence of nucleoid reorganization on the altered transcription pattern, we measured the expression of the hlyE gene in the context of no transcription-activator protein, excess transcription-repressor protein, and a combination of both (Fig. 6B). In the absence of cAMP(Δcrp), hemolysin expression in the mutant was similar to that in the wild type, confirming that, contrary to the wild-type situation, cAMP-CRP does not play a major role in the constitutive expression of hlyE in the mutant. In SK3842 bearing a multicopy plasmid expressing H-NS, even overnight induction of H-NS could reduce the level of hemolysin to only ≈50% of that of SK3842, showing that H-NS, which causes total silencing of hlyE under normal circumstances, failed to turn off transcription, even at excess concentrations. In the hupA mutant with both crpA deletion and over-expressing H-NS, the level of hemolysin was reduced slightly more but was not turned off totally. However, when provided with excess wild-type HUα from a multicopy plasmid, the hlyE gene was completely repressed. We performed primer-extension studies on RNA extracted from the hupA(Δcrp) mutant carrying the plasmid pHNS42 to study the expression of hlyE (Fig. 6C). Consistent with the liquid-hemolysin assay result, the expression of hlyE decreased gradually with time and reached a plateau at ≈50% of its original level. Expression of the wild-type HUα, however, completely turned off hlyE transcription. Although the stimulatory effect of HUαE38K,V42L on hlyE was rather independent of transcription regulators, we envision that the overall effect of HUαE38K,V42L on most promoters would be more complex and ramified. Because global transcription regulators are a part of the cellular transcriptome, their expression levels would also be affected significantly in the mutant and contribute to the overall changes in transcription pattern.
Fig. 6.
Direct influence of HUαE38K,V42L-mediated nucleoid reorganization on specific changes in transcription profile. (A) Western blot analysis of CRP and H-NS concentration in midlog-phase cultures of wild-type and hupA mutant cells. (B) Hemolytic activity of hupA mutant strains with altered levels of transcription regulators for hlyA. From left to right, the strains used were SK3842, SK3842 Δcrp, SK3842 (pHNS42), SK3842 Δcrp (pHNS42), and SK3842 Δcrp (pHNS42 and pHU-GFPuv). Assays were done 16 h after the addition of IPTG. (C) Primer extension analysis of hlyE mRNA expression in SK3842 Δcrp pHNS42 (lanes 1, 3, 5, 7, 9, and 11) and MG1655 Δcrp pHNS42 (lanes 2, 4, 6, 8, 10, and 12) at different time points after induction of H-NS. Lane 13 shows the hlyE mRNA expression in SK3842 Δcrp pHNS42, pHU-GFPuv at 16 h after IPTG addition. (D) GFP fluorescence intensities from PproU–GFP translation fusions at the proU, attP, and lamB chromosomal loci in the wild-type and hupA mutant strains.
If the nucleoid architecture in the mutant determined the fate of individual promoters, at least partially, then the context of chromosomal location should also play a role in determining the sensitivity of individual promoters. We inserted a PproU–gfp translational fusion at different chromosomal locations in the wild type and mutant, replacing the existing native promoters. In the mutant, PproU activity was highest at its original chromosomal locus, decreased by ≈50% at the attP locus, and decreased even further at the lamB locus (Fig. 6D). PproU was repressed at all three loci in the wild-type strain. The differential activity of a single promoter at different chromosomal locations further confirmed that the global topological state of the chromosome determined the expression of individual genes.
Discussion
Nucleoid Condensation and Global Changes in Gene Expression by a Mutant HUα. We describe a mutant bacterial histone-like protein, HUαE38K,V42L, which profoundly affected the morphology, growth, and physiology in wild-type E. coli. HUαE38K,V42L and made the bacterial nucleoid undergo a radical transformation from a loosely organized structure into a densely condensed globular configuration, without disruption of growth, chromosome segregation, and cell division. Overexpression of some histone-like proteins is known to induce strong chromosomal condensation in E. coli. Cyt1Aa of Bacillus thuringiensis (17), chlamydial histone H1-like protein, Hc1 (18), or E. coli nucleoid protein H-NS (19), when overexpressed, causes a global downshift in transcription and even cell death. The nucleoid condensation by the Dps protein during stationary phase (20) or the absence of the SeqB protein in wild-type E. coli (21) causes growth and metabolic downshifts. Condensed nucleoids are not always symbolic of functional inefficiency; the naturally occurring, highly compacted nucleoids of Deinoccoccus (22) or the metabolically active, condensed nucleoids induced by certain mitochondrial and chloroplast proteins (23, 24) are prime examples. The hupA mutant also exhibited an increase in DNA supercoiling in vivo. DNA supercoiling is known to be modified during many environmental challenges, including host infection, thereby coordinating the outputs of the gene-regulatory networks in response to environmental cues (25). Hence, the change in degree of global supercoiling in the hupA mutant was not unexpected, given the widespread changes in its transcription program. It is not clear whether the increased superhelicity was generated because of changes in cellular topoisomerase activities or an altered mode of supercoil constraint by the mutant HUα. It is, however, clear that the combination of HUαE38K,V42L-mediated chromosome compaction and altered superhelical tension resulted in unique functional remodeling of the nucleoid structure, causing a global shift in the transcription pattern.
HUαE38K,V42L-Mediated Cellular Changes Define a Shift in E. coli Behavior. There was a functional directionality in the global changes of the transcription profile of SK3842; many of the genes, which are redundant or unfavorable to E. coli K-12 for its laboratory existence, were activated, whereas other genes that are routinely expressed were turned off. The morphogenic differentiation of E. coli into coccoid form serves a vital role in its response to the host environment in certain cases. E. coli BJ4, during growth in the rat intestine, differentiates from rods to cocci, and the coccoid form is selected for in vivo (26). During the multistage differentiation and maturation program in vivo, uropathogenic E. coli converts from rods to cocci as a natural process of infection (27). The hupA mutant was more efficient in using substrates involved in glycolytic and Entner–Doudoroff pathways and the phosphotransferase-uptake systems. Substrates from the tricarboxylic acid cycle, glyoxalate shunt, and gluconeogenesis pathways were used very poorly. E. coli utilizes gluconate as the main carbon source and exploits only a small number of other carbohydrates to grow and colonize in the small intestine (28, 29). It appears significant that SK3842 could efficiently metabolize only certain carbon sources, most of which corresponds to the substrates preferred by E. coli in vivo. It has also been shown that most of the amino acid biosynthetic genes in E. coli are repressed in vivo (29). Finally, the extensive biofilm formation in the hupA mutant and the expression of hemolysin and curli fibers all point to coordinated and directional changes in the transcription profile of the hupA mutant, more suitable for survival inside a host environment.
Normal Architecture and Transcriptional Program in Bacteria. HUαE38K,V42L brought about a qualitatively different nucleoid configuration that engendered a functional shift in the transcription pattern. The altered nucleoid architecture and the resulting changes in the gene-expression profile could be reversed back to their incipient states by the introduction of a large excess of wild-type HUα, revealing that mutant HUα acted as a master switch to coordinate the entire spectrum of physical and physiological changes. Bacteria can switch their gene-expression program rapidly and precisely in response to environmental changes. We propose that nucleoid structural reorganization could serve as an efficient mechanism to synchronize the genetic response to external conditions. Certain effector molecules present in pathogenicity-inducing environments probably elicit similar changes in nucleoid condensation, leading to swift and concerted changes in the basal transcription program.
The Nature and Possible Evolutionary Significance of HUαE38K,V42L Mutant. HUαE38K,V42L had two amino acid substitutions, E38K and V42L, both of which were essential for the phenotypic changes in the hupA mutant. Wild-type HUα has a lysine at position 37. So, the presence of lysine at position 38 created a lysine–lysine motif in this highly basic protein. It is noteworthy that K38 is one of the critical amino acids responsible for the thermostability of BstHU (30). We have also observed that chromosomal integration of BstHU into the ΔhupA E. coli strain manifested many of the characteristics of SK3842 (S.K. and S.A., unpublished data).
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Author contributions: S.K. and S.A. designed research; S.K. and R.E. performed research; S.K. and S.A. analyzed data; and S.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviation: IPTG, isopropyl β-d-thiogalactoside.
References
- 1.Hobot, J. A., Villiger, W., Escaig, J., Maeder, M., Ryter, A. & Kellenberger, E. (1985) J. Bacteriol. 162, 960-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinow, C. & Kellenberger, E. (1994) Microbiol. Rev. 58, 211-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broyles, S. S. & Pettijohn, D. E. (1986) J. Mol. Biol. 187, 47-60. [DOI] [PubMed] [Google Scholar]
- 4.Struhl, K. (1999) Cell 9, 1-4. [DOI] [PubMed] [Google Scholar]
- 5.Drlica, K. & Rouviere-Yaniv, J. (1987) Microbiol. Rev. 51, 301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouviere-Yaniv, J. & Gros, F. (1975) Proc. Natl. Acad. Sci. USA 72, 3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dame, R. T. & Goosen, N. (2002) FEBS Lett. 529, 151-156. [DOI] [PubMed] [Google Scholar]
- 8.Sagi, D., Friedman, N., Vorgias, C., Oppenheim, A. B. & Stavans, J. (2004) J. Mol. Biol. 341, 419-428. [DOI] [PubMed] [Google Scholar]
- 9.Kar, S. & Adhya, S. (2001) Genes Dev. 15, 2273-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmerman, S. B. & Murphy, L. D. (2001) J. Bacteriol. 183, 5041-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Castillo, F. J., Leal, S. C., Moreno, F. & del Castillo, I. (1997) Mol. Microbiol. 25, 107-115. [DOI] [PubMed] [Google Scholar]
- 12.Olsen, A., Jonsson, A. & Normark, S. (1989) Nature 338, 652-655. [DOI] [PubMed] [Google Scholar]
- 13.Kellenberger, E. & Arnold-Schulz-Gahmen, B. (1992) FEMS Microbiol. Lett. 79, 361-370. [DOI] [PubMed] [Google Scholar]
- 14.Lewis, D. E., Geanacopoulos, M. & Adhya, S. (1999) Mol. Microbiol. 2, 451-461. [DOI] [PubMed] [Google Scholar]
- 15.Gowrishankar, J. & Manna, D. (1996) Genetica 3, 363-378. [DOI] [PubMed] [Google Scholar]
- 16.Westermark, M., Oscarsson, J., Mizunoe, Y., Urbonaviciene, J. & Uhlin, B. E. (2000) J. Bacteriol. 182, 6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manasherob, R., Zaritsky, A., Metzler, Y., Ben-Dov, E., Itsko, M. & Fishov, I. (2003) Microbiology 149, 3553-3564. [DOI] [PubMed] [Google Scholar]
- 18.Barry, C. E., III, Brickman, T. J. & Hackstadt, T. (1993) Mol. Microbiol. 9, 273-283. [DOI] [PubMed] [Google Scholar]
- 19.Spurio, R., Durrenberger, M., Falconi, M., La Teana, A., Pon, C. L. & Gualerzi, C. O. (1992) Mol. Gen. Genet. 231, 201-211. [DOI] [PubMed] [Google Scholar]
- 20.Frenkiel-Krispin, D., Ben-Avraham, I., Englander, J., Shimoni, E., Wolf, S. G. & Minsky, A. (2004) Mol. Microbiol. 2, 395-405. [DOI] [PubMed] [Google Scholar]
- 21.Weitao, T., Nordstrom, K. & Dasgupta, S. (2000) EMBO Rep. 1, 494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin-Zaidman, S., Englander, J., Shimoni, E., Sharma, A. K., Minton, K. W. & Minsky, A. (2003) Science 299, 254-256. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, T., Takahara, M., Miyagishima, S. Y., Kuroiwa, H., Sasaki, N., Ohta, N., Matsuzaki, M. & Kuroiwa, T. (2002) Plant Cell 14, 1579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki, N., Kuroiwa, H., Nishitani, C., Takano, H., Higashiyama, T., Kobayashi, T., Shirai, Y., Sakai, A., Kawano, S., Murakami-Murofushi, K., et al. (2003) Mol. Biol. Cell 14, 4758-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Travers, A. & Muskhelishvili, G. (2005) Nat. Rev. Microbiol. 3, 157-169. [DOI] [PubMed] [Google Scholar]
- 26.Krogfelt, K. A., Poulsen, L. K. & Molin, S. (1993) Infect. Immun. 61, 5029-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Justice, S. S., Hung, C., Theriot, J. A., Fletcher, D. A., Anderson, G. G., Footer, M. J. & Hultgren, S. J. (2004) Proc. Natl. Acad. Sci. USA 101, 1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweeney, N. J., Laux, D. C. & Cohen, P. S. (1996) Infect. Immun. 64, 3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang, D. E., Smalley, D. J., Tucker, D. L., Leatham, M. P., Norris, W. E., Stevenson, S. J., Anderson, A. B., Grissom, J. E., Laux, D. C., Cohen, P. S., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 7427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawamura, S., Abe, Y., Ueda, T., Masumoto, K., Imoto, T., Yamasaki, N. & Kimura, M. (1998) J. Biol. Chem. 273, 19982-19987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.