Abstract
RNA interference silencing of up to 90% of Arp3 protein expression, a major subunit of the Arp2/3 complex, proportionately decreases the intracellular motility of Listeria monocytogenes and actin nucleation activity ascribable to the Arp2/3 complex in mouse embryonic fibroblasts. However, the Arp2/3-deficient cells exhibit unimpaired lamellipodial actin network structure, translational locomotion, spreading, actin assembly, and ruffling responses. In addition, Arp3-silenced cells expressing neural Wiskott-Aldrich syndrome protein-derived peptides that inhibit Arp2/3 complex function in wild-type cells retained normal PDGF-induced ruffling. The Arp2/3 complex can be dispensable for leading-edge actin remodeling.
Keywords: RNA interference, motility, ruffling
Activation of the seven-subunit Arp2/3 protein complex causes Listeria monocytogenes and other intracellular pathogens to form actin “rocket tails” (1) and generates certain actin filament morphologies in yeast (2), Drosophila (3), and Caenorhabditis elegans (4) as well as in podosomes (5) and integrin adhesion sites (6) of mammalian cells. A prevalent theory holds that the Arp2/3 complex assembles actin into an expanding array of actin filaments branching off one another at a 70° angle that radially pushes out the plasma membrane to extend actin-rich protrusions such as lamellipodia and ruffles at the leading edges of migrating cell (7). If this mechanism is responsible for leading-edge membrane protrusion, reducing the cellular content of the Arp2/3 complex ought to have an adverse effect on cell morphology and motility. We tested this prediction by suppressing Arp2/3 complex expression and by combining this suppression with the introduction of Arp2/3 function-inhibitory polypeptides in fibroblasts.
Materials and Methods
Construction of RNA Interference Vectors. A blast search, using the human Arp3 cDNA sequence (AF006083 chr.2 Homo sapiens), retrieved the Mus musculus Arp3 gene sequence (clone MGC:7062) on the Celera database (www.ensembl.org/Mus_musculus). Short hairpin RNAs (shRNAs) were directed against exon 3, exon 9, and the 3′ UTR of mouse Arp3 mRNA. Expression of the shRNAs was driven by RNA polymerase III, by amplifying the corresponding DNA hairpins under the control of the U6 promoter with a PCR-SHAGging strategy (8). The last 21 nt of the oligonucleotides corresponded to the U6 promoter, and the sequences directed against exon 3, exon 9, and the 3′ UTR, respectively, are 5′-AAAAAAGCTCCACCACCAATGAAGAAGCCTAGGCCCAAGCTTCGACCTAGACTTCTTCATTGGTGATGAAGCGGTGTTTCGTCCTTTCCACAA-3′, 5′-AAAAAAGCAATTCTGAATAACCTCATCCACAACCTC AAGCTTCAAGTTGTAGATGAAGTCATTCAGAATTGCG GTGTTTCGTCCTTTCCACAA-3′, and 5′-AAAAAAGCTCTCCTCCACATCCGCATGCTGACACACAAGCTTCTGTGTCAACATGCAGATGTAGAAGAGAGCGGTGTTTCGTCCTTTCCACAA-3′.
The synthesized oligonucleotides (Invitrogen) were used in a PCR reaction in combination with the pENTR/D-Topo SP6 primer (5′-CACCGATTTAGGTGACACTATAG-3′) with a pGEM/U6 (PCR-SHAG, Promega) plasmid as template (8). The amplified product was placed in the pENTR/D-Topo (Invitrogen) plasmid by TOPO cloning. The vectors were subsequently shuffled into the mammalian expression vector pMSCV puro, HpaI-Gateway (Clontech).
A mutant DNA oligonucleotide (5′-AAAAAAGCACTAGAGACGTGGGACAAGCCCGCAAGCTTCCAGGCTTGTCCCCATCACGCCTCTAATGCGGTGTTTCGTCCTTTCCACAA-3′) was expressed as a control plasmid (pMh vector) for specificity of silencing. All of the construct sequences were verified, using forward (5′-CCCTTGAACCTCCTCGTTCGAC-3′) and reverse (5′-GAGACGTGCTACTTCCATTTGT-3′) pMSCV oligonucleotide primers.
Cell Culture and Transfection. Immortalized mouse embryonic fibroblasts were cultured in DMEM, supplemented with 10% bovine serum (FBS), 10 mM Hepes, 1 mM MEM nonessential amino acids, 100 units/ml penicillin, and 100 mg/ml streptomycin (Gibco/Invitrogen). Transfections were carried out by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 8-10 days of selection in 1 μg/ml puromycin, clones were picked. Mock transfections were carried out with pMSCV and pMh vectors.
Antibodies and Reagents. For immunoblot and immunofluorescence analyses, we used rabbit polyclonal anti-gelsolin (9); anti-p34 from Upstate Biotechnology (Lake Placid, NY) and anti-p21 from M. Welch (University of California, Berkeley); mouse monoclonal anti-α-tubulin from Sigma; and goat anti-Arp3, rabbit polyclonal anti-Arp2, goat anti-actin, and secondary HRP-conjugated antibodies from Santa Cruz Biotechnology.
Cell Extract Actin Assay. Cells (3.5 × 105) were harvested by trypsinization in 0.1 ml and, after washing with PBS, permeabilized by adding 0.1 vol of PHEM buffer (60 mM Pipes/25 mM Hepes/10 mM EGTA/2 mM MgCl2, pH 6.9) containing 1% Triton X-100 and a protease inhibitor mixture (Roche Diagnostics). The number of F-actin pointed and barbed ends per cell were calculated from pyrene-actin polymerization rates in the presence or absence of the neural Wiskott-Aldrich syndrome protein (N-WASP) peptide and 2 μM cytochalasin B (Sigma) (10).
L. monocytogenes Analyses. Cells, plated on 25-mm glass coverslips, were infected for 3 h with a 1:10 dilution of an overnight Listeria culture (strain 10403S; provided by F. Southwick, University of Florida, Gainesville). Listeria, grown in brain infusion medium (Difco), were centrifuged at 2,000 × g for 2 min and rinsed twice in DMEM before adding to the cells. Extracellular bacteria were killed by replacing the media with fresh DMEM supplemented with 5 μg/ml gentamicin and 20 mM Hepes for 4 h. Cells were then fixed and processed for imaging of bacteria associated with F-actin. Infection rates were assessed by counting cells with internalized bacteria (100 cells per experiment). Pathogens were visualized by rabbit anti-Listeria O Antiserum Poly types 1 and 4 (Difco) antibody or by Hoechst 33342 (Molecular Probes) staining of the DNA. The number of bacteria with actin comet tails was scored in three independent experiments by counting 100-130 pathogens after TRITC-phalloidin staining. Lengths of the actin comet tails were measured by using the imagej software (http://rsb.info.nih.gov/ij) line function.
Wound Healing, PDGF Stimulation, and Video Microscopy. Cells were seeded in six-well plates and grown to confluence. After overnight serum starvation, a wound scratch was applied and healing was monitored every 4 h over a period of 12 h in three different wounds in the presence of 10 ng/ml of PDGF (Sigma). Wound area was measured by using the selection tool of the imagej software, and healing was expressed as closure per hour. For PDGF stimulation, after overnight serum-starvation in DMEM, cells were treated with 10 ng/ml PDGF for 8 min and processed for immunofluorescence. F-actin structures were visualized by staining with TRITC-phalloidin. Quantification of cells with dorsal ruffles was done from time-lapse images in the bright field from a charge-coupled device camera (CCD-1300-Y, Roper Scientific, Trenton, NJ) for 20 min at 15-s intervals with a ×10 objective. For the quantification of total F-actin increase, serum-starved and PDGF-stimulated cells were fixed and stained with TRITC-phalloidin. F-actin content was quantified by measuring the total pixel intensity of single cells tracked with imagej software.
Electron Microscopy. Cells, seeded onto 5-mm round coverslips as above, were serum-starved overnight, stimulated with 10 ng/ml PDGF for 8 min, and permeabilized in Triton-PHEM, and the cytoskeletons were fixed, washed, rapidly frozen, metal-coated, and examined with an electron microscope as described in ref. 11.
Cloning of N-WASP CA and VCA Domains and Expression in Cells. CA (residues 450-505) and VCA (residues 392-505) domains of N-WASP that, respectively, inhibit or activate the Arp2/3 complex were PCR amplified from bovine GST-CA and GST-VCA plasmids (12). The same reverse primer was used for the amplification of both CA and VCA (5′-GTCAGTCAGTCACGATGAATTCC-3′), and the following 5′ primers were used as forward primers for the CA and the VCA, respectively: 5′-GCTAGATCTGTAACTGATGCCCCAGAGTC-3′ and 5′-GCTAGATCTCTCCCTTCTGATGGTGACC-3′. The PCR products were digested with BglII and EcoRI, gel-purified, and cloned in the pECFP-C1 vector (Clontech) to generate plasmids CFP-CA and CFP-UCA. Constructs were sequence-verified.
Cells were seeded on 35-mm glass-bottom dishes (MatTek, Ashland, MA) and microinjected with ≈1 pg of CFP-CA or CFP-VCA vector DNA by using a MO-150 micromanipulator and an IM200 pressure injector (Narishige, Tokyo). Protein expression was allowed to occur for 30 min followed by 2 h of serum-starvation. Ruffling responses were assessed by time-lapse videos of cells stimulated with PDGF (17 ng/ml) for 8 min followed by immunofluorescence with TRITC-phalloidin.
Results
RNA Interference of Arp3 Expression. We engineered three stable cell lines with reduced Arp3 expression (hereafter denoted Arp3-si cells): cl.4-3 (with construct si-ex9) and cl.7-4 and cl.7-14 (with si-ex3, si-ex9, and si-3′ UTR). Western blot analysis showed markedly reduced Arp3 expression compared with untransfected and mock-transfected cells (Fig. 1A). As shown in Fig. 6, and described in Supporting Materials and Methods, which are published as supporting information on the PNAS web site, Arp3 protein levels, quantified by densitometric scanning of Western blots, were reduced by 75%, 85%, and 90% in lines cl.4-3, cl.7-4, and cl.7-14, respectively. A relative decrease in the expression of other subunits of the Arp2/3 complex, Arp2, p34, and p21, correlated with the extent of interference of Arp3 expression, suggesting that reduction in the production of one component compromises the stability of the complex as a whole. Immunostaining with p34 antibody was markedly reduced in the Arp3-si cells (see Fig. 7, which is published as supporting information on the PNAS web site). Residual stain is mainly cytoplasmic, similar to controls, and nearly absent from the leading edges. At lower magnification, this subunit appears to be homogeneously reduced in all Arp3-si cells (see Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 1.
Arp3 expression levels, nucleation activity, and Listeria movement in Arp3-si cells. (A) Immunoblot analysis of total cell lysates containing identical protein amounts for Arp3 and several controls (gelsolin, actin, tubulin, and Akt) from control lines wild-type (WT), pMSCV (line expressing the empty vector), and pMh (line expressing a vector harboring a mutated siRNA-hairpin), and from Arp3-si cells: cl.4-3 (si-ex9), cl.7-4, and cl.7-14 (si-ex3, si-ex9, and si-3′ UTR). (B) Actin barbed-end nucleation sites in permeabilized control and Arp3-si cells in the absence or in the presence of VCA peptide (means ± SD of three experiments). (C) Listeria motility in Arp3-si cell lines shows a striking reduction in the number of Listeria with actin comet tails and in the length of the tails (Table 1). The boxes show enlarged frames of intracellular bacteria. All images are at the same magnification. (Scale bar: 10 μm.)
Silencing of the Arp3 Gene Interferes with Established Arp2/3 Complex Functions in Vitro and in Vivo. The VCA domain of N-WASP activates the Arp2/3 complex, and its CA domain inhibits the stimulation of actin polymerization by the activated Arp2/3 complex (12). VCA induced actin polymerization in control cell lysates with an increase of 20-25,000 barbed ends per cell (Fig. 1B). In contrast, the actin polymerization response to VCA in lysates of Arp3-si cells was 10% of the control cells (increase of 2-5,000 barbed ends), concordant with the reduction in the expression of Arp3 protein. To examine Arp2/3 complex function in vivo, we monitored the formation of actin comet tails induced by L. monocytogenes in control and Arp3-si cells. As shown in Fig. 1C, numerous Listeria organisms with actin tails reside within wild-type, pMSCV, and pMh cell lines. The infection rate of Arp3-si cells was comparable to that of the control lines (Table 1). However, the number of Listeria with an actin tail was markedly reduced in the Arp3-si cells compared with the control lines (11-23% vs. 42-58%, respectively; pairwise comparisons, P ≤ 0.04), and this reduction was more pronounced in cl.7-4 and cl.7-14 with lower Arp3 expression. The lengths of comet tails in the Arp3-si cells were significantly reduced compared with controls (4.1-5.9 μm vs. 7.8-10.7 μm, respectively; all pairwise, P < 0.0001). Compared with control cells, the total F-actin content associated with Listeria was reduced in Arp3-si cl.4-3 by 71% and in Arp3-si cl.7-4 and cl.7-14 by 90%.
Table 1. Listeria-induced actin assembly.
| Cell line | Infection rate, % | Bacteria with actin tail, % | Tail length, μm | Total Listeria associated F-actin |
|---|---|---|---|---|
| WT | 45 ± 1.4 | 42 ± 2.9 | 9.7 ± 0.3 | 407.4 |
| pMSCV | 46 ± 6.8 | 48.3 ± 4.2 | 10.7 ± 0.4 | 516.8 |
| pMh | 59 ± 3 | 58 ± 6 | 7.8 ± 0.3 | 452.4 |
| cl.4-3 | 59 ± 6.8 | 22.5 ± 6.5 | 5.9 ± 0.2 | 132.75 |
| cl.7-4 | 27.6 ± 2.8 | 9.8 ± 1 | 4.4 ± 0.2 | 43.1 |
| cl.7-14 | 42.5 ± 3.5 | 11 ± 0.6 | 4.1 ± 0.3 | 45.1 |
Quantification of Listeria infection rates (means ± SD of three experiments, 100 cells counted per experiment), percentage of bacteria with actin tails (100–130 bacteria counted per experiment), and lengths of the actin comet tails (110–190 bacteria counted per experiment). Total F-actin associated with Listeria was estimated in arbitrary units in each cell line by multiplying the average number of bacteria with an actin comet tail and the average tail length. There was a 71% (cl.4-3) and a 90% (cl.7-4 and cl.7-14) reduction in F-actin associated with Listeria in comparison with the control cell lines.
Arp3-si Cells Have Normal Actin Structures and Respond to PDGF by Migrating and Ruffling. Despite marked reductions in Arp2/3 complex protein content and function, F-actin-rich structures such as stress fibers and lamellipodia appeared to be identical in Arp3-si and control cells (Fig. 2A). Cell perimeters and areas were comparable in all lines (Table 2, which is published as supporting information on the PNAS web site). The total F-actin content and the lamellipodia index [percentage of the cell perimeter occupied by lamellipodia (13)] were also similar among these cells. The number of filopodia per cell was slightly higher in the Arp3-si lines in comparison with control lines. The leading edges of control and the Arp3-si cells had similar actin-filament networks visualized by electron microscopy (Fig. 2B).
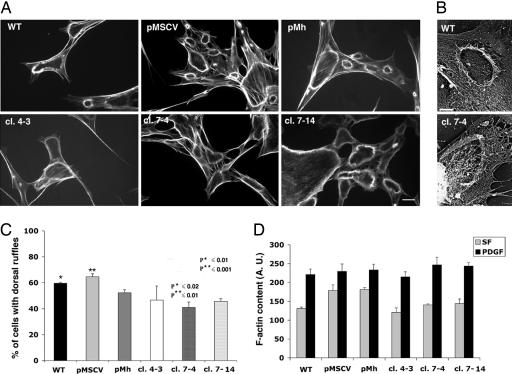
Fig. 2.
F-actin organization in control and Arp3-si cells. (A) Actin filament structures were visualized by staining with TRITC-phalloidin. (Scale bar: 10 μm.) (B) Organization of F-actin in lamellipodia visualized by electron microscopy from a representative control and Arp3-si cl.7-4 cytoskeleton. (Scale bar: 0.1 μm.)
Wound healing rates of Arp3-si cells in the presence of 10 ng/ml PDGF were comparable to those of the representative control cell line pMSCV (Fig. 3 A and B). Serum-starved Arp3-si and control cells both responded to PDGF stimulation with formation of large dynamic dorsal ruffles (Fig. 4A). The actin filament ultrastructure of ruffles and lamellipodia of wild-type and Arp3-si cells was indistinguishable (Fig. 4B). Movies 1 and 2, which are published as supporting information on the PNAS web site, show that apart from a slight reduction in speed in the remodeling of the leading edge of the Arp3-si cl.7-4, overall the response to PDGF stimulation is comparable to that in the control line pMSCV. Quantification of the fraction of cells with dorsal ruffles in response to PDGF shows a slight reduction (≈52-65% in controls vs. 42-46% in the Arp3-si lines) in the two lowest-expressing Arp3-si cells (Fig. 4C). However, the total F-actin content in the presence of PDGF increased to the same extent in Arp3-si and control cells (Fig. 4D). Extents and rates of spreading of control and Arp3-si cells on fibronectin substrate varied by ≈2-fold but did not correlate with Arp2/3 expression (see Fig. 9, which is published as supporting information on the PNAS web site). Cell migration toward PDGF (50 ng/ml) as chemoattractant was also assessed by using a modified Boyden chamber assay, and comparable migration rates were observed in control line pMSCV and pMh and Arp3-si cells cl.7-4 and cl.7-14 (data not shown).
Fig. 3.
Wound healing by Arp3-si and control cells. (A) Time-lapse phase contrast images of cells grown at times 0, 4, 8, and 12 h after wounding in the presence of 10 ng/ml PDGF. (Scale bar: 100 μm.) The boxes display individual migrating cells. (B) Wound closure plotted as area closed per hour. Values are means ± SD of three wounds.
Fig. 4.
Ruffling responses of cells serum starved for 12 h and stimulated with 10 ng/ml PDGF. (A) TRITC-phalloidin staining of F-actin. (Scale bar: 20 μm.) (B) Ultra structure of F-actin in ruffles and lamellipodia from representative control and Arp3-si cell. (Scale bar: 5 μm.) (C) Percent of cells with ruffles 20 min after PDGF addition. Values represent the average ± SD of three experiments (100 cells counted per experiment). The percentage of Arp3-si cells with dorsal ruffles is only slightly reduced in comparison with control cell lines. (D) F-actin content of cells before and after 8 min of PDGF stimulation. The value are means ± SD of three experiments (50 cells per experiment).
Expression of CA and VCA Domains of N-WASP in Arp3-si Cells Does Not Alter the Ruffling Responses. To suppress residual Arp2/3 complex activity further, we expressed VCA and CA fusion proteins in Arp3-si cells. As expected (14), actin tail formation was reduced by 70% in Listeria-infected wild-type cells transiently expressing a CFP-CA in comparison with cells expressing CFP alone (Fig. 10 A and B, which is published as supporting information on the PNAS web site). Control and Arp3-si cells transiently expressing CFP-CA responded to PDGF stimulation by making dorsal actin ruffles (Fig. 5A). Silencing 75-90% of Arp3 subunit expression did not interfere with recruitment of residual p34 to actin-rich structures. This finding is consistent with the observation that a partial complex consisting of a p34/p20 heterodimer retains the ability to bind F-actin, although this association is not functional for promotion of actin assembly (15). CFP-CA distributed widely through these cells, with enrichment in the ruffles. Expression of CA had no detectable effect on the ruffling response to PDGF of either control line pMSCV (9 of 11 injected cells ruffled) or Arp3-si cl.4-3 (19 of 21 injected cells ruffled) and cl.7-4 cells (16 of 20 injected cells ruffled). In contrast, when CFP-VCA was expressed, neither CFP-VCA nor the p34 Arp2/3 subunit localized to actin-rich structures (Fig. 5B). VCA expression did decrease ruffling in comparison with uninjected cells, but it was not abolished and was comparable in control and Arp3-si cells [wild type, 11/39; pMSCV, 6/21; pMh, 6/14; Arp3-si cl.4-3, 36/96; cl.7-4, 11/36; cl.7-14, 20/55 (ruffling/total number of injected cells for all)]. The diminished ruffling in all cells is probably a result of competitive sequestration of actin monomers by VCA-induced Arp2/3 activation leading to unregulated actin polymerization.
Fig. 5.
Control and Arp3-si cell lines still ruffle after microinjection with CFP-CA or CFP-VCA DNA. (A) Triplicate images illustrate CFP-CA expression (cyan panel), F-actin (TRITC-phalloidin staining), and p34 distribution in cells microinjected with CFP-CA and stimulated with 10 ng/ml PDGF. Localization of p34 in ruffles indicates that CFP-CA does not interfere with recruitment of the Arp2/3 complex to the actin structures (white arrow). All images are at the same magnification. (Scale bar: 10 μm.) (B) Cells injected with the CFP-VCA DNA show displacement of p34 from actin-rich structures (white arrow) although actin ruffles still form in response to PDGF in both control and Arp3-si cells. All images are the same magnification. (Scale bar: 10 μm.)
Discussion
Marked reduction in Arp2/3 complex content and functional activity in fibroblasts by RNA interference silencing had no detectable effects on the surface motility and locomotion of the depleted cells. Even challenging the silenced cells with N-WASP peptides that inhibit or delocalize Arp2/3 actin polymerization activity failed to inhibit PDGF-mediated ruffling activity. These cells, deficient in Arp2/3 complex function, might possibly have impaired protrusive responses to agonists not tested here. In addition, other cell types might lack compensatory actin-assembly mechanisms possessed by the silenced cells we tested and exhibit obvious abnormalities. These explanations might account for abnormalities described in previous reports of Arp2/3 complex component-depleted cell lines. Drosophila S2 cells depleted of the p20-Arc subunit of the Arp2/3 complex as part of an extensive screen of cytoskeletal proteins had abnormal shapes and defects in spreading (16). Cultured carcinoma cells depleted by RNA interference of the p34-Arc subunit of the Arp2/3 complex, although generally normal in appearance, had impaired protrusion of actin-rich nodules (”invadopodia”) associated with metastatic behavior (17). HeLa cells depleted of the p21-Arc subunit of the Arp2/3 demonstrated defective cell growth (18). Apart from a slight reduction in the percentage of the mostly depleted Arp3-si cells with dorsal ruffles in response to PDGF stimulation, overall actin remodeling appears to be preserved. The silenced gene, Arp3, is the major subunit of the Arp2/3 complex. Although there is a second Arp3 isoform, it is expressed only in neuronal cells (19). Therefore, the findings we report here, taken together with our previous observations in platelets and melanoma cells (20, 21) suggest that the Arp2/3 complex might contribute by enhancing actin assembly but is not essential for leading-edge protrusion.
For example, the Arp2/3 complex does not localize at the leading edge of advancing nerve growth cones or of migrating fibroblasts overexpressing tropomyosin, and inhibition of Arp2/3 complex function stimulates rather than inhibits growth-cone protrusion (14, 22). Introduced into intact cells by microinjection or genetic transfection, the N-WASP-derived Arp2/3 complex-activating peptide VCA inhibits intracellular motility of Listeria (23), leading-edge protrusion (24), and phagocytosis (25). However, this reagent induces massive intracellular actin assembly, thereby causing generalized cytoplasmic disruption and delocalization of more components than just the Arp2/3 complex from sites of normal actin elongation. Therefore, in contrast to previous reports (24), the effect of VCA expression was analyzed within 2 1/2 h of microinjection. VCA depleted p34 from the leading edge of Arp2/3-silenced cells but still failed to inhibit PDGF-mediated ruffling activity. Introduction into cells of the more specifically targeted CA peptide, which also inhibits Listeria motility (ref. 14 and this work), had no effect on leading-edge function in this study or in a previous one (24).
Arp2/3 complex silencing does not appear to affect leading-edge actin ultrastructure. The appearance of actin filaments in electron micrographs of leading edges of spreading or translocating cells depends in part on preparative methods, but leading-edge actin networks are highly complex, exhibiting many varieties of interfilament interactions. The possibility that the Y-shaped actin filament branch structures generated by activated Arp2/3 complexes in vitro are accurate representations of actin networks within the leading lamellae of migrating cells or have the mechanical capacity to extend leading-edge membranes have been questioned previously (21, 26, 27).
Arp2/3-mediated de novo nucleation or branching amplification of actin filament assembly is the most recent of multiple mechanisms proposed for regulating leading-edge actin assembly. These mechanisms, including removal at the plasma membrane of barbed-end actin-capping proteins from preexisting actin filaments, exposure of free barbed ends by actin-filament severing, uncapping of actin oligomers, and de novo assembly of actin filaments from sequestered monomers, are not mutually exclusive, and all have experimental support (28, 29). The fact that cells seem capable of withstanding, without obvious impairments, the absence or near absence of proteins with powerful actions on actin remodeling (30, 31) is a testimony to the importance of this physiological process to cell survival and the richness of the strategies cells use to sustain actin remodeling under widely different environmental conditions.
Supplementary Material
Acknowledgments
We thank Dr. A. Lader (Brigham and Women's Hospital) for providing the immortalized mouse embryonic fibroblasts used in this study, Dr. L. Vidali for helpful discussions, Dr. P. Allen for the use of the Leica DM RXA inverted microscope, and Karen Vengerow for editorial assistance.
Author contributions: A.D.N., G.C., H.F., J.H.H., T.P.S., and D.J.K. designed research; A.D.N., G.C., H.F., and J.H.H. performed research; A.D.N., G.C., H.F., J.H.H., T.P.S., and D.J.K. contributed new reagents/analytic tools; A.D.N., G.C., H.F., J.H.H., T.P.S., and D.J.K. analyzed data; and A.D.N., T.P.S., and D.J.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviation: CFP, cyan fusion peptide.
References
- 1.Gouin, E., Welch, M. D. & Cossart, P. (2005) Curr. Opin. Microbiol. 8, 35-45. [DOI] [PubMed] [Google Scholar]
- 2.Engqvist-Goldstein, A. E. & Drubin, D. G. (2003) Annu. Rev. Cell Dev. Biol. 19, 287-332. [DOI] [PubMed] [Google Scholar]
- 3.Hudson, A. M. & Cooley, L. (2002) J. Cell Biol. 156, 677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawa, M., Suetsugu, S., Sugimoto, A., Miki, H., Yamamoto, M. & Takenawa, T. (2003) J. Cell Sci. 116, 1505-1518. [DOI] [PubMed] [Google Scholar]
- 5.Linder, S., Nelson, D., Weiss, M. & Aepfelbacher, M. (1999) Proc. Natl. Acad. Sci. USA 96, 9648-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMali, K. A., Barlow, C. A. & Burridge, K. (2002) J. Cell Biol. 159, 881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rafelski, S. M. & Theriot, J. A. (2004) Annu. Rev. Biochem. 73, 209-239. [DOI] [PubMed] [Google Scholar]
- 8.Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmeister, K. M., Falet, H., Toker, A., Barkalow, K. L., Stossel, T. P. & Hartwig, J. H. (2001) J. Biol. Chem. 276, 24751-24759. [DOI] [PubMed] [Google Scholar]
- 10.Falet, H., Hoffmeister, K. M., Neujahr, R. & Hartwig, J. H. (2002) Blood 100, 2113-2122. [PubMed] [Google Scholar]
- 11.Hartwig, J. H., Bokoch, G. M., Carpenter, C. L., Janmey, P. A., Taylor, L. A., Toker, A. & Stossel, T. P. (1995) Cell 82, 643-653. [DOI] [PubMed] [Google Scholar]
- 12.Rohatgi, R., Ma, L., Miki, H., Lopez, M., Kirchhausen, T., Takenawa, T. & Kirschner, M. W. (1999) Cell 97, 221-231. [DOI] [PubMed] [Google Scholar]
- 13.Verma, S., Shewan, A. M., Scott, J. A., Helwani, F. M., den Elzen, N. R., Miki, H., Takenawa, T. & Yap, A. S. (2004) J. Biol. Chem. 279, 34062-34070. [DOI] [PubMed] [Google Scholar]
- 14.Strasser, G. A., Rahim, N. A., VanderWaal, K. E., Gertler, F. B. & Lanier, L. M. (2004) Neuron 43, 81-94. [DOI] [PubMed] [Google Scholar]
- 15.Gournier, H., Goley, E. D., Niederstrasser, H., Trinh, T. & Welch, M. D. (2001) Mol. Cell 8, 1041-1052. [DOI] [PubMed] [Google Scholar]
- 16.Rogers, S. L., Wiedemann, U., Stuurman, N. & Vale, R. D. (2003) J. Cell Biol. 162, 1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi, H., Lorenz, M., Kempiak, S., Sarmiento, C., Coniglio, S., Symons, M., Segall, J., Eddy, R., Miki, H., Takenawa, T. & Condeelis, J. (2005) J. Cell Biol. 168, 441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harborth, J., Elbashir, S. M., Bechert, K., Tuschl, T. & Weber, K. (2001) J. Cell Sci. 114, 4557-4565. [DOI] [PubMed] [Google Scholar]
- 19.Jay, P., Berge-Lefranc, J. L., Massacrier, A., Roessler, E., Wallis, D., Muenke, M., Gastaldi, M., Taviaux, S., Cau, P. & Berta, P. (2000) Eur. J. Biochem. 267, 2921-2928. [DOI] [PubMed] [Google Scholar]
- 20.Falet, H., Hoffmeister, K. M., Neujahr, R., Italiano, J. E., Jr., Stossel, T. P., Southwick, F. S. & Hartwig, J. H. (2002) Proc. Natl. Acad. Sci. USA 99, 16782-16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flanagan, L. A., Chou, J., Falet, H., Neujahr, R., Hartwig, J. H. & Stossel, T. P. (2001) J. Cell Biol. 155, 511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupton, S. L., Anderson, K. L., Kole, T. P., Fischer, R. S., Ponti, A., Hitchcock-DeGregori, S. E., Danuser, G., Fowler, V. M., Wirtz, D., Hanein, D. & Waterman-Storer, C. M. (2005) J. Cell Biol. 168, 619-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.May, R. C., Hall, M. E., Higgs, H. N., Pollard, T. D., Chakraborty, T., Wehland, J., Machesky, L. M. & Sechi, A. S. (1999) Curr. Biol. 9, 759-762. [DOI] [PubMed] [Google Scholar]
- 24.Machesky, L. M. & Insall, R. H. (1998) Curr. Biol. 8, 1347-1356. [DOI] [PubMed] [Google Scholar]
- 25.May, R. C., Caron, E., Hall, A. & Machesky, L. M. (2000) Nat. Cell Biol. 2, 246-248. [DOI] [PubMed] [Google Scholar]
- 26.Small, J. V., Stradal, T., Vignal, E. & Rottner, K. (2002) Trends Cell Biol. 12, 112-120. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura, F., Osborn, E., Janmey, P. A. & Stossel, T. P. (2002) J. Biol. Chem. 277, 9148-9154. [DOI] [PubMed] [Google Scholar]
- 28.Schafer, D. A. & Cooper, J. A. (1995) Annu. Rev. Cell Dev. Biol. 11, 497-518. [DOI] [PubMed] [Google Scholar]
- 29.Condeelis, J. (1993) Annu. Rev. Cell Biol. 9, 411-444. [DOI] [PubMed] [Google Scholar]
- 30.Andre, E., Brink, M., Gerisch, G., Isenberg, G., Noegel, A., Schleicher, M., Segall, J. E. & Wallraff, E. (1989) J. Cell Biol. 108, 985-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witke, W., Schleicher, M. & Noegel, A. A. (1992) Cell 68, 53-62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.