Abstract
Iron (Fe) is an essential element for most organisms which must be obtained from the local environment. In the case of pathogenic bacteria, this fundamental element must be acquired from the fluids and tissues of the infected host. A variety of systems have evolved in bacteria for efficient acquisition of host-bound Fe. The gram-negative bacterium Bordetella avium, upon colonization of the avian upper respiratory tract, produces a disease in birds that has striking similarity to whooping cough, a disease caused by the obligate human pathogen Bordetella pertussis. We describe a B. avium Fe utilization locus comprised of bhuR and six accessory genes (rhuIR and bhuSTUV). Genetic manipulations of B. avium confirmed that bhuR, which encodes a putative outer membrane heme receptor, mediates efficient acquisition of Fe from hemin and hemoproteins (hemoglobin, myoglobin, and catalase). BhuR contains motifs which are common to bacterial heme receptors, including a consensus FRAP domain, an NPNL domain, and two TonB boxes. An N-terminal 32-amino-acid segment, putatively required for rhuIR-dependent regulated expression of bhuR, is present in BhuR but not in other bacterial heme receptors. Two forms of BhuR were observed in the outer membrane of B. avium: a 91-kDa polypeptide consistent in size with the predicted mature protein and a smaller 82-kDa polypeptide which lacks the 104 amino acids found at the N terminus of the 91-kDa form. A mutation in hemA was engineered in B. avium to demonstrate that the bacterium transports heme into the cytoplasm in a BhuR-dependent manner. The role of BhuR in virulence was established in turkey poults by use of a competitive-infection model.
Iron (Fe) is an essential element for most organisms. In the case of bacterial pathogens, Fe must be obtained from the tissues, cells, and fluids of the infected vertebrate host. Since failure to acquire Fe limits the capability of the bacterial pathogen to colonize and proliferate, Fe acquisition is often considered to be an essential property of virulence (19, 56, 59, 62, 66). In animals, including humans, Fe is generally not present in the free elemental form but is sequestered in high-affinity Fe-binding complexes, such as hemoglobin, hemopexin, transferrin, and lactoferrin. These and other Fe-binding molecules maintain the concentration of free Fe within a prospective host at approximately 10−18 M, a concentration below the minimal level required by bacterial pathogens for survival and proliferation (5). In response to this selective pressure, efficient Fe acquisition systems have evolved in bacterial pathogens to “steal” the element from the various host Fe sequestration mechanisms. Expression of these bacterial Fe uptake systems is often regulated in response to the level of Fe in the microenvironment (32).
Heme, a tetrapyrolic molecule which coordinately binds Fe, is a component of many biologically active molecules. Hemoglobin, myoglobin, cytochromes, catalases, and peroxidases all contain heme as a prosthetic group. The biological utility of heme depends upon the propensity of the Fe moiety to alternate between its two oxidation states (Fe2+ and Fe3+). Due to the toxic (i.e., oxidative) nature of heme, the molecule is not allowed to circulate in its free form within the vertebrate host. When heme-containing molecules are degraded, either as a natural process of metabolism or by inadvertent damage, the heme that is released is rapidly bound by molecules such as hemopexin and albumin. Due to the action of these scavenging molecules, the concentration of free heme within the vertebrate host is maintained at exceedingly low levels (39). Nonetheless, the abundance of heme within the cells and tissues of animals provides invading bacteria with a large potential reservoir of nutrient Fe. Pathogenic bacteria, including Bordetella bronchiseptica (4), Bordetella pertussis (4), Escherichia coli (17), Yersinia spp. (45, 54), Neisseria meningitidis (15), Neisseria gonorrhoeae (15), Haemophilus influenzae (46), Vibrio cholerae (53), Corynebacterium diphtheriae (50), and Shigella spp. (28), have been shown to utilize heme as a source of nutrient Fe.
Bordetella avium, a gram-negative bacterium, is the causative agent of coryza (bordetellosis), an avian upper respiratory tract disease which afflicts turkeys and chickens. While this highly infectious disease can be clinically mild, the mortality of infected flocks may be as high as 75% due to secondary infections (51). Symptoms of B. avium infection include anorexia, exudative conjunctivitis, sneezing, and serous discharge from the nares (22). Pathological changes include rhinitis, sinusitis, bronchopneumonia, and, in severe cases, tracheal collapse (22). Coryza exhibits many similarities in disease presentation to whooping cough, an upper respiratory tract infection caused by B. pertussis, an obligate human pathogen. For that reason, B. avium can be utilized as a model system to study the virulence mechanisms of B. pertussis.
We report the identification of an outer membrane protein (OMP), BhuR, which has amino acid sequence homology to bacterial outer membrane heme receptors and whose expression is essential for the utilization of heme and hemoproteins as sources of nutrient Fe by B. avium. In this study, the capacity of B. avium to acquire and utilize heme and hemoproteins as Fe sources is defined. Furthermore, the role of BhuR in heme transport and the importance of the receptor in virulence are firmly established.
MATERIALS AND METHODS
Strains, antibiotics, and reagents.
The bacterial strains used in this study are listed in Table 1. E. coli strains were grown on Luria-Bertani agar, while B. avium strains were maintained on brain heart infusion (BHI) agar (Difco Laboratories, Detroit, Mich.). Bacteria grown under Fe-replete conditions were cultured in BHI broth supplemented with FeSO4 at a concentration of 144 μM, while bacteria grown under Fe-stressed conditions were cultured in BHI containing 100 μM ethylenediamine di-o-hydroxy-phenylacetic acid [BHI(100 μM EDDHA)]. For growth analysis in BHI broth, addition of 300 μM EDDHA to the medium was used to create conditions of extreme Fe stress, under which B. avium cannot replicate. Unless otherwise noted, ampicillin was used at a concentration of 200 μg/ml, kanamycin was used at 50 μg/ml, tetracycline was used at 10 μg/ml, and rifampin was used at 10 μg/ml. All antibiotics were obtained from Sigma Biochemicals (St. Louis, Mo.). Reagents were purchased from Life Technologies, Inc. (Frederick, Md.) and Sigma Biochemicals. Restriction enzymes and DNA-modifying enzymes were obtained from Fermentas (Hanover, Md.). Deionized water with a resistance of >18 MΩ was used for all solutions.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| B. avium | ||
| 4169 | Wt | 49 |
| 4169rif | Spontaneous rifampin-resistant mutant of 4169 | This study |
| Pho20 | TnphoA insertion into bhuR of 4169rif | 7 |
| 4169rif(bhuR::kan) | 4169rif with a nonpolar kan cassette in bhuR | This study |
| 4169rifhemA | 4169rif with chromosomal deletion in hemA | This study |
| 4169rif(bhuR::kan)hemA | 4169rif(bhuR::kan) with a chromosomal deletion in hemA | This study |
| E. coli | ||
| DH5α | F− ϕ80lacZM15 Δ(lacZTA-argF)I169 deoR recA1 endA1phoA hsdR17 (rK− mK+) supE44 λ−thi-1 gyrA96 relA1 | Life Technologies |
| DH5αmcr | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 phoA supE44 λ−thi-1 gyrA96relA1 | Life Technologies |
| DH5αF′tet | DH5α transconjugate with [F′ proAB lacqZΔM15 Tn10(Tet)r] | Life Technologies |
| SM10λpir | Conjugation helper strain; RP4 plasmid integrated into the chromosome | Life Technologies |
| RK1065 | hemA | 37 |
| RK1065nal | Spontaneous nalidixic acid-resistant mutant of RK1065 | This study |
| Plasmids | ||
| pBluescript KS(+) | Cloning vector; Ampr | Stratagene |
| pBluescript KSII(+) | Stratagene | |
| pMECA | Cloning vector; Ampr | 61 |
| pUFR047 | Cloning vector; Ampr Genr | 13 |
| pCVD442 | Positive selection suicide vector; Ampr | 14 |
| pCVD442tet | pCVD442 derivative containing a tet cassette cloned into SmaI site; Ampr Tetr | This study |
| pCOS5 | Cloning vector; Ampr Chlr | 7 |
| pRK415 | Broad-host-range expression vector; Tetr | 24 |
| p1016 | 45-kbp fragment of 4169 chromosome encoding the rhuIRbhuRSTUV operon in pCOS5 | This study |
| p31 | 30-kbp fragment of 4169 chromosome encoding the rhuIR/bhuRSTUV operon in pCOS5 | This study |
| pL1 | pCOS5 containing 4169 chromosomal fragment encoding wt hemA of B. avium | This study |
| pAD3 | pBluescript KS(+) containing the 5′ terminus of B. avium bhuR and 7.4 kbp of upstream sequence | This study |
| pERM1 | 3-kbp EcoRI fragment of p1016 ligated into pMECA | This study |
| pERM11 | pBluescript KS(+) encoding the 5′ terminus of bhuR of B. avium | This study |
| pERM11.1 | pERM11 with a nonpolar kan cassette insertion into the partial bhuR ORF of B. avium | This study |
| pERM16 | pCVD442 containing the 5′ terminus of bhuR of B. avium interrupted by the kan cassette | This study |
| pERM23 | pBluescript KS(+) encoding wt bhuR of B. avium | This study |
| pERM25 | pRK415 encoding wt B. avium bhuR | This study |
| pERM32 | pBluescript KS(+) encoding the rhuIRbhuRSTUV operon | This study |
| pERM33 | pURF047 containing the rhuIRbhuRSTUV operon | This study |
| pERM34 | pBluescript KS(+) encoding wt hemA of B. avium | This study |
| pERM36 | pCVD442 containing the ΔhemA mutant gene | This study |
Hybridizations.
Colony blot hybridizations were performed using routine protocols (35). Briefly, colonies were replica plated onto appropriate solid agar media. After overnight incubation at 37°C, one set of replicated colonies was transferred to an Optitran BA-S 85 nitrocellulose filter (pore size, 0.45 μm) (Schleicher & Schuell, Keene, N.H.), which was sequentially incubated at room temperature (RT) for 5 min on 3-mm-diameter Whatman paper disks saturated with solutions of (i) 0.1% sodium dodecyl sulfate (SDS), (ii) 1.5 M NaCl plus 0.5 M NaOH, (iii) 1.5 M NaCl plus 0.5 M Tris (pH 8.0), and (iv) 2× SSPE solution (180 mM NaCl, 10 mM NaH2PO4 [pH 7.4], 1 mM EDTA [pH 7.4]). The air-dried filters were baked at 80°C under vacuum (NAPCO model 5831 vacuum oven; Precision Scientific, Chicago, Ill.) for 3 h. For hybridization, filters were incubated at RT in a prehybridization buffer (6× SSPE, 0.5% SDS, 5× Denhardt's solution, 5.0 mM sodium pyrophosphate [NaPPi], and 0.01 M EDTA) for 1 h. The prehybridization solution was replaced by hybridization buffer (6× SSPE, 0.5% SDS, 5× Denhardt's solution, 5.0 mM NaPPi and 0.01 M EDTA) containing a heat-denatured [α-32P]dATP-labeled (DuPont New England Nuclear, Boston, Mass.) polynucleotide probe. The filters were hybridized overnight at 65°C in a shaking water bath (Bellco Glass, Inc., Vineland, N.J.). To remove unhybridized probe, the filters were washed two times for 15 min each in 2× SSPE solution (2× SSPE, 0.5% SDS, 5 mM NaPPi) at RT, for 60 min in 0.1× SSPE solution (0.1× SSPE, 0.5% SDS, 5 mM NaPPi) at 65°C, and for 30 min in 0.1× SSPE solution at 65°C. Each filter was exposed overnight at −80°C to Biomar blue-sensitive autoradiographic film (Marsh Biomedical Products Inc., Rochester, N.Y.). The autoradiographs were developed using Kodak developer and fixative solutions (Sigma Biochemicals).
For Southern blot hybridization, DNA fragments were resolved by electrophoresis in a 0.8% agarose gel and stained with ethidium bromide. The DNA was transferred from the agarose gel to a Nytran membrane filter (pore size, 0.2 μm) (Schleicher & Schuell) by a capillary transfer method (35). DNA was irreversibly bound to the filter using a UVC-308 UV cross-linker (Ultra·L̄um, Carson, Calif.). After incubation in prehybridization buffer for 1 h at RT, the filters were hybridized overnight at 65°C in a shaking water bath (Bellco Glass, Inc.) with a heat-denatured [α-32P]dATP-labeled polynucleotide probe. Following hybridization, the filters were washed two times for 15 min each time with 2× SSPE solution at RT, for 60 min in 0.1× SSPE solution at 65°C, and for 30 min in 0.1× SSPE solution at 65°C. DNA fragments with homology to the radiolabeled probe were detected by autoradiography, as described above.
Isolation of total B. avium RNA.
Total RNA was isolated as previously detailed (6). Briefly, B. avium was cultured to mid-log phase in 12.5 ml of BHI broth containing rifampin (10 μg/ml), EDDHA (100 μM), and hemin (25 μM). Following centrifugation at 16,000 × g for 6 min, the pelleted bacteria were resuspended in 250 μl of 2× lysis buffer (20 mM Tris-HCl [pH 8.0], 20 mM NaCl, 400 μg of proteinase K/ml) and transferred to a chilled microcentrifuge tube containing 300 μl of 10% SDS. Samples were then vortexed, incubated at 37°C for 5 min, and chilled on ice prior to the addition of 300 μl of ice-cold 5 M NaCl. The samples were incubated on ice for 10 min and then centrifuged for 10 min at 16,000 × g. Aliquots (330 μl) were transferred to prechilled microcentrifuge tubes containing 900 μl of ethanol (EtOH). The samples were vortexed and incubated at −80°C for 1 h. Following incubation, the samples were brought to 0°C, and the total nucleic acids were pelleted by centrifugation at 16,000 × g at 4°C for 15 min. The pellets containing RNA and DNA were resuspended in 100 μl of 1 mM EDTA. RNA-DNA solutions were extracted twice with equal volumes of phenol, twice with equal volumes of phenol-chloroform (1:1), and twice with equal volumes of chloroform prior to the addition of 40 μl of 3 M sodium acetate (pH 5.2) and 1 ml of 95% EtOH to precipitate the nucleic acids. Following overnight precipitation at −80°C, the nucleic acid mixture was pelleted by centrifugation at 16,000 × g for 15 min at 4°C. The nucleic acid pellets were washed with ice-cold 75% EtOH, followed by centrifugation at 16,000 × g for 10 min at 4°C. After removal of the EtOH and drying, the nucleic acid pellets were resuspended in 160 μl of water containing 0.05% diethylpyrocarbonate, to which was subsequently added 20 μl of 10× DNase 1 buffer, 132 U of RNasin, and 16 U of DNase 1. The mixture was incubated at 37°C for 20 min prior to the addition of 200 μl of 1 mM EDTA. The reaction mixtures were extracted with an equal volume of phenol-chloroform (1:1), followed by an equal volume of chloroform. One-tenth volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of 95% EtOH were added to the RNA solutions, and the RNA was precipitated for 1 h at −80°C. After being washed with 75% EtOH, the total RNA was stored at −80°C.
RNase protection assay (RPA).
The synthetic oligonucleotide pairs pAD3-35-pAD3-36 (5′-GGA ATT CCG TCT GAG TAC TGC GAT GGC G-3′ [the EcoRI site is underlined] and 5′-CCT CTA GAT ATG GTG TGC AGG TGT TGT CGC CC-3′ [the XbaI site is underlined]) and pAD3-33-pAD3-34 (5′-CGA ATT CCA TTA TTA GTC GGT AGA CTG GTA TA-3′ [the EcoRI site is underlined] and 5′-CCT CTA GAG TAA CCT ATA AAC CCA CTC GGC CTC-3′ [the XbaI site is underlined] were designed to be used as primers in a PCR to amplify the 5′ terminus of bhuR and the intergenic region between bhuR and bhuS from cosmid p1016, respectively (PCR conditions: denaturation for 45 s at 92°C, annealing for 45 s at 50°C, and extension for 60 s at 72°C for 30 cycles in a Perkin-Elmer [Norwalk, Conn.] DNA Thermal Cycler 480). The amplified products were digested with EcoRI and XbaI prior to ligation into the corresponding endonuclease restriction sites of pBluescript SKII(+) (Stratagene, La Jolla, Calif.). The plasmids were designated pERM29 and pERM30, respectively. Radiolabeled RNA hybridization probes 1 and 2 were produced by in vitro transcription using pERM29 and pERM30 as templates (Ambion [Austin, Tex.] T3 MAXIscript transcription kit), respectively.
RPAs were performed using a Hybspeed RPA kit (Ambion) following the vendor's guidelines. Protected fragments were visualized using a PhosphorImager (Bio-Rad, Hercules, Calif.).
Outer membrane isolation.
A modification of the protocol of Leyh and Griffith (31) was used to purify B. avium outer membranes. Briefly, B. avium was cultured at 37°C to stationary phase in 250 ml of BHI broth containing either 100 μM EDDHA (Fe stressed) or 36 μM FeSO4 (Fe replete). The cells were pelleted by centrifugation at 3,000 × g for 20 min at 4°C and resuspended in 15 ml of ice-cold HEPES buffer (10 mM HEPES [pH 7.4] containing 0.1 mM phenylmethylsulfonyl fluoride). The cells were frozen at −80°C overnight, thawed in an ice bath, and immediately stored on ice. Cells packed in ice were sonicated three time for 2 min each using a microtip and a Sonifier 450 ultrasonicator (Branson Ultrasonics Corp., Danbury, Conn.) at a setting of 6 and at a 50% cycle. The sonicate was centrifuged at 3,000 × g for 20 min at 4°C to remove unbroken cells. Total membranes were obtained from the clarified supernatant by centrifugation at 100,000 × g for 60 min at 4°C. Total membranes were resuspended in 2 ml of extraction buffer (1% N-lauroyl-sarcosine in 10 mM HEPES [pH 7.4]) by gentle agitation at RT for 60 min. The insoluble outer membranes were pelleted by centrifugation at 100,000 × g for 60 min at 4°C. The extraction was repeated to remove residual amounts of contaminating cytoplasmic proteins. The OMPs were resuspended in 200 μl of deionized water and stored at −80°C. The total protein concentration of the OMP suspensions was determined with the Bio-Rad protein assay using bovine serum albumin as the standard.
SDS-PAGE analysis of OMPs.
OMPs were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) (27). For each sample, 8 μg of total OMPs was solubilized by being boiled for 10 min in loading buffer (31 mM Tris [pH 6.8]-2% SDS-2.5% 2-mercaptoethanol-10% glycerol). The solubilized proteins were resolved in 7.5% polyacrylamide gels and visualized by staining them with colloidal Coomassie brilliant blue (40).
Cloning and sequencing of bhuR.
The bhuR gene was initially identified by characterizing the insertion site of TnphoA in Pho20. TnphoA contains a unique SalI site which is located asymmetrically to the kanamycin-resistance gene of the transposon. To clone the SalI fragment comprising the fusion joint and flanking regions of the transposon insertion of Pho20, chromosomal DNA of the mutant was digested with SalI. The resulting restriction fragments were ligated into the SalI site of pBluescript KS(+). The ligated plasmids were transformed by osmotic shock into E. coli DH5αF′tet (Life Technologies, Inc.), and transformants were selected for the ability to grow in the presence of kanamycin. One kanamycin-resistant transformant was chosen for further study. The plasmid in this transformant, which contained a 12-kbp SalI insert of B. avium chromosomal DNA, was designated pAD3. The 12-kbp insert of pAD3 was comprised of the leftmost portion of TnphoA, including the Kanr cassette, the phoA gene, the transposon insertion site, and 7.4 kbp of Pho20 chromosomal DNA (data not shown).
A partial sequence of the insertionally mutated gene of Pho20 was obtained by sequencing from the point of the transposon insertion into the B. avium chromosomal DNA of Pho20 in pAD3. A synthetic oligonucleotide homologous to TnphoA (Tnphoterm: 5′-CTG AGC AGC CCG GTT TT-3′) was used to prime a sequencing reaction of pAD3. Using that partial sequence, the synthetic oligonucleotides pAD3-2 (5′-CGTGCGCGGTTCCGA-3′) and pAD3-3 (5′-GGCCGAAGCGGTGGC-3′) were engineered for use as primers in PCR to amplify a 358-bp fragment from pAD3 which was homologous to the chromosomal sequences located 5′ to the transposon fusion joint (PCR conditions: denaturation for 30 s at 92°C, annealing for 45 s at 51°C, and extension for 60 s at 72°C for 30 cycles in a Perkin-Elmer DNA Thermal Cycler 480). The 358-bp fragment was radiolabeled with [α32-P]ATP by random priming (Random Primers DNA-labeling system; Life Technologies) and used as a colony blot hybridization probe to screen by colony blot hybridization a cosmid-based B. avium genomic library for clones containing a cosmid with homology to the insertionally mutated gene of Pho20. A single colony, containing a cosmid designated p1016, was chosen for further study.
To subclone the region containing homology to the insertionally mutated gene of Pho20, p1016 was digested with EcoRI and the restriction fragments were ligated into the EcoRI site of pMECA (61). The ligation reaction was transformed by osmotic shock into E. coli strain DH5αF′tet, and transformants were selected for the ability to grow in the presence of ampicillin. The transformants were screened by colony blot hybridization for homology to the 358-bp radiolabeled hybridization probe to identify a single clone containing a plasmid designated pERM1.
Sequence walking was performed to sequence the insert of pERM1, beginning at the point identified to be the site of the transposon insertion in Pho20 and continuing in a 5′ direction. After a partial open reading frame (ORF) had been identified, the remaining 972 bp of the ORF were obtained by sequencing the insert of p1016.
To isolate the bhuR gene for cloning, p1016 was digested with XhoI and EcoRV. The nucleotide fragments were directionally cloned into pBluescript KS(+) and transformed into E. coli strain DH5αF′tet. Transformants selected for the ability to grow in the presence of ampicillin were screened by colony blot hybridization for homology to bhuR using the bhuR-specific 358-bp radiolabeled hybridization probe. Plasmid pERM23, containing a 2.7-kbp insert comprising the entire 2,565-bp ORF of wild-type (wt) B. avium bhuR, 120 bp of 5′ flanking DNA, and 67 bp of DNA flanking the 3′ terminus of the gene, was obtained. pERM25 was engineered by ligating a KpnI/SstI fragment of pERM23, which included the bhuR ORF, into the corresponding restriction sites of the broad-host-range vector pRK415 (24). Insertion of the KpnI/SstI DNA fragment into pRK415 placed the bhuR gene under the control of the vector's lac promoter.
Cloning the rhuIR/bhuRSTUV locus.
Sequence analysis revealed that the insert of cosmid p1016 included six of the seven predicted genes of the heme uptake locus (63) (i.e., rhuIR and bhuRSTU, but not bhuV). To obtain a cosmid clone which included the complete locus, a 309-bp nucleotide hybridization probe homologous to the 5′ terminus of bhuS, which was obtained by EcoRV digestion of p1016, was used in a colony blot hybridization to screen a cosmid-based B. avium genomic library for a cosmid clone with homology to bhuS. The 30-kbp insert of the bhuS-positive cosmid p31 was confirmed by sequence analysis to include all seven genes of the uptake locus. An 11-kbp DNA fragment containing the locus, isolated by digestion of p31 with BglII and SalI endonucleases, was ligated into the BamHI and SalI sites of pBluescript KS(+) to produce pERM32. For subsequent expression experiments in B. avium, the 11-kbp SalI/SmaI insert was removed from pERM32 and ligated into pUFR047 (13) at the XbaI (blunted by use of the Klenow fragment of DNA polymerase) and SalI sites. The pUFR047-based plasmid including the heme uptake locus was designated pERM33.
Engineering a chromosomal nonpolar bhuR mutation in 4169rif.
A nonpolar mutation of the bhuR gene of B. avium was introduced by allelic exchange into the chromosome of 4169rif, a spontaneous rifampin-resistant mutant of 4169. In brief, a 1.7-kbp truncated copy of bhuR was amplified by PCR from B. avium 4169rif chromosomal DNA using a synthetic oligonucleotide primer homologous to the 5′ terminus of the bhuR ORF (pAD3-11; 5′-TTGGATCCTCATCGATAAAAC-3′ [the BamHI site is underlined]) and a second oligonucleotide primer homologous to internal sequence of the bhuR ORF (pAD3-13; 5′-GACTGCAGGTGAATTCATCCTGGC-3′ [the PstI site is underlined]) (PCR conditions: denaturation for 30 s at 92°C, annealing for 45 s at 44°C, and extension for 2 min 15 s at 72°C for 30 cycles). The amplified DNA fragment including the truncated bhuR gene was directionally cloned into pBluescript KS(+) to create pERM11. A nonpolar kanamycin resistance cassette obtained from Tn5 (T. D. Connell, unpublished data) was ligated into a unique EcoNI site located within the bhuR gene fragment of pERM11 to produce pERM11.1. The DNA fragment of pERM11.1 containing bhuR::kan that was isolated by digestion of the plasmid with BamHI and EcoRI was subsequently directionally cloned into pCVD442, a sacB-based positive-selection suicide vector (14), to produce pERM16. Allelic exchange to replace the wt bhuR with the mutant bhuR::kan allele in 4169rif was performed using a two-step process. In the initial step, pERM16 was mobilized by conjugation into 4169rif using E. coli SM10λpir (Life Technologies, Inc.) as the helper strain (35). Cointegrates were selected for the ability to grow on medium containing ampicillin. Due to the substantial number of false positives produced by the high rate of spontaneous ampicillin resistance, a screening method was employed to identify a positive clone. Colony blot hybridizations of 1,500 first-stage cointegrates were screened for the presence of the kanamycin cassette of pERM16 using the radiolabeled Tn5-derived Kanr cassette as a hybridization probe. The cointegrates were analyzed by Southern hybridization to ensure that pERM16 had integrated properly into the bhuR locus of 4169rif (data not shown). One cointegrate with the proper hybridization pattern was cultured to stationary phase in BHI broth supplemented with rifampin. One milliliter of the stationary-phase culture was inoculated onto BHI agar supplemented with rifampin and 20% sucrose. Of the 92 colonies which grew in the presence of 20% sucrose, 89 were sensitive to ampicillin, an indication of the loss of the vector sequences. One clone, designated 4169rif(bhuR::kan), in which the wt bhuR gene had been replaced with the bhuR::kan allele, was chosen for further study.
Complementation of bhuR mutants.
pRK415- and pUFR047-based recombinants were mobilized into B. avium by conjugation using E. coli SM10λpir as a helper strain. Strains containing only pRK415 or pUFR047 were used as negative controls in all complementation growth assays.
Growth assays.
Initial growth assays in broth cultures were performed using 300-ml acid-washed sidearm flasks (Bellco Glass, Inc.) containing 50 ml of BHI broth. The cultures were inoculated with 500 μl of Fe-stressed BHI(100 μM EDDHA) bacterial cultures which had been cultured to an optical density at 600 nm (OD600) of 1.0 (DU 640B spectrophotometer; Beckman Instruments Inc., Fullerton, Calif.). The cultures were supplemented, as appropriate, with an Fe source or with an Fe chelator. All cultures were incubated at 37°C in an Innova 4330 incubator shaker (New Brunswick Scientific, Edison, N.J.). The cell densities of the cultures at various time points were determined using a Klett-Summerson photoelectric colorimeter (Klett Manufacturing Co., Inc, New York, N.Y.). Other broth culture growth assays and complementation growth assays were performed using 15-ml sterile culture tubes (Laboratory Products Sales, Rochester, N.Y.) containing 3 ml of BHI broth. Each culture was inoculated with 30 μl of an Fe-stressed [BHI(100 μM EDDHA)] seed culture which had been cultured to an OD600 of 1.0 (Beckman DU 640B spectrophotometer). At various time points, 100 μl of each sample was diluted with 100 μl of the appropriate medium within the wells of a 96-well ELISA plate, and the OD600 of the culture was determined using a Titertek Multiskan Plus enzyme-linked immunosorbent assay plate reader (ICN, Irvine, Calif.). In the initial hemin utilization assays, hemin was added to the culture at a final concentration of between 5 and 50 μM. While no growth difference was observed, a concentration of 5 μM hemin was chosen for all subsequent growth experiments.
Engineering a hemA mutation in B. avium.
A cosmid-based genomic library of B. avium 4169, packaged into lambda phage heads using a GigapakII kit (Stratagene) (Connell, unpublished), was transfected into E. coli RK1065nal, a spontaneous nalidixic acid-resistant hemA mutant of E. coli RK1065 (37) which requires supplementation of culture medium with the heme precursor δ-aminolevulinic acid (ALA) for growth. Transfectants expressing hemA were selected by growth on Luria-Bertani agar in the absence of ALA. Of the six positive transfectants, clone pL1 was chosen for further study. A 2-kbp EcoRI fragment of pL1 which included hemA was ligated into the EcoRI recognition site of pBluescript KS(+) to produce pERM34. To confirm that the plasmid conferred a hemA phenotype, pERM34 was transformed into E. coli RK1065 and the recombinants were tested for growth in the absence of ALA.
Gene splicing by overlapping ends (23) was used to engineer a mutant hemA gene for ligation into the suicide vector pCVD442tet, a derivative of pCVD442 (14). pCVD442tet contains a tetracycline resistance cassette cloned into the SmaI site of pCVD442. Using plasmid pERM34 as a template, 450 bp of DNA located 5′ of the ATG start codon and 500 bp of the 3′ terminus and downstream DNA of B. avium hemA were amplified by PCR. For that purpose, the synthetic oligonucleotide primer pairs hemA-6-hemA-5 and hemA-4-hemA-3 were used, respectively (hemA-3, 5′-CCG AGC TCG TCG ACG GTA TAC GAT AAG CT-3′ [the SacI site is underlined]; hemA-4, 5′-AGC GTT TCC CTA CTC CTG CAG GCT GAG GCC ATT ATT GAA AC-3′; hemA-5, 5′-CAG GAG TAG GGA AAC GCT G-3′; and hemA-6, 5′-GTG AGC TCC GCT CTA GAA CTA GTG GAT CAG-3′ [the SacI site is underlined]). The PCR conditions were as follows: denaturation for 45 s at 95°C, annealing for 45 s at 42°C, and extension for 1 min at 72°C for 30 cycles. Each PCR product was purified and mixed together, and the mixture was used as the template in PCR using the oligonucleotides hemA-6 and hemA-3 as primers for a secondary round of DNA amplification (gene splicing by overlapping ends). To produce pERM34, a 1.0-kbp final product, consisting of 500 bp of 5′ flanking DNA to the ATG start codon of hemA, the 3′-terminal 162 bp of the gene, and 162 bp of downstream flanking DNA, was digested with SacI endonuclease, and the fragment was ligated into the unique SacI site of pCVD442tet. pERM36 was mobilized into 4169rif and 4169rif(bhuR::kan) by conjugation as described above (35). Selection of primary cointegrates was accomplished by plating the transconjugates onto BHI agar containing tetracycline (10 μg/ml). Southern hybridizations confirmed that pERM36 had properly integrated into the hemA locus of both 4169rif and 4169rif(bhuR::kan) (data not shown). One cointegrate from each strain with the proper hybridization pattern was cultured to stationary phase in BHI broth supplemented with rifampin. Bacteria were inoculated onto BHI agar supplemented with 20% sucrose and 60 sucrose-resistant colonies from each strain were screened by colony blot hybridization for the loss of the 5′ region of hemA (data not shown). One potential hemA mutant clone from each initial B. avium strain was further characterized by Southern blot hybridization to confirm that wt hemA had been replaced by the mutant hemA allele (data not shown). These hemA mutants were designated 4169rifhemA and 4169rif(bhuR::kan)hemA, respectively.
B. avium virulence studies.
The virulence of wt and mutant B. avium strains was determined using both direct-challenge and competitive-infection assays.
In the direct-challenge infection, cultures of 4169rif and Pho20 were grown in BHI broth supplemented with the metal chelator EDDHA (100 μM) and with 200 μg of streptomycin/ml. Overnight cultures of the bacteria were diluted to 109 CFU/ml. Serial dilutions of B. avium cultures were plated on blood agar plates containing streptomycin on the day of challenge to confirm the inoculum size. Two-day-old turkey poults (n = 30 per experimental group) were challenged with 104 CFU (0.1 ml at 105 CFU/ml) of bacteria per inoculum by droplet instillation in the right nare. After challenge, the poults were observed daily during a 2-week experimental period and scored for clinical signs of coryza.
For the competition infection, cultures of 4169rif and Pho20 were grown as described above. Two-day-old turkey poults were challenged with a mixture of 104 CFU of 4169rif and 104 CFU of Pho20 by instillation into the right nare (0.1 ml per instillation; n = 10 poults per experiment). After euthanasia on day 14, tracheal tissue samples isolated from each poult were weighed and homogenized. Serial 10-fold dilutions of tracheal homogenates were cultured on blood agar plates containing rifampin (10 μM) and streptomycin (200 μM) to select for total B. avium (4169rif plus Pho20) and on blood plates containing rifampin, streptomycin, and kanamycin (50 μM) to select solely for Pho20. The numbers of recovered bacteria were determined by use of a colony counter (MiniCount; Imaging Products International, Simi Valley, Calif.). The number of wt bacteria was calculated by subtracting the number of Pho20 CFU from the total number of CFU. The relative numbers of wt and mutant B. avium CFU were reported as percentages of total B. avium CFU. The percentage for each strain was calculated by calculating the ratio of either wt or mutant CFU isolated to the total number of B. avium CFU isolated and multiplying the value by 100.
Nucleotide sequencing.
Nucleotide sequencing was performed by the CAMBI Nucleotide Sequencing Facility at The University at Buffalo, State University of New York, and by the Biopolymer Facility at Roswell Park Cancer Institute, Buffalo, N.Y.
Nucleotide sequence accession number.
The nucleotide sequence of the rhuIR/bhuRSTUV locus was assigned GenBank accession number AY095952.
RESULTS
B. avium utilizes hemin as an Fe source.
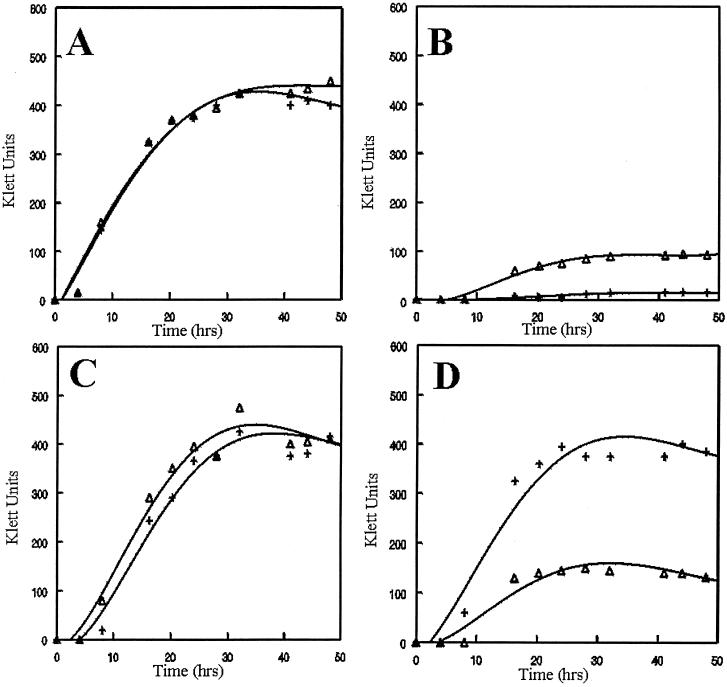
Earlier studies reported that B. pertussis utilizes heme and hemin, the oxidized form of heme, as sources of Fe to support growth (4). Whether this trophic capacity was shared by B. avium was unknown. Growth analysis of wt B. avium 4169rif in medium containing hemin as a sole Fe source was performed to address this question. EDDHA is a high-affinity chelator of Fe and other divalent cations. For this reason, EDDHA has been widely used as a reagent for sequestering Fe in culture media. Empirical studies demonstrated that the addition of 300 μM EDDHA to BHI broth [BHI(EDDHA)] was sufficient to decrease the final culture density of wt B. avium 4169rif from 400 to <50 Klett units (8) (compare Fig. 1A and B). To demonstrate that the inhibition of growth was due to Fe limitation, Fe in the form of ferrous sulfate (FeSO4) was added back to BHI(EDDHA). Addition of 144 μM FeSO4 restored the growth of 4169rif in BHI(EDDHA) to levels similar to that observed for growth in BHI (Fig. 1C). These results indicate that the decrease in growth of the bacterium in BHI(EDDHA) is due to the sequestration of the available Fe within BHI broth into biologically inaccessible chelation complexes. To determine if B. avium has the capacity to utilize hemin as a source of Fe for growth, 4169rif was cultured in BHI(EDDHA) to which hemin was added as the sole source of Fe (EDDHA is unable to remove the coordinately bound Fe from hemin). Culture densities of 4169rif in BHI(EDDHA) supplemented with hemin were similar to the culture density of the bacterium grown either in BHI broth or in BHI(EDDHA) broth supplemented with FeSO4 (Fig. 1D; ≈400 Klett units). These results demonstrated that 4169rif has the potential to utilize hemin as a sole source of Fe for growth.
FIG. 1.
Growth curves demonstrating the growth of 4169rif and Pho20 in culture media containing various Fe sources. (A) BHI broth; (B) BHI(EDDHA); (C) BHI(EDDHA) plus 144 μM FeSO4; (D) BHI(EDDHA) plus 50 μM hemin. While the data in the curves are derived from a single experiment, the growth trends were identical for each strain in two additional replicates. +, 4169rif(pRK415); ▵, Pho20(pRK415).
In a previous study, a mutant library of B. avium 4169rif was produced by introducing the transposon TnphoA into the strain (8). Mutants in the library were originally screened to identify those with TnphoA insertions into Fe-regulated genes. Pho20, one of the several mutants which exhibited an Fe-regulated fusion to phoA, was shown to have a decreased growth rate when cultured in Fe-depleted medium supplemented with turkey serum as an undefined source of biological Fe (8). Since heme, mainly in the form of hemoproteins, was likely a significant source of Fe in turkey serum, Pho20 was subsequently evaluated for the capacity to utilize hemin as a sole source of Fe for growth. The growth rates of Pho20 and 4169rif cultured in BHI broth were indistinguishable (Fig. 1A). The addition of EDDHA to the BHI broth greatly reduced the growth of each strain (Fig. 1B). Supplementation of BHI(EDDHA) broth with FeSO4 rescued the growth of both stains, indicating that the lack of growth seen in BHI(EDDHA) was due to Fe limitation (Fig. 1C). Supplementation of BHI(EDDHA) with hemin rescued the growth of 4169rif but not that of Pho20 (Fig. 1D). These data indicate that the transposon insertion into the Fe-regulated gene of Pho20 significantly impairs the ability of the mutant to utilize hemin as a sole source of nutrient Fe.
Cloning the B. avium heme receptor gene.
To identify the gene of Pho20 into which the transposon had been inserted and to provide genetic tools to clone the wt gene, the fragment of the Pho20 chromosome containing the transposon was cloned and sequenced. Sequencing of the wt gene revealed a 2,565-bp ORF, designated bhuR (for Bordetella heme utilization), which had a coding capacity for a predicted polypeptide of 93 kDa with significant amino acid sequence similarity to bacterial outer membrane heme receptor proteins, including BhuR of B. pertussis (63), PfhR of Pseudomonas fluorescens (44), PhuR of Pseudomonas aeruginosa (43), HupA of Vibrio vulnificus (33), HmbR of N. meningitidis (56), and HutA of V. cholerae (20) (Table 2). A typical signal peptide and an Ala-X-Ala cleavage site at amino acid positions 20 to 22 are evident in the polypeptide sequence, which would be expected for a protein destined for transport to the outer membrane. The projected cleavage of the signal peptide would produce a mature polypeptide of 91 kDa.
TABLE 2.
Amino acid homology of BhuR to other bacterial heme receptors
| Protein | % Identitya | Reference |
|---|---|---|
| B. pertussis BhuR | 64 | 63 |
| P. fluorescens PfhR | 31 | 44 |
| P. aeruginosa PhuR | 30 | 43 |
| V. vulnificus HupA | 28 | 33 |
| N. meningitidis HmbR | 28 | 56 |
| V. cholerae HutA | 27 | 20 |
Percent identity to B. avium BhuR is indicated for each protein. Amino acid homology analysis was performed using the National Center for Biotechnology Information database (http://www.ncbi.nlm.nib.gov) and a TBLAST search strategy.
Domain structure of BhuR.
Additional analysis of the predicted amino acid sequence of BhuR revealed the presence of several structural motifs, each of which is conserved among TonB-dependent outer membrane receptors (2, 34). Most bacterial receptors involved in heme uptake contain consensus amino acid sequences that are required for interaction with TonB, an energy-transducing protein. A motif with homology to the consensus “TonB box,” (D/E/N/F)(S/T)(L/I/V/M/F)(L/I/V/S/T/E/Q)VX(A/G/P)(S/T/A/N/E/Q/P/K) (34),is located in BhuR between amino acid positions 118 and 125 (GSVSQLAP; boldface denotes the conserved amino acids). An additional TonB motif, located between amino acid positions 485 and 502 of BhuR (SDGYDNCPTIAPNLPAPF) matches 6 of 10 conserved amino acids of the amino acid sequence of a secondary TonB motif having the consensus sequence (L/Y/G/S/T/A/N/E)XXX(G/S/T/A/E/N/Q)X(P/G/E)RX (L/I/V/F/Y/W/A)X(L/I/V/M/F/T/A)(S/T/A/G/N/Q)(L/I/V/M/F/Y/G/T/A)X(L/I/V/M/F/Y/W/G/T/A/D/Q)XF (34).
FRAP and NPNL motifs are also conserved among bacterial outer membrane heme receptors (2). Amino acid sequences consistent with a FRAP box (YRAP) were distinguished between amino acids 604 and 607 of BhuR, while an amino acid sequence with homology to an NPNL motif (NPNLKPETSKGWE) was identified between amino acid positions 627 and 639. Unlike other bacterial outer membrane heme receptors, a conserved histidine which is commonly found between the FRAP and NPNL domains was absent in the BhuR amino acid sequence. Rather, a tyrosine (Tyr-616) occupied the position usually held by the histidine. Since tyrosine and histidine have similar biological properties, it is conceivable that the tyrosine at amino acid position 616 is a conservative substitution for heme utilization. Furthermore, BhuR contains a tyrosine residue at the extreme C terminus. The presence of a hydrophobic aromatic residue is highly conserved among proteins which form a β-barrel within the bacterial outer membrane (58).
A unique feature of BhuR in comparison to other bacterial heme receptors is the presence of an extension of 36 amino acids at the N terminus of the protein. While this portion of the protein has no homology to other bacterial heme receptors, with the exception of the closely related heme receptor protein of B. pertussis (63), the region does have homology to the N terminus of FecA, an outer membrane siderophore receptor of E. coli (64). It has been established that the N-terminal domain of FecA interacts in a manner to promote siderophore-dependent regulatory activity of the fec operon (25). It is feasible that a similar heme-dependent regulatory role in the controlled expression of the bhu operon may be inherent in the N-terminal extension of BhuR (26).
bhuR resides in a heme utilization locus.
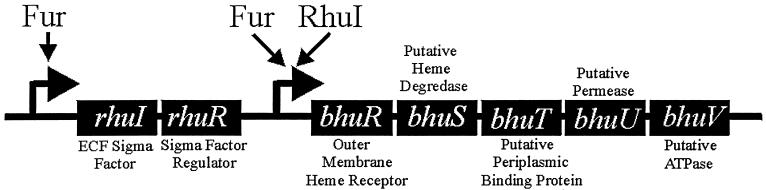
Sequence analysis of DNA flanking bhuR revealed that the gene resides within a heme utilization locus comprising seven cistrons. Genes located downstream of bhuR encode proteins with amino acid homology to a putative heme degradase (bhuS), a periplasmic binding protein (bhuT), a permease (bhuU), and an ATPase (bhuV). Located immediately upstream of bhuR are rhuI and rhuR, two ORFs which encode proteins with homology to FecI and FecR of E. coli (26). RhuI, an extracytoplasmic-function sigma factor, and RhuR, a sigma factor regulator, have been shown to regulate the expression of bhuR in response to the local concentrations of both heme and Fe (26). A locus with similar genetic structure has been reported in B. pertussis (63). An inspection of the arrangement of the genes in the bhu locus indicated that the genes likely reside in a polycistronic operon. Consensus promoter sequences were identified in the DNA located immediately 5′ of rhuIR and in the intergenic region between rhuR and bhuR. Promoter sequences were not evident elsewhere in the locus. A schematic of the B. avium heme utilization locus is depicted in Fig. 2.
FIG. 2.
Schematic representation of the entire B. avium heme utilization locus. Each identified promoter is indicated, along with the regulatory proteins known to mediate expression from the given promoter (26).
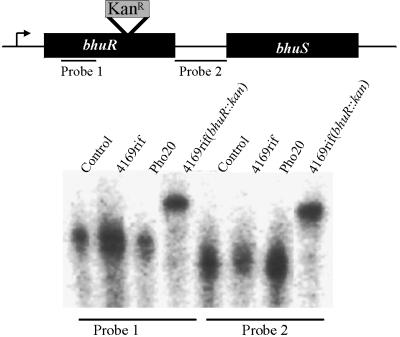
Transposon insertions into operons are known to produce polar effects on genes located downstream of the site of insertion. When it was observed that bhuR resides in an operon, it was possible that the inability of Pho20 to utilize hemin as a source of nutrient Fe was due not to disruption of bhuR but to the alteration in expression of a gene located downstream of the transposon insertion. To ensure that the diminished capacity of Pho20 to utilize hemin was solely a result of a genetic defect in bhuR and not a result of pleiotropic effects caused by polar effects of the transposon insertion on downstream genes, the wt copy of bhuR in the chromosome was replaced with bhuR::kan, an insertionally inactivated, putatively nonpolar allele of the gene. In further demonstration that the insertion of the Kanr cassette did not eliminate the expression of the downstream genes, a transcriptional analysis of bhuR and the downstream bhuS gene was performed using an RPA. Probe 1, corresponding to the 5′ terminus of bhuR, generated and radiolabeled by in vitro transcription, was hybridized with total RNA isolated from 4169rif, Pho20, and 4169rif(bhuR::kan) to confirm that the insertion of the transposon or the kanamycin cassette did not alter transcription of bhuR from the upstream promoter. In both Pho20 and 4169rif(bhuR::kan), a protected RNA fragment of identical size was detected (Fig. 3, probe 1). This fragment was equivalent in size to the fragment protected from 4169rif, indicating that the insertional mutation of bhuR in the two mutants did not interfere, in any dramatic way, with initiation of transcription from a promoter(s) (26) located upstream of bhuR (PbhuR). To evaluate whether polar effects on expression of downstream genes were present in either Pho20 or 4169rif(bhuR::kan), total RNAs from 4169rif and the two mutants were hybridized with a 287-bp DNA fragment (probe 2) corresponding to sequence located immediately downstream of bhuR but upstream of bhuS. In all strains, mutant and wt, the DNA fragment protected an equivalently sized fragment of RNA (Fig. 3, probe 2). These data are consistent with a model in which transcription of a polycistronic mRNA is undisturbed by the insertion of either TnphoA or the Kan cassette into bhuR and the failure of Pho20 and 4169rif(bhuR::kan) (see Fig. 5) to thrive in heme-supplemented medium is due solely to inactivation of bhuR.
FIG. 3.
RNase protection assay demonstrating that the insertions into bhuR in Pho20 and 4169rif(bhuR::kan) are nonpolar. Total RNA isolated from each strain was hybridized to probe 1 or probe 2. Probe 1 was homologous to DNA sequences located 5′ of the insertion site of TnphoA in Pho20 and of the insertion of the Kan resistance cassette in 4169(bhuR::kan); probe 2 was homologous to DNA sequences located in the bhuR-bhuS intergenic region. The control lanes contained only the radiolabeled hybridization probes.
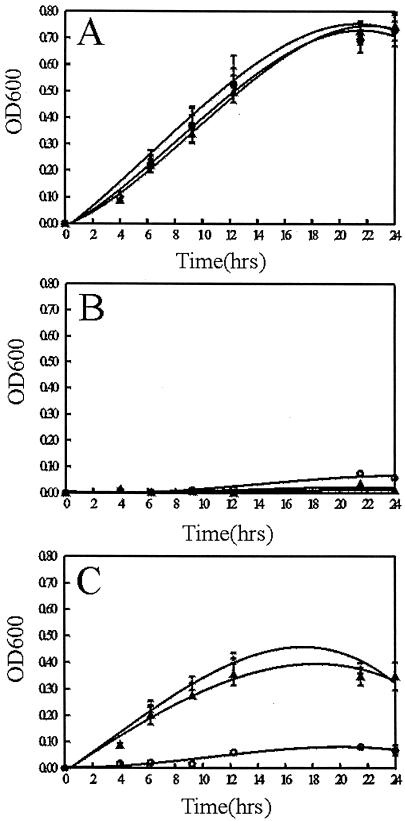
FIG. 5.
Growth curve analysis to determine the role of bhuR in heme utilization. (A) BHI broth; (B) BHI(EDDHA); (C) BHI(EDDHA) plus 5 μM hemin. The error bars represent the standard deviations obtained during three independent growth experiments. +, 4169rif(pUFR047); ○, 4169rif(bhuR::kan)(pUFR047); ▴, 4169rif(bhuR::kan)(pERM33).
BhuR is an OMP.
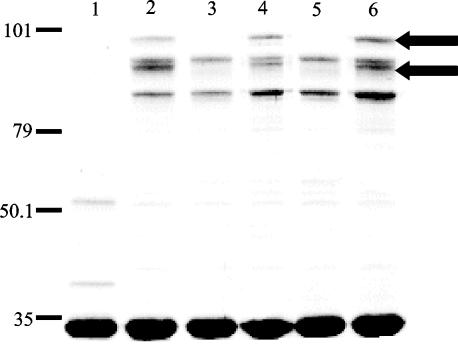
When cultured under Fe-stressed conditions, 4169rif expresses five Fe-regulated OMPs (FeRPs) (8). Insertion of the transposon into bhuR of Pho20 abrogated expression of two of the five FeRPs, specifically the 91- and 82-kDa FeRPs (previously reported as FeRPs with molecular masses of 93 and 84 kDa) (8) (Fig. 4, lane 3). While the 91-kDa polypeptide was likely to be BhuR, as the size of that protein correlated well with the molecular mass of the predicted mature polypeptide encoded by the bhuR gene, the molecular mass of the 82-kDa polypeptide did not correspond to the molecular mass of any of the predicted polypeptides encoded by bhuS, bhuT, bhuU, or bhuV (38, 29, 35, and 28 kDa, respectively). Nevertheless, to determine if the loss of the 82-kDa FeRP was attributable to the transposon insertion, outer membranes isolated from Pho20 were compared to those isolated from 4169(bhuR::kan). SDS-PAGE analysis revealed that Pho20 (Fig. 4, lane 3) and 4169rif(bhuR::kan) (Fig. 4, lane 5) exhibit identical patterns of FeRP expression. Furthermore, complementation with pERM25, a plasmid which encodes wt bhuR, restored expression of the 91- and 82-kDa proteins in both Pho20 and 4169rif(bhuR::kan) (Fig. 4, lanes 4 and 6).
FIG. 4.
SDS-PAGE of the OMPs from B. avium. Lanes: 1, 4169rif cultured in BHI broth containing 144 μM FeSO4; 2, 4169rif cultured in BHI(100 μM EDDHA); 3, Pho20 cultured in BHI(100 μM EDDHA); 4, Pho20(pERM25) cultured in BHI(100 μM EDDHA); 5, 4169rif(bhuR::kan) cultured in BHI(100 μM EDDHA); 6, 4169rif(bhuR::kan)(pERM25) cultured in BHI(100 μM EDDHA). Each lane contained 8 μg of total protein. Molecular masses are indicated in kilodaltons on the left. The positions of the 91-kDa FeRP and the 82-kDa FeRP are indicated by arrows. The gel was stained with colloidal Coomassie brilliant blue.
The loss of expression of the 91- and 82-kDa FeRPs upon inactivation of bhuR and the subsequent complementation of both proteins with bhuR was an unexpected observation that could not be explained by polar effects on expression of downstream genes in the operon. Two alternative models were feasible: (i) expression of the 82-kDa protein is, in some manner, dependent upon expression of the 91-kDa BhuR polypeptide or (ii) bhuR encodes both the 91- and 82-kDa polypeptides. To distinguish between the two models, the 91- and 82-kDa FeRPs were subjected to internal and N-terminal amino acid sequencing (ProSeq, Boxford, Mass.) (data not shown). Analysis of the amino acid sequences confirmed that both the 91- and 82-kDa polypeptides are encoded by bhuR. The 91-kDa form derives from translation of nucleotide sequences comprising the entire bhuR gene; the 82-kDa form is an internal portion of the mature protein whose N terminus is Ser-105 of mature BhuR. It is not yet clear whether the two forms of BhuR are biologically relevant or that the elimination of the 104 amino acids is the result of a programmed proteolytic cleavage. Nonetheless, it cannot be overlooked that this 104-amino-acid segment of BhuR contains the N-terminal 36-amino-acid extension with homology to the FecA domain involved in ligand-dependent signal transduction (25).
Utilization of heme and hemoproteins by B. avium requires bhuR expression.
While bhuR has been shown to have homology to heme receptors and Pho20 has been demonstrated to lack the ability to grow with hemin as the sole source of Fe, the importance of bhuR in heme uptake had not been confirmed using the defined mutant 4169rif(bhuR::kan). To determine if expression of bhuR is required for the utilization of hemin as an Fe source by B. avium, the growth of 4169rif (pUFR047) was compared to the growth of 4169rif(bhuR::kan) and to the growth of 4169rif(bhuR::kan) after the introduction of pERM33, a pURF047-derived plasmid encoding the complete rhu/bhu locus of B. avium. When cultured in BHI broth, 4169rif(pUFR047), 4169rif(bhuR::kan)(pURF047), and 4169rif(bhuR::kan)(pERM33) exhibited equivalent growth (Fig. 5A). Supplementation of BHI broth with the Fe chelator EDDHA at 300 μM [BHI(EDDHA)] inhibited the growth of all three strains (Fig. 5B). Supplementation of BHI(EDDHA) broth with hemin rescued the growth of 4169rif(pURF047) but did not restore the growth of 4169rif(bhuR::kan)(pUFR047). In contrast, when BHI(EDDHA) was supplemented with hemin, the culture densities of 4169rif(bhuR::kan)(pERM33) were equivalent to that of 4169rif(pUFR047) (Fig. 5C). These data are consistent with the model that expression of bhuR is essential for the utilization of hemin as a sole source of Fe by B. avium.
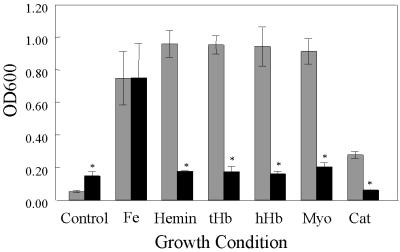
Heme is rarely found in its free form in the body. Rather, the molecule is usually coordinately bound to proteins such as hemoglobin, myoglobin, and catalase. To determine whether B. avium has the capacity to utilize hemoproteins as sources of nutrient Fe, 4169rif and 4169rif(bhuR::kan) were compared in an end point growth assay (8) in which the culture medium had been supplemented with either hemin or a hemoprotein. The relative growth potential of each strain was determined by measuring the OD600 of individual stationary-phase cultures (Fig. 6). At 300 μM, EDDHA inhibits growth of both 4169rif and 4169rif(bhuR::kan) (Fig. 6, Control). To ensure that the lack of growth observed in BHI(EDDHA) was due to the chelation of Fe, the chelated broth was supplemented with FeSO4 (Fig. 6, Fe). Under these conditions, 4169rif and 4169rif(bhuR::kan) displayed equivalent growth. Supplementation of BHI(EDDHA) with either hemin (Fig. 6, Hemin), human hemoglobin (Fig. 6, hHb), turkey hemoglobin (Fig. 6, tHb), myoglobin (Fig. 6, Myo), or catalase (Fig. 6, Cat) supported significantly less growth of mutant 4169rif(bhuR::kan) than of 4169rif. These data support the conclusion that expression of bhuR is essential for the utilization of heme and, furthermore, establish that efficient acquisition of Fe from hemoproteins requires expression of bhuR.
FIG. 6.
End point growth analysis of 4169rif and 4169rif(bhuR::kan) in Fe-depleted media supplemented with various heme-containing Fe sources. Control, BHI (EDDHA); Fe, BHI(EDDHA) plus 144 μM FeSO4; Hemin, BHI(EDDHA) plus 5 μM heme; tHb, BHI(EDDHA) plus 1.25 μM turkey hemoglobin; hHb, BHI(EDDHA) plus 1.25 μM human hemoglobin; Myo, BHI(EDDHA) plus 5 μM myoglobin; Cat, BHI(EDDHA) plus 5 μM catalase. With the exception of FeSO4, the concentration of each Fe source was equivalent to 5 μM Fe. The mean (±1 standard deviation from the mean, as indicated by the error bars) is shown for each value. An asterisk indicates that the growth of 4169rif(bhuR::kan) was significantly different (P < 0.05) from the growth of 4169rif cultured under identical conditions, as determined by Student's t test.
B. avium transports heme into the cytoplasm of the cell.
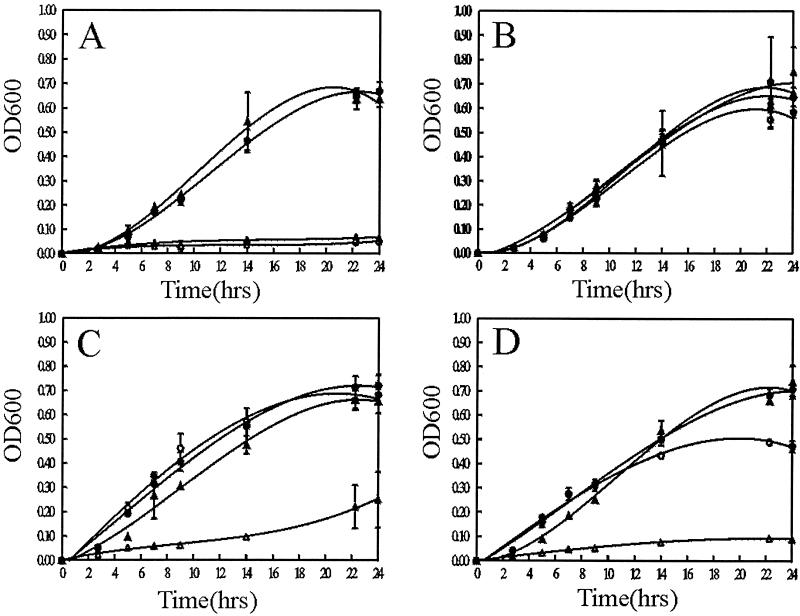
Although previous experiments established that BhuR is critical for heme utilization by B. avium, the mechanism by which the receptor and its accessory factors internalized Fe from heme was not evident. One possibility was that heme, after binding to BhuR, is degraded at the cell surface. After destruction of the heme ring, the freed Fe is transported into the cell. In contrast, binding of heme to BhuR may trigger a process in which the intact heme molecule is transported into the cytoplasm of the cell. In the latter case, the heme could be utilized as an intermediate in various heme-dependent metabolic pathways. Intracellular uptake of intact heme has been established in several bacteria, including N. meningitidis (30), H. influenzae (11), and Yersinia enterocolitica (54, 55). To evaluate whether B. avium has the capacity to internalize heme in a BhuR-dependent manner, hemA mutants which lacked the capacity to synthesize heme were engineered in 4169rif and 4169rif(bhuR::kan) (Fig. 7A and B). Growth of the mutants could be rescued only by supplementation of the culture medium with ALA, the product of the HemA enzyme (Fig. 7A and B). To test whether B. avium had the capacity to internalize heme, 4169rif, 4169rifhemA, 4169rif(bhuR::kan), and 4169rif(bhuR::kan)hemA were cultured in BHI(100 μM EDDHA) broth with and without hemin supplementation. These experiments were performed in BHI(100 μM EDDHA) broth, a condition of Fe stress under which wt B. avium will grow moderately and under which BhuR is optimally expressed. As expected, only 4169rif and 4169rif(bhuR::kan) thrived in BHI(100 μM EDDHA) (Fig. 7A). When ALA was added to the medium, all four strains grew well (Fig. 7B). Addition of hemin to the BHI(100 μM EDDHA) broth rescued the growth of 4169hemA but did not rescue the growth of 4169rif(bhuR::kan)hemA (Fig. 7C). These experiments established that B. avium had the capacity to internalize heme and that bhuR was required for the process. Similar experiments were conducted using hemoglobin to determine if the molecular context in which the heme was present would influence the bhuR-dependent intracellular uptake of the molecule. Again, while 4169rif, 4169rifhemA, and 4169rif(bhuR::kan) grew well in BHI(100 μM EDDHA) supplemented with hemoglobin, 4169rif(bhuR::kan)hemA failed to thrive (Fig. 7D).
FIG. 7.
B. avium transports heme into the cell. (A) BHI(100 μM EDDHA); (B) BHI(100 μM EDDHA) supplemented with 50 μM ALA; (C) BHI(100 μM EDDHA) plus 5 μM hemin; (D) BHI(100 μM EDDHA) plus 10 μM turkey hemoglobin. The error bars represent the standard deviations obtained during three independent growth experiments. •, 4169rif; ▴, 4169rif(bhuR::kan); ○, 4169rifhemA; ▵, 4169rif(bhuR::kan)hemA.
BhuR is a virulence-associated protein.
The capability of utilizing heme and hemoglobin as sources of nutrient Fe (or nutrient heme) is an important virulence determinant in several gram-negative bacteria, including N. meningitidis (56), Haemophilus ducreyi (52), and E. coli (62). To determine whether bhuR is essential for pathogenesis, experiments to compare the virulence of the parent strain to that of the bhuR mutant Pho20 were performed using a turkey challenge model. Mutant strain Pho20 was utilized for the virulence studies since, unlike 4169rif(bhuR::kan), this mutant can be distinguished from wt 4169rif by growth in the presence of kanamycin. Furthermore, Pho20 has been shown to be phenotypically identical to the fully characterized mutant 4169rifbhuR::kan in each in vitro analysis (data not shown). Turkey poults were inoculated intranasally with 4169rif or Pho20. Over a period of 2 weeks, the birds were scored daily for the presentation of clinical signs of coryza. Unexpectedly, the birds challenged with either strain presented with equivalent levels of clinical symptoms. These data indicated that inactivation of bhuR alone did not abolish the ability of B. avium to produce upper respiratory tract disease in turkeys.
As a more sensitive means to evaluate survival of the organism in vivo, birds were also infected with mixtures of wt and mutant strains in a competitive-challenge model. In this model, turkey poults were simultaneously challenged intranasally with equal mixtures of 4169rif and Pho20 (104 CFU each). After 14 days, 90.15% (±19.8 standard deviation from the mean) of B. avium cells recovered from the tracheas of infected poults were the wt strain. Only 9.95% (±19.8; statistical difference [P < 0.05] from the percentage of the total obtained for 4169rif; Student's t test) of the total B. avium cells recovered from the infected birds were the bhuR mutant. These data strongly indicated that the bhuR mutant is competitively inferior to its wt parent in its capacity to colonize the tracheas of poults.
DISCUSSION
Without a continuous source of Fe, invading bacterial cells cannot proliferate within a host. Without proliferation, disease cannot ensue, nor can the pathogen survive. Thus, there is a strong selective pressure for bacterial pathogens to evolve efficient mechanisms to extract Fe from the various complexes into which the host has sequestered the element. Heme is one of the most abundant forms of organic Fe in animals. It is not unreasonable that pathogenic bacteria would have devised efficient systems to utilize this rich source of an essential nutrient. In this study, a genetic locus in B. avium was identified that encodes an efficient heme-hemoprotein acquisition and utilization system. BhuR, an OMP whose expression is regulated in response to the local concentrations of Fe and heme (26), was established as an essential component of this multigenic uptake system and, furthermore, was shown to be indispensable for transport of the heme molecule into the bacterial cell. Inactivation of bhuR diminished the capacity of the bacterium to compete with a bhuR-proficient strain of B. avium in a turkey model of infection, thus establishing its role in virulence.
The molecular mechanism by which heme receptors translocate heme-bound Fe into the cell has not been elucidated to any great degree. A common trend, however, is that heme receptors are present in the outer membranes of gram-negative bacteria as a single polypeptide moiety. In contrast, BhuR may exist in two forms on the surface of B. avium. The outer membranes of Fe-stressed B. avium cells harbor two forms of BhuR: a 91-kDa form and an 82-kDa form. A striking difference between the two forms of the protein is that the smaller form lacks a 36-amino-acid N-terminal extension which is present in the 91-kDa form. Notably, this 36-amino-acid segment is absent from the amino acid sequences of other bacterial heme receptors. The segment does, however, have amino acid homology with the N terminus of FecA, an outer membrane ferric citrate receptor of E. coli (64). In a manner similar to that of the expression of bhuR (26), expression of fecA is controlled by an extracytoplasmic-function sigma factor (FecI) and an anti-sigma factor (FecR) which respond simultaneously to the local concentrations of Fe and ferric citrate, the specific receptor ligand of FecA (25, 43, 48). To respond to the presence of ferric citrate in the local environment, the N terminus of FecA must interact with some component of the FecI-FecR signal cascade. When ferric citrate is detected by the FecA-FecI-FecR complex, expression of the polycistronic FecABCDE locus is enhanced (25). It is provocative to speculate that the 36-amino-acid segment of BhuR has a role in BhuR-RhuI-RhuR-dependent, heme-induced expression of the bhu locus (bhuRSTUV). Experiments to evaluate this regulatory model are being vigorously pursued (T. D. Connell and A. Kirby, unpublished results).
In contrast to the unelucidated function of the N-terminal extension, BhuR harbors other conserved motifs whose potential biological functions are discernible from structural comparisons to other bacterial heme receptors. Two structural motifs that are conserved among proteins which functionally interact with TonB, an energy transducer and common component of Fe uptake machinery in bacterial cells (3), are also conserved in BhuR. The acquisition and utilization of heme as a source of nutrient Fe has been shown to be a TonB-dependent process in various bacteria (1, 16, 37, 42, 47, 54, 60, 62). The presence of two conserved TonB motifs within BhuR, in combination with the observation that TonB is required for heme utilization by B. pertussis (41, 47), suggests that a functional interaction between BhuR and TonB may be necessary for the operation of the bhu-encoded heme utilization system. This hypothesis is further strengthened by the observation that introduction of the genes encoding BhuR or the genes constituting the entire locus (bhuRSTUV) did not complement RK1065, a tonB-proficient hemA mutant of E. coli (37) (data not shown). Failure of bhuR or the bhuRSTUV locus to confer heme uptake upon RK1065 may be explained by the inability of the B. avium heme uptake system to interact in a productive manner with the TonB system of E. coli. Other studies confirmed that heterologous combinations of tonB genes and genes encoding Fe uptake systems from different species are often dysfunctional (20). Experiments are ongoing to engineer a tonB mutant of B. avium to directly evaluate the functional dependence of BhuR on TonB.
Although bhuR is essential for efficient utilization of heme and hemoproteins as sole sources of Fe by B. avium, growth experiments confirmed that disruption of the gene in Pho20 and in 4169rif(bhuR::kan) did not totally abrogate heme utilization (Fig. 1D and 5C). Several bacterial pathogens, including H. influenzae (9, 10, 18, 38), N. meningitidis (30, 57), P. aeruginosa (29, 43, 44), and V. cholerae (20, 36), have been shown to express multiple heme utilization systems. It is likely that the residual growth observed for Pho20 and 4169rif(bhuR::kan) in culture medium containing hemin as the sole Fe source is promoted by expression of a secondary, albeit lower-affinity, heme utilization system.
Due to potent toxicity caused by its oxidative potential, accumulation of unsequestered heme within a cell is lethal. Cells, therefore, have evolved methods to inhibit inordinate uptake of heme. For example, genes encoding heme acquisition proteins are coordinately regulated to prevent the lethal accumulation of heme. Disruption in the stoichiometric levels of expression of a gene within a bacterial heme utilization regulon has been demonstrated to have dire effects on cell survival (21, 55). The introduction of pERM25, a multicopy expression plasmid encoding recombinant BhuR, into 4169rif(bhuR::kan) was correlated with a decreased rate of growth of the cell in culture medium in which heme was the sole source of available Fe (data not shown). In contrast, normal growth was reestablished for 4169rif(bhuR::kan) when a plasmid containing the entire rhuIR-bhuRSTUV locus was introduced into the mutant (Fig. 5C). These results supported the model that stoichiometric expression of all seven bhu genes is necessary to avoid heme toxicity of the cell. Supporting data was obtained when p2025 was obtained from compensatory mutants which arose in the culture. p2025 harbors a spontaneous mutation within the vector sequences of pERM25. Compared to the level of expression of BhuR in 4169rif(bhuR::kan)(pERM25), expression of the heme receptor was dramatically less in 4169rif(bhuR::kan)(p2025). Yet, in contrast to 4169rif(bhuR::kan)(pERM25), the growth rate of 4169rif(bhuR::kan)(p2025) in heme-containing culture medium was closer to that observed for 4169rif, the wt parent (data not shown). A similar pattern of expression and growth inhibition (e.g., toxicity) was reported for B. pertussis when a chromosomal bhuR mutant was complemented with a recombinant plasmid expressing only bhuR (63); the bhuR mutant of B. pertussis could be complemented only by the introduction of a plasmid containing the entire bhuRSTUV locus. In that study, the authors proposed that the failure to complement the mutant with a recombinant bhuR plasmid was due to polar effects of the chromosomal bhuR mutation that altered the expression of downstream bhuSTUV genes (63). In the case of B. avium, RNase protection experiments demonstrated that polar effects on downstream genes were not significant in either Pho20 or 4169(bhuR::kan). Without similar analysis of the expression of the downstream genes in the bhuR mutants of B. pertussis, it cannot be ascertained whether the inability to complement with recombinant bhuR can be attributed to polar effects. From the data obtained in this study, it is more likely that the failure to complement the bhuR mutant of B. pertussis with bhuR was the result of lethal events caused by excessive uptake of heme in the absence of adequate levels of “detoxifying” components encoded by the accessory genes in the locus.
Several bacterial outer membrane heme receptors have been shown to directly bind heme and/or hemoglobin (12, 37, 42, 57, 65, 67). While a physical interaction between B. avium BhuR and heme (or hemoglobin) could not be demonstrated in this study, the experiments reported here using the hemA mutant provide strong evidence that BhuR is essential for the transport of heme into the bacterial cell. The inability to demonstrate heme binding to BhuR in vitro may indicate that the binding affinity for the molecule is low or that the conditions used for binding were inadequate. It is intuitive that there is at least some transient interaction of heme with BhuR. The present model, based on the hemA data, the comparative homology of BhuR to other heme receptors, and the outer membrane localization of the protein, is that BhuR binds extracellular heme prior to initiating transport of the molecule into the cytoplasm of the cell.
The ability to acquire Fe from the host is considered to be an important virulence determinant for most bacterial pathogens (66). This belief has been substantiated in a variety of bacteria, including Streptococcus pneumoniae (59), V. cholerae (19), N. meningitidis (56), and E. coli (62). In like manner, heme uptake systems in B. avium appear to be a factor contributing to virulence. Inactivation of the bhuR gene was correlated with a reduced ability of the mutant strain to compete with its wt parent for colonization of the trachea during infection. Nevertheless, inactivation of bhuR did not absolutely abrogate virulence in B. avium. These data suggest that the Bhu system is not the sole means by which B. avium acquires heme during infection. It is apparent that for many bacterial pathogens the need to acquire heme during infection must be an inordinately strong selective pressure. Other bacterial pathogens encode multiple heme uptake systems (9, 10, 18, 20, 29, 30, 36, 38, 43, 44, 57), which is likely a reflection of the importance of heme uptake in their life cycles. Thus, it is not unreasonable to hypothesize that B. avium encodes a second heme utilization system for acquisition of heme in the absence of BhuR and that deletion of both systems may be required before virulence is totally abrogated.
A gene whose product has amino acid homology to BhuR has been identified in B. pertussis and in B. bronchiseptica (63). The high degree of conservation throughout the pathogenic members of the Bordetellae suggests an essential role for bhuR in survival and pathogenesis.
Acknowledgments
We thank Amy E. Kirby, Jason P. Folster, and Anthony J. Serrio for their creative input and critical review of the manuscript. We thank Shelley M. Payne for generously supplying us with E. coli strain RK1065.
This work was supported by funds made available to E.R.M. by training grant AI07614-01 from the National Institutes of Health; by funds made available to T.D.C. by the School of Medicine and Biomedical Sciences at the University at Buffalo, State University of New York; and by funds made available to P.E.O. and R.E.S. by the U.S. Department of Agriculture.
Editor: J. T. Barbieri
REFERENCES
- 1.Biswas, G. D., J. E. Anderson, and P. F. Sparling. 1997. Cloning and functional characterization of Neisseria gonorrhoeae tonB, exbB and exbD genes. Mol. Microbiol. 24:169-179. [DOI] [PubMed] [Google Scholar]
- 2.Bracken, C. S., M. T. Baer, A. Abdur-Rashid, W. Helms, and I. Stojiljkovic. 1999. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J. Bacteriol. 181:6063-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 4.Brickman, T. J., and S. K. Armstrong. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J. Bacteriol. 181:5958-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullen, J. J., H. J. Rogers, and E. Griffiths. 1978. Role of iron in bacterial infection. Curr. Top. Microbiol. Immunol. 80:1-35. [DOI] [PubMed] [Google Scholar]
- 6.Chauhan, S., D. E. Titus, and M. R. O'Brian. 1997. Metals control activity and expression of the heme biosynthesis enzyme delta-aminolevulinic acid dehydratase in Bradyrhizobium japonicum. J. Bacteriol. 179:5516-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connell, T. D., A. J. Martone, and R. K. Holmes. 1995. A new mobilizable cosmid vector for use in Vibrio cholerae and other gram-negative bacteria. Gene 153:85-87. [DOI] [PubMed] [Google Scholar]
- 8.Connell, T. D., A. Dickenson, A. J. Martone, K. T. Militello, M. J. Filiatraut, M. L. Hayman, and J. Pitula. 1998. Iron starvation of Bordetella avium stimulates expression of five outer membrane proteins and regulates a gene involved in acquiring iron from serum. Infect. Immun. 66:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cope, L. D., R. Yogev, U. Muller-Eberhard, and E. J. Hansen. 1995. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177:2644-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cope, L. D., S. E. Thomas, Z. Hrkal, and E. J. Hansen. 1998. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect. Immun. 66:4511-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulton, J. W., and J. C. S. Pang. 1983. Transport of haemin by Haemophilus influenzae type b. Curr. Microbiol. 9:93-98. [Google Scholar]
- 12.Dashper, S. G., A. Hendtlass, N. Slakeski, C. Jackson, K. J. Cross, L. Brownfield, R. Hamilton, I. Barr, and E. C. Reynolds. 2000. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J. Bacteriol. 182:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeFeyter, R., C. I. Kado, and D. W. Gabriel. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65-72. [DOI] [PubMed] [Google Scholar]
- 14.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyer, D. W., E. P. West, and P. F. Sparling. 1987. Effects of serum carrier proteins on the growth of pathogenic neisseriae with heme-bound iron. Infect. Immun. 55:2171-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elkins, C., P. A. Totten, B. Olsen, and C. E. Thomas. 1998. Role of the Haemophilus ducreyi Ton system in internalization of heme from hemoglobin. Infect. Immun. 66:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths, E. 1987. The iron-uptake systems of pathogenic bacteria, p. 69-137. In J. J.Bullen and E. Griffiths (ed.), Iron and infection, molecular, clinical and physiological aspects. John Wiley, London, United Kingdom.
- 18.Hanson, M. S., C. Slaughter, and E. J. Hansen. 1992. The hbpA gene of Haemophilus influenzae type b encodes a heme-binding lipoprotein conserved among heme-dependent Haemophilus species. Infect. Immun. 60:2257-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, D. P., and S. M. Payne. 1994. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun. 62:5120-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, D. P., and S. M. Payne. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 176:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, D. P., E. E. Wyckoff, C. E. Rashidi, H. Verlei, and A. L. Oldham. 2001. Characterization of the Plesiomonas shigelloides genes encoding the heme iron utilization system. J. Bacteriol. 183:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinz, K. H., G. Glunder, and H. Luders. 1978. Acute respiratory disease in turkey poults caused by Bordetella bronchiseptica-like bacteria. Vet. Rec. 103:262-263. [DOI] [PubMed] [Google Scholar]
- 23.Horton, R. M. 1997. In vitro recombination and mutagenesis of DNA, p. 141-149. In B. A. White (ed.), PCR cloning protocols from molecular cloning to genetic engineering. Humana Press, Totowa, N.J.
- 24.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 25.Kim, I., A. Stiefel, S. Plantor, A. Angerer, and V. Braun. 1997. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol. Microbiol. 23:333-344. [DOI] [PubMed] [Google Scholar]
- 26.Kirby, A. E., D. J. Metzger, E. R. Murphy, and T. D. Connell. 2001. Heme utilization in Bordetella avium is regulated by RhuI, a heme-responsive extracytoplasmic function sigma factor. Infect. Immun. 69:6951-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lawlor, K. M., P. A. Daskaleros, R. E. Robinson, and S. M. Payne. 1987. Virulence of iron transport mutants of Shigella flexneri and utilization of host iron compounds. Infect. Immun. 55:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letoffe, S., V. Redeker, and C. Wandersman. 1998. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol. Microbiol. 28:1223-1234. [DOI] [PubMed] [Google Scholar]
- 30.Lewis, L. A., M. H. Sung, M. Gipson, K. Hartman, and D. W. Dyer. 1998. Transport of intact porphyrin by HpuAB, the hemoglobin-haptoglobin utilization system of Neisseria meningitidis. J. Bacteriol. 180:6043-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyh, R., and R. W. Griffith. 1992. Characterization of the outer membrane proteins of Bordetella avium. Infect. Immun. 60:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litwin, C. M., and B. L. Byrne. 1998. Cloning and characterization of an outer membrane protein of Vibrio vulnificus required for heme utilization: regulation of expression and determination of the gene sequence. Infect. Immun. 66:3134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundrigan, M. D., and R. J. Kadner. 1986. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB. J. Biol. Chem. 261:10797-10801. [PubMed] [Google Scholar]
- 35.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 37.Mills, M., and S. M. Payne. 1995. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J. Bacteriol. 177:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morton, D. J., P. W. Whitby, H. Jin, Z. Ren, and T. L. Stull. 1999. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect. Immun. 67:2729-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller-Eberhard, U. 1970. Hemopexin. N. Engl. J. Med. 283:1090-1094. [DOI] [PubMed] [Google Scholar]
- 40.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 41.Nicholson, M. L., and B. Beall. 1999. Disruption of tonB in Bordetella bronchiseptica and Bordetella pertussis prevents utilization of ferric siderophores, haemin and haemoglobin as iron sources. Microbiology 145:2453-2461. [DOI] [PubMed] [Google Scholar]
- 42.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 43.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 93:4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185-198. [DOI] [PubMed] [Google Scholar]
- 45.Perry, R. D., and R. R. Brubaker. 1979. Accumulation of iron by yersiniae. J. Bacteriol. 137:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pidcock, K. A., J. A. Wooten, B. A. Daley, and T. L. Stull. 1988. Iron acquisition by Haemophilus influenzae. Infect. Immun. 56:721-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pradel, E., N. Guiso, F. D. Menozzi, and C. Locht. 2000. Bordetella pertussis TonB, a Bvg-independent virulence determinant. Infect. Immun. 68:1919-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pressler, U., H. Staudenmaier, L. Zimmermann, and V. Braun. 1988. Genetics of the iron dicitrate transport system of Escherichia coli. J. Bacteriol. 170:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rimler, R. B., and D. G. Simmons. 1983. Differentiation among bacteria isolated from turkeys with coryza (rhinotracheitis). Avian Dis. 27:491-500. [PubMed] [Google Scholar]
- 50.Schmitt, M. P. 1997. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simmons, D. G., L. P. Rose, F. J. Fuller, L. C. Maurer, and G. H. Luginbuhl. 1986. Turkey coryza: lack of correlation between plasmids and pathogenicity of Bordetella avium. Avian Dis. 30:593-597. [PubMed] [Google Scholar]
- 52.Stevens, M. K., S. Porcella, J. Klesney-Tait, S. Lumbley, S. E. Thomas, M. V. Norgard, J. D. Radolf, and E. J. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoebner, J. A., and S. M. Payne. 1988. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect. Immun. 56:2891-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stojiljkovic, I., and K. Hantke. 1992. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 11:4359-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stojiljkovic, I., and K. Hantke. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 56.Stojiljkovic, I., V. Hwa, M. L. de Saint, P. O'Gaora, X. Nassif, F. Heffron, and M. So. 1995. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531-541. [DOI] [PubMed] [Google Scholar]
- 57.Stojiljkovic, I., J. Larson, V. Hwa, S. Anic, and M. So. 1996. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J. Bacteriol. 178:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Struyve, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 59.Tai, S. S., C. J. Lee, and R. E. Winter. 1993. Hemin utilization is related to virulence of Streptococcus pneumoniae. Infect. Immun. 61:5401-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Requirement of the Pseudomonas aeruginosa tonB gene for high-affinity iron acquisition and infection. Infect. Immun. 68:4498-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomson, J. M., and W. A. Parrott. 1998. pMECA: a cloning plasmid with 44 unique restriction sites that allows selection of recombinants based on colony size. BioTechniques 24:922-924. [DOI] [PubMed] [Google Scholar]
- 62.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagegg, W., and V. Braun. 1981. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein FecA. J. Bacteriol. 145:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 66.Weinberg, E. D. 1978. Iron and infection. Microbiol. Rev. 42:45-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyckoff, E. E., D. Duncan, A. G. Torres, M. Mills, K. Maase, and S. M. Payne. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28:1139-1152. [DOI] [PubMed] [Google Scholar]