Abstract
Oxidative stress plays a central role in many human diseases and in aging. In Caenorhabditis elegans the SKN-1 protein induces phase II detoxification gene transcription, a conserved oxidative stress response, and is required for oxidative stress resistance and longevity. Oxidative stress induces SKN-1 to accumulate in intestinal nuclei, depending on p38 mitogen-activated protein kinase signaling. Here we show that, in the absence of stress, phosphorylation by glycogen synthase kinase-3 (GSK-3) prevents SKN-1 from accumulating in nuclei and functioning constitutively in the intestine. GSK-3 sites are conserved in mammalian SKN-1 orthologs, indicating that this level of regulation may be conserved. If inhibition by GSK-3 is blocked, background levels of p38 signaling are still required for SKN-1 function. WT and constitutively nuclear SKN-1 comparably rescue the skn-1 oxidative stress sensitivity, suggesting that an inducible phase II response may provide optimal stress protection. We conclude that (i) GSK-3 inhibits SKN-1 activity in the intestine, (ii) the phase II response integrates multiple regulatory signals, and (iii), by inhibiting this response, GSK-3 may influence redox conditions.
Keywords: longevity, p38 mitogen-activated protein kinase, stress response, Phase II gene
Oxidative stress is a major etiologic factor that affects numerous human diseases, including diabetes, atherosclerosis, and cancer, and aging (1–6). Eukaryotic cells respond to oxidative or xenobiotic stress by inducing expression of the phase II detoxification genes, which encode enzymes that synthesize glutathione, scavenge free radicals, and detoxify reactive products of the phase I (P450) system (7–10). This detoxification response appears to be conserved from yeast to humans.
In Caenorhabditis elegans, this oxidative stress response is orchestrated by the transcription factor SKN-1 (11). During postembryonic stages, the SKN-1 protein is present primarily in the ASI chemosensory neurons and the digestive system (the intestine). In the ASI neurons, SKN-1 localizes to nuclei and constitutively induces phase II gene expression. In contrast, in the intestine, oxidative stress is required to direct SKN-1 to accumulate in nuclei and activate gene expression. This intestinal response to oxidative stress is mediated through posttranscriptional regulation of SKN-1 and can occur within minutes, indicating that it involves relocalization of a cytoplasmic SKN-1 pool. The skn-1 mutant is sensitive to oxidative stress, and its life span is shortened by 25–30%, demonstrating the importance of SKN-1 and this oxidative stress defense for C. elegans.
SKN-1 binds to DNA through a unique monomeric mechanism but is related to its functional counterparts in mammals, the two dimeric NF-E2-related factor (Nrf) basic-leucine zipper proteins (Nrf1 and Nrf2) (12). Like SKN-1, Nrf1 and Nrf2 accumulate in cell nuclei in response to oxidative stress and are required for oxidative stress resistance (7, 10, 13). In the absence of stress, Nrf2 is bound and targeted for degradation by the cytoplasmic actin-binding protein Keap1 (14–16), a direct ortholog of which is not present in C. elegans. Nrf protein regulation otherwise is not well understood, although certain cell signaling pathways appear to be involved (9).
In addition to mediating this ancient stress response function during postembryonic stages, SKN-1 also initiates development of the entire digestive tract in the early embryo (17). SKN-1 that is expressed maternally then establishes the fate of the EMS blastomere and its descendants but is prevented from inappropriately establishing the fate of a different cell (the C blastomere) by gsk-3 (also called sgg-1), the C. elegans ortholog of mammalian gsk-3 α/β (18). Glycogen synthase kinase-3 (GSK-3) is broadly conserved among eukaryotes and was originally identified as a regulator of glycogen synthesis but has since been shown to control a diverse array of additional cell functions (19, 20). In C. elegans, gsk-3 influences C blastomere differentiation independently of its well documented role in Wnt signaling (18). It has not been determined whether GSK-3 signaling then acts on the SKN-1 protein directly or through a different mechanism.
Here we report that gsk-3 prevents SKN-1 from accumulating in nuclei and constitutively inducing phase II gene expression in the postembryonic intestine. This inhibition is mediated through GSK-3 phosphorylation of the SKN-1 protein. We conclude that GSK-3 is an important regulator of the phase II detoxification response in C. elegans and possibly in other organisms.
Materials and Methods
Strains and RNA Interference (RNAi). The following strains were maintained at 20°C by standard methods unless otherwise noted (21): WT N2 (Bristol Laboratories), N2 Is007[SKN-1::GFP], N2 Ex007[SKN-1::GFP], N2 Ex007[SKN-1::GFP S74, 340A], sek-1(km4) Is007[SKN-1::GFP], gcs-1::gfp, gcs-1::gfp Δ2, and gcs-1::gfp Δ2 mut3 (gcs-1, γ-glutamylcysteine synthetase). RNAi experiments were performed by feeding the worms as described in ref. 22. Most eggs produced by gsk-3(RNAi) worms failed to hatch, but many escapers developed into adults. These F1 escaper progeny were used in SKN-1::GFP localization and gcs-1::gfp induction experiments as described in ref. 11.
Plasmid Constructions and Transgenesis. We created SKN-1::GFP mutant constructs (see Fig. 2 A) by using the QuikChange method (Stratagene). All PCRs were performed by using Pfu polymerase (Stratagene). Each mutant transgene maintained the original intron/exon structure with the exception of T386A, which lacks intron 5 because Thr-386 is encoded by the junction of exons 5 and 6. Transgenic strains were generated by injecting DNA into young adult animals as described in ref. 23. SKN-1::GFP mutant constructs were injected at 10 ng/μl along with the rol-6 marker (pRF4) at 100 ng/μl. At least two independent extrachromosomal lines were analyzed for each construct. Differential interference contrast microscopy and fluorescence images were acquired with a Zeiss AxioSKOP2 microscope and an AxioCam cooled color digital camera. To discriminate intestinal autofluorescence from SKN-1::GFP epifluorescence, we used a triple-band emission filter set (Chroma 61000) in conjunction with a narrow-band excitation filter (484/14 nm), as reported in ref. 11. This combination allowed autofluorescence to be detected as yellow/orange fluorescence deriving from a combined green and red signal, with GFP remaining green.
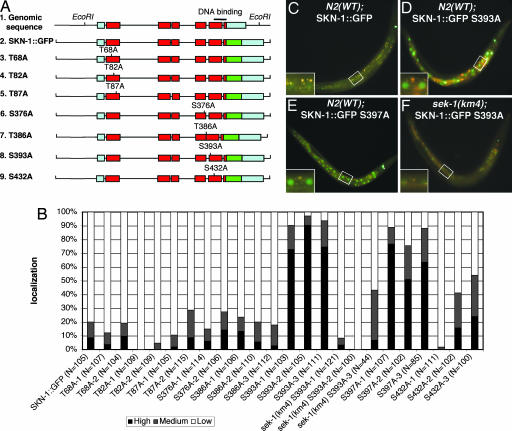
Fig. 2.
A predicted GSK-3 phosphorylation site prevents constitutive accumulation of SKN-1::GFP in intestinal nuclei. (A) The SKN-1 coding region and mutant transgenic constructs. Predicted coding regions are indicated by red boxes, untranslated regions are indicated by blue boxes, and GFP is indicated by a green box. Potential GSK-3 sites in SKN-1 were predicted by the scansite program (Table 1) (32). These predicted phosphorylated residues and the priming site Ser-397 (see Results and Discussion) were individually substituted with alanine within SKN-1::GFP. (B–E) Analysis of transgenic animals. (B) Individual transgenic lines were generated by injecting the transgenes diagrammed in A along with the marker rol-6 (pRF4). These extrachromosomal lines were scored for presence of SKN-1 in intestinal nuclei, as in Fig. 1B. Nuclear SKN-1::GFP was dramatically increased in SKN-1::GFP S393A lines in the WT but not sek-1(km4) background and in SKN-1::GFP S397A lines. (C–F) Fluorescent images of representative transgenic animals are shown, as in Fig. 1 A.
Kinase Assay. In the in vitro kinase assay, the peptides (shown in Fig. 3B) were incubated with rabbit GSK-3β (New England Biolabs) in buffer containing 20 mM Tris (pH 7.5), 5 mM DTT, 20 mM MgCl2, cold 200 μM ATP, and 0.4 μCi (1 Ci = 37 GBq) [γ-32P]ATP at 30°C for 1 h. The reaction was stopped by addition of 100 μg of BSA and cold 20% trichloroacetic acid, incubated on ice, then spun down. Supernatants were spotted on P81-phosphocellulose discs (Upstate Biotechnology, Lake Placid, NY), which were washed several times with 0.3% phosphoric acid and counted in a scintillation counter.
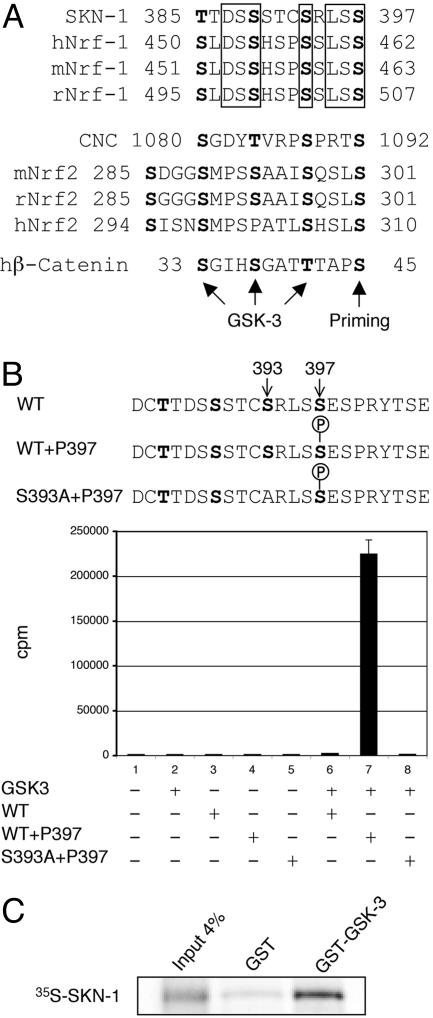
Fig. 3.
Phosphorylation of SKN-1 by GSK-3. (A) A GSK-3 phosphorylation motif in C. elegans SKN-1 is conserved in mammalian Nrf1 proteins. This motif, which has three predicted GSK-3 sites, is similar in structure to the GSK-3 target motif in β-catenin, in which priming phosphorylation at Ser-45 by casein kinase-1α is followed by sequential GSK-3 phosphorylation at Thr-41, Ser-37, and Ser-33 (26). Predicted compound GSK-3 phosphorylation motifs are also present in mammalian Nrf2 and the orthologous Drosophila protein CNC. Conserved residues in each group are boxed. h, human; m, mouse; r, rat. (B) Phosphorylation of SKN-1 Ser-393 by GSK-3β in vitro. GSK-3β phosphorylates the SKN-1 peptide shown provided that phosphorylated serine is present at position 397. This phosphorylation depends on the presence of Ser-393. cpm incorporated in a representative phosphorylation assay are graphed. (C) SKN-1 binds specifically to a C. elegans GST–GSK-3 fusion protein in vitro.
Protein Interaction Experiments. GST and a fusion of GST with full length C. elegans GSK-3 were expressed in Escherichia coli BL21 and purified on glutathione-Sepharose 4B. Full-length SKN-1 was translated in vitro by using the TNT SP6 coupled reticulocyte lysate transcription/translation system (Promega) and labeled with [35S]methionine (Amersham Biosciences, which is now GE Healthcare) as described in ref. 11. GST and GST–GSK-3 were immobilized on Sepharose beads and incubated with these reticulocyte lysates in phosphate buffer (pH 7.4) containing 137 mM NaCl, 2.7 mM KCl, and 1% Triton X-100 overnight at 4°C. Beads were washed 15 times with phosphate buffer (pH 7.4) containing 274 mM NaCl, 5.4 mM KCl, and 1% Triton X-100. After the last wash, samples were denatured and separated by a Novex Tris-glycine gradient gel (Invitrogen), which was dried and exposed on a PhosphorImager (Molecular Dynamics).
Oxidative Stress-Resistance Assay. SKN-1::GFP, SKN-1::GFP S393A, and rol-6 extrachromosomal arrays were genetically transferred from the N2 background into skn-1(zu67) (17) as previously described in ref. 11. skn-1(zu67) is balanced with DnT1, which carries a dominant Unc phenotype so that skn-1 homozygotes segregate as WT. Each of these arrays transmitted the corresponding SKN-1::GFP transgene along with rol-6 in essentially all animals. In each case, the levels of transgenic SKN-1 detected in the intestine were approximately proportional to the original amounts of injected transgene DNA (2.5 or 10 ng/μ1). To investigate sensitivity to oxidative stress, young adults were transferred onto plates that included 6.2 mM t-butyl hydroperoxide (Sigma) in nematode growth medium agar. Worms were incubated on these plates at 20°C and scored as dead when they did not respond to repeated prodding with a pick. P values were calculated by Student's t test using Microsoft excel.
Results and Discussion
gsk-3 Inhibits SKN-1 Localization and Target Gene Induction. To investigate whether GSK-3 inhibits SKN-1 during postembryonic stages, we have tested whether gsk-3 is required to prevent SKN-1 from localizing to nuclei and activating phase II gene expression in the intestine under normal conditions. When gsk-3 expression is reduced by RNAi, the most penetrant phenotype is embryonic lethality that involves abnormal C blastomere differentiation and Wnt signaling (18, 24). It is possible, however, to investigate postembryonic gsk-3 functions by examining the gsk-3(RNAi) F1 progeny that complete embryonic development and generally survive into adulthood.
In these gsk-3(RNAi) larvae and young adults, we examined localization of a previously described transgenic protein in which full-length SKN-1 is fused at its C terminus to GFP (11). Under normal conditions, SKN-1::GFP was present at low levels in intestinal nuclei, as shown in ref. 11. In contrast, SKN-1 was constitutively present in intestinal nuclei in the absence of oxidative stress after RNAi knockdown of gsk-3 but not of various control genes (Fig. 1 A and B and data not shown). In the ASI neurons, the levels of SKN-1 were not detectably affected by gsk-3 RNAi, however (data not shown). The data indicate that gsk-3 is required to prevent SKN-1 from accumulating in nuclei constitutively in the postembryonic intestine.
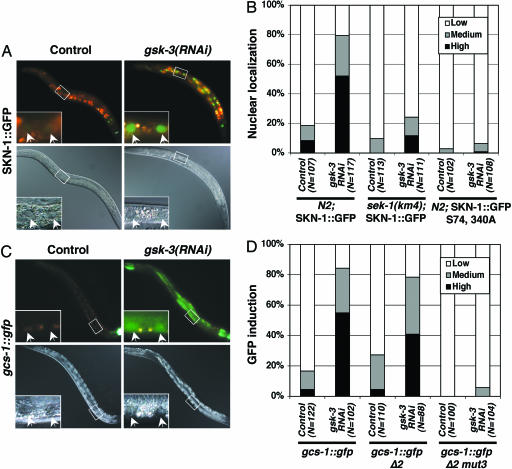
Fig. 1.
gsk-3 inhibits SKN-1 localization and SKN-1 target gene expression. (A and B) SKN-1 is localized to intestinal nuclei in gsk-3(RNAi) animals in the absence of stress. N2 Is007[SKN-1::GFP], sek-1(km4) Is007[SKN-1::GFP], and N2 Ex[SKN-1::GFP S74, 340A] L4 animals were placed onto nematode growth medium plates containing E. coli HT115 that carried either gsk-3 dsRNA or control (L4440) feeding vectors. The animals were then incubated for 24 h at 20°C and allowed to lay eggs. Their surviving progeny were scored as L4 larvae and young adults for accumulation of SKN-1::GFP in intestinal nuclei as described in ref. 11. “High” indicates that a strong SKN-1::GFP signal was present in all intestinal nuclei, as in the gsk-3(RNAi) animal shown. “Medium” refers to animals in which nuclear SKN-1::GFP was present at high levels anteriorly or anteriorly and posteriorly but was barely detectable midway through the intestine. sek-1 encodes a C. elegans p38 MAPK kinase and is required for SKN-1 function in the intestine (see Results and Discussion) (28). (C and D) gcs-1::gfp is expressed constitutively in gsk-3(RNAi) animals. gsk-3 RNAi was performed in gcs-1::gfp, gcs-1::gfp Δ2, and gcs-1::gfp Δ2-mut3 strains as in A and B, then intestinal gcs-1::gfp expression was scored in F1 L4 larvae and young adults (11). “High” indicates that gcs-1::gfp was present at high levels anteriorly and was detectable throughout most of the intestine, as in the gsk-3(RNAi) image shown. “Medium” refers to animals in which gcs-1::gfp was present at high levels anteriorly and possibly posteriorly but was not detected in between. The gcs-1::gfp fluorescence apparent in the control image derives from SKN-1-independent pharyngeal expression (11). In A and C, upper images show fluorescence, lower images show Nomarski imaging, and pairs of intestinal nuclei (arrows) are shown in each inset.
In the intestine, SKN-1 directly regulates and is required for the expression of gcs-1, a well characterized phase II gene and Nrf protein target (11). The enzyme encoded by gcs-1 is rate-limiting for glutathione biosynthesis. As described in ref. 11, in the intestine a transgene in which the gcs-1 promoter region is linked to GFP (gcs-1::gfp) was expressed at high levels in response to oxidative stress but at low levels under normal conditions (Fig. 1 C and D and data not shown). In contrast, in gsk-3(RNAi), animals' intestinal gcs-1::gfp expression was dramatically elevated without stress (Fig. 1 C and D). gsk-3 RNAi similarly results in expression of the gcs-1::gfp Δ2 transgene, which lacks SKN-1-independent pharyngeal expression (data not shown) but not the gcs-1::gfp Δ2 mut3 transgene, in which an essential SKN-1 binding site in its promoter has been mutated (11) (Fig. 1D). This result suggests that SKN-1 is required for gsk-3 RNAi knockdown to increase gcs-1::gfp expression. We conclude that, in the absence of stress, gsk-3 prevents SKN-1 from constitutively inducing phase II gene expression in the intestine.
A GSK-3 Phosphorylation Site Required to Exclude SKN-1 from Intestinal Nuclei. The SKN-1 amino acid sequence includes seven predicted potential GSK-3 phosphorylation sites (Fig. 2A and Table 1, which is published as supporting information on the PNAS web site). To investigate whether these residues are important for SKN-1 localization in the intestine, we mutated them individually to alanine (Fig. 2 A). Remarkably, the SKN-1::GFP S393A mutant was constitutively localized to intestinal nuclei in almost all animals examined (Fig. 2 B and D), indicating that Ser-393 is essential for SKN-1 regulation and may be a functionally critical GSK-3 site. The S432A mutation modestly increased presence of SKN-1::GFP in intestinal nuclei, but none of the other individual residues examined appeared to influence SKN-1 localization (Fig. 2B). Ser-393 and Ser-432 were the most strongly predicted GSK-3 sites in SKN-1 (Table 1), supporting the idea that they may be true GSK-3 substrates.
Phosphorylation of a substrate by GSK-3 usually depends on prior action of a priming kinase, which phosphorylates the amino acid that is located four positions C-terminal to the GSK-3 site (Fig. 3A) (19, 25). For example, priming phosphorylation of β-catenin by casein kinase-1α at Ser-45 precedes sequential GSK-3β phosphorylation at Ser-41, Ser-37, and Ser-33 (Fig. 3A) (26). This model predicts that priming phosphorylation of SKN-1 at Ser-397 would be required to allow GSK-3 to phosphorylate Ser-393 and, thus, to prevent SKN-1 from localizing to intestinal nuclei (Fig. 3A). Accordingly, in an in vitro assay, GSK-3 robustly phosphorylated a SKN-1 peptide in which phosphorylated serine was present at position 397. This GSK-3 phosphorylation was abolished when unphosphorylated Ser-397 was present or when Ser-393 was substituted with alanine (Fig. 3B). Further supporting the idea that GSK-3 targets SKN-1 directly, SKN-1 strongly and specifically bound C. elegans GSK-3 in vitro (Fig. 3C).
A transgenic SKN-1 protein in which Ser-397 had been substituted with alanine (SKN-1::GFP S397A) was constitutively present in intestinal nuclei comparably with SKN-1::GFP S393A (Fig. 2 B and E). The GSK-3 site Ser-393 and its predicted priming site, Ser-397, thus were each required to prevent SKN-1 from accumulating in intestinal nuclei in the absence of stress, strongly supporting the idea that GSK-3 phosphorylates and inhibits SKN-1 directly in vivo. It is intriguing that mammalian Nrf1 proteins include a sequence that is related to the SKN-1 motif that contains Ser-393 and Ser-397 (Fig. 3A). Additional predicted GSK-3 sites, including similar compound motifs, are present in Nrf1, Nrf2, and the related Drosophila protein CNC (Fig. 3A and data not shown). The presence of these sequences suggests the exciting possibility that GSK-3 phosphorylation might also be involved in Nrf protein regulation.
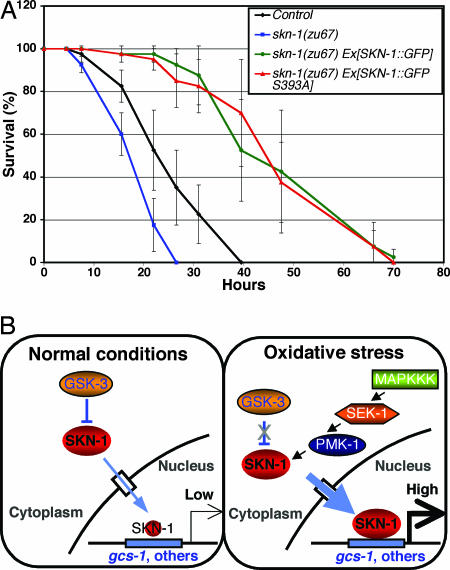
WT and Constitutively Nuclear SKN-1 Comparably Protect Against Oxidative Stress. We previously showed that skn-1 mutants are hypersensitive to oxidative stress and that the SKN-1::GFP (Fig. 2A) transgene can rescue their embryonic developmental defect (11). We have now found that introduction of SKN-1::GFP renders skn-1(zu67) homozygotes significantly more resistant to oxidative stress than WT (Fig. 4A), indicating that SKN-1 overexpression dramatically enhances stress resistance. This finding raises the question of whether a version of SKN-1 that is constitutively nuclear in the intestine might provide even more effective stress protection. However, the SKN-1::GFP S393A transgene (Fig. 2B) increased oxidative stress resistance comparably with WT SKN-1::GFP when these transgenes were introduced in parallel at two different dosages of DNA (Fig. 4A and data not shown). Constitutively nuclear SKN-1 is therefore no more effective than WT at conferring stress resistance. Although it cannot be ruled out that additional GSK-3-inhibited signals are required for full activity of SKN-1::GFP S393A, our findings suggest that an inducible phase II response may provide optimal protection against oxidative stress. Presence of a robust inducible oxidative response system may be advantageous because it would avoid potential complications of continuous detoxification gene expression. For example, in mice that lack Keap1, constitutive Nrf2 activity causes developmental abnormalities in the digestive system (27).
Fig. 4.
SKN-1 protects against oxidative stress. (A) SKN-1::GFP overexpression confers oxidative stress resistance. Transgenically expressed WT and S393A forms of SKN-1::GFP comparably rescue the oxidative stress sensitivity of skn-1(zu67) mutants and provide additional protection against oxidative stress. All animals analyzed carry the rol-6 transgenic marker and were derived from worms that are heterozygous for the DnT1 balancer. Control thus refers to N2;rol-6 progeny that were derived from N2;rol-6;DnT1/+ animals. Individual worms were scored for survival at the times shown after they had been placed on nematode growth medium plates containing 6.2 mM t-butyl hydroperoxide. Under these stress conditions, SKN-1::GFP accumulated in intestinal nuclei and gcs-1::gfp expression was increased (data not shown). This representative experiment involved 40 worms in each group. Error bars indicate standard deviations. P values for skn-1(zu67), skn-1(zu67) Ex[SKN-1::GFP], and skn-1(zu67) Ex[SKN-1::GFP S393A] compared with control were 0.034, 0.007, and 0.009, respectively. (B) A model for regulation of SKN-1 in the intestine. Under normal conditions, SKN-1 is phosphorylated constitutively by GSK-3 and prevented from accumulating in the nucleus. This inhibition depends on the prior priming phosphorylation of SKN-1, which represents an additional negative signal (see Results and Discussion). SKN-1 target genes, represented by gcs-1, are then expressed at very low levels. Under oxidative stress conditions, p38 pathway signaling and PMK-1 phosphorylation of SKN-1 are dramatically increased (28). This signal counteracts inhibition of SKN-1 by GSK-3 and is independently required for SKN-1 to accumulate in nuclei at high levels and induce phase II gene expression. These phosphorylation events are arbitrarily shown as occurring in the cytoplasm. MAPKKK, MAPK kinase kinase.
SKN-1 Directly Integrates Multiple Regulatory Signals. Recently, we have determined that in the intestine SKN-1 function depends on signaling through the p38 mitogen-activated protein kinase (MAPK) pathway (28). In C. elegans, signaling through this conserved pathway has previously been shown to be essential for some neuronal asymmetries, and for innate immunity (29, 30). Oxidative stress results in activation of a p38 MAPK, PMK-1, which is a C. elegans p38 ortholog (28). SEK-1, the MAPK kinase that phosphorylates PMK-1, is required for SKN-1 to accumulate in intestinal nuclei and activate phase II gene expression, and sek-1(km4)-null mutants are hypersensitive to oxidative stress. SKN-1 is phosphorylated by PMK-1 on Ser-74 and Ser-340, and SKN-1::GFP fails to accumulate in nuclei or rescue the stress sensitivity of skn-1(zu67) mutants when these residues are substituted with alanine.
Our finding that GSK-3 prevents SKN-1 from acting constitutively in the intestine raises the question of whether p38 signaling to SKN-1 function might be needed simply to overcome inhibition by GSK-3. To investigate this possibility, we blocked p38 phosphorylation of SKN-1 and asked whether SKN-1 still appeared in intestinal nuclei constitutively when its phosphorylation by GSK-3 was inhibited. In sharp contrast to results obtained in the WT background, in sek-1(km4) animals, gsk-3 RNAi did not detectably alter SKN-1::GFP localization (Fig. 1B), and the GSK-3 phosphorylation mutant SKN-1(S393A) accumulated in nuclei only weakly (Fig. 2 B and F). In addition, in the WT background, gsk-3 RNAi did not result in nuclear localization of the SKN-1(S74A, S340A) mutant, which is defective in phosphorylation by PMK-1 (28) (Fig. 1B). These results demonstrate that sek-1 and p38 signaling are epistatic to gsk-3 with respect to SKN-1 regulation, thereby excluding the possibility that p38 signaling simply regulates GSK-3 or otherwise blocks its inhibition of SKN-1. In the intestine, the phase II detoxification response thus not only requires that inhibition of SKN-1 by gsk-3 be overcome but also depends on p38 phosphorylation of SKN-1 providing an independent positive stimulus (Fig. 4B). By inhibiting SKN-1, GSK-3 may set a threshold that prevents background levels of p38 signaling (28) from maintaining continuous SKN-1 activity, and it may provide a buffering effect that allows SKN-1 to respond proportionately to a broader range of stress signals.
This inducible regulation of SKN-1 in the postembryonic intestine contrasts sharply with how SKN-1 is localized to nuclei constitutively in the embryo and in the ASI neurons (11, 31). Our findings indicate that, in the intestine, SKN-1 directly integrates multiple independent signals, which combine either to restrain or activate the phase II response (Fig. 4B). Thus, our results reveal a paradigm through which GSK-3 signaling “converts” this otherwise nuclear transcription factor into a signal-responsive protein in this particular tissue, possibly by promoting cytoplasmic retention or degradation of SKN-1. A lack of signaling from GSK-3 or its priming kinase could therefore explain why SKN-1 is constitutively nuclear and active in the ASI neurons (11). An intriguing possibility suggested by our results is that GSK-3 might also inhibit SKN-1 through direct phosphorylation in the embryonic C blastomere.
The observation that GSK-3 regulates this conserved oxidative stress response in C. elegans reveals a function for this versatile and important kinase, which has been implicated in cellular proliferation, differentiation, motility, and survival (19, 20). This finding also predicts that signals or agents that reduce GSK-3 activity analogously to gsk-3(RNAi) would have similar effects on phase II gene expression and, thus, would provide a means of activating this detoxification response. GSK-3 is regulated by multiple cellular signals in mammals and is involved in human disease states that include cancer, diabetes, and Alzheimer's disease (20). If Nrf proteins are regulated by GSK-3 analogously to SKN-1, then alterations in GSK-3 activity could influence cellular redox conditions in many mammalian contexts.
Supplementary Material
Acknowledgments
We thank Craig Mello and Yanxia Bei (both from University of Massachusetts Medical School, Worcester) for providing the gsk-3RNAi and GST-GSK-3 constructs, Elizabeth Veal and Riva Oliveira for critically reading this manuscript and establishing the t-butyl hydroperoxide resistance assay, and Rosanna Baker for analyzing sequence relationships among C. elegans Keap-related genes. This work was supported by National Institutes of Health Grant GM62891 (to T.K.B.), the Iacocca Foundation (T.K.B.), and by special grants for Core Research for Evolutional Science and Technology and Advanced Research on Cancer from the Ministry of Education, Culture, and Science of Japan (to N.H. and K.M.).
Author contributions: T.K.B. designed research; J.H.A., K.V., and M.L. performed research; H.I., N.H., and K.M. contributed new reagents/analytic tools; J.H.A., K.M., and T.K.B. analyzed data; and J.H.A. and T.K.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: GSK-3, glycogen synthase kinase-3; RNAi, RNA interference; GCS-1, γ-glutamylcysteine synthetase; Nrf, NF-E2-related factor; MAPK, mitogen-activated protein kinase.
References
- 1.Brownlee, M. (2001) Nature 414, 813-820. [DOI] [PubMed] [Google Scholar]
- 2.Sheetz, M. J. & King, G. L. (2002) J. Am. Med. Assoc. 288, 2579-2588. [DOI] [PubMed] [Google Scholar]
- 3.Droge, W. (2002) Physiol. Rev. 82, 47-95. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, J. K. (2004) Nat. Med. 10, S18-S25. [DOI] [PubMed] [Google Scholar]
- 5.Balaban, R. S., Nemoto, S. & Finkel, T. (2005) Cell 120, 483-495. [DOI] [PubMed] [Google Scholar]
- 6.Katic, M. & Kahn, C. R. (2005) Cell Mol. Life Sci. 62, 320-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes, J. D. & McMahon, M. (2001) Cancer Lett. 174, 103-113. [DOI] [PubMed] [Google Scholar]
- 8.Toone, W. M., Morgan, B. A. & Jones, N. (2001) Oncogene 20, 2336-2346. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen, T., Sherratt, P. J. & Pickett, C. B. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 233-260. [DOI] [PubMed] [Google Scholar]
- 10.Motohashi, H. & Yamamoto, M. (2004) Trends Mol. Med. 10, 549-557. [DOI] [PubMed] [Google Scholar]
- 11.An, J. H. & Blackwell, T. K. (2003) Genes Dev. 17, 1882-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker, A. K., See, R., Batchelder, C., Kophengnavong, T., Gronniger, J. T., Shi, Y. & Blackwell, T. K. (2000) J. Biol. Chem. 275, 22166-22171. [DOI] [PubMed] [Google Scholar]
- 13.Xu, Z., Chen, L., Leung, L., Yen, T. S., Lee, C. & Chan, J. Y. (2005) Proc. Natl. Acad. Sci. USA 102, 4120-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh, K., Wakabayashi, N., Katoh, Y., Ishii, T., Igarashi, K., Engel, J. D. & Yamamoto, M. (1999) Genes Dev. 13, 76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, D. D. & Hannink, M. (2003) Mol. Cell. Biol. 23, 8137-8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi, A., Kang, M. I., Okawa, H., Ohtsuji, M., Zenke, Y., Chiba, T., Igarashi, K. & Yamamoto, M. (2004) Mol. Cell. Biol. 24, 7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowerman, B., Eaton, B. A. & Priess, J. R. (1992) Cell 68, 1061-1075. [DOI] [PubMed] [Google Scholar]
- 18.Maduro, M. F., Meneghini, M. D., Bowerman, B., Broitman-Maduro, G. & Rothman, J. H. (2001) Mol. Cell 7, 475-485. [DOI] [PubMed] [Google Scholar]
- 19.Cohen, P. & Frame, S. (2001) Nat. Rev. Mol. Cell. Biol. 2, 769-776. [DOI] [PubMed] [Google Scholar]
- 20.Jope, R. S. & Johnson, G. V. (2004) Trends Biochem. Sci. 29, 95-102. [DOI] [PubMed] [Google Scholar]
- 21.Brenner, S. (1974) Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timmons, L. & Fire, A. (1998) Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- 23.Mello, C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. (1991) EMBO J. 10, 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlesinger, A., Shelton, C. A., Maloof, J. N., Meneghini, M. & Bowerman, B. (1999) Genes Dev. 13, 2028-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harwood, A. J. (2002) Dev. Cell 2, 384-385. [DOI] [PubMed] [Google Scholar]
- 26.Liu, C., Li, Y., Semenov, M., Han, C., Baeg, G. H., Tan, Y., Zhang, Z., Lin, X. & He, X. (2002) Cell 108, 837-847. [DOI] [PubMed] [Google Scholar]
- 27.Wakabayashi, N., Itoh, K., Wakabayashi, J., Motohashi, H., Noda, S., Takahashi, S., Imakado, S., Kotsuji, T., Otsuka, F., Roop, D. R., et al. (2003) Nat. Genet. 35, 238-245. [DOI] [PubMed] [Google Scholar]
- 28.Inoue, H., Hisamoto, N., An, J. H., Oliveira, R. P., Nishida, E., Blackwell, T. K. & Matsumoto, K. (2005) Genes Dev. 19, 2278-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sagasti, A., Hisamoto, N., Hyodo, J., Tanaka-Hino, M., Matsumoto, K. & Bargmann, C. I. (2001) Cell 105, 221-232. [DOI] [PubMed] [Google Scholar]
- 30.Kim, D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., Tanaka-Hino, M., Hisamoto, N., Matsumoto, K., Tan, M. W., et al. (2002) Science 297, 623-626. [DOI] [PubMed] [Google Scholar]
- 31.Bowerman, B., Draper, B. W., Mello, C. & Priess, J. (1993) Cell 74, 443-452. [DOI] [PubMed] [Google Scholar]
- 32.Yaffe, M. B., Leparc, G. G., Lai, J., Obata, T., Volinia, S. & Cantley, L. C. (2001) Nat. Biotechnol. 19, 348-353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.