Abstract
Chlamydia pneumoniae is an obligate intracellular prokaryotic human pathogen that causes community-acquired respiratory infection and has been associated with atherosclerosis and cardiovascular disease. Unexpected results from genomic sequencing indicate that significant intrastrain polymorphism exists for some C. pneumoniae isolates. These polymorphisms could reflect genotypes with differing disease-causing characteristics. A definitive means to test this hypothesis is to obtain genetically homogeneous clonal populations of the pathogen and test them in models of infection and disease. To date, methods for cloning C. pneumoniae have not been reported. In this study, we describe the isolation of clonal variants with genetic differences in the tyrP locus from a polymorphic respiratory isolate, using a novel focus-forming assay. These results now allow investigations on the biology and pathogenesis of C. pneumoniae clonal genovars that could lead to new insights into the pathogenesis of this important human pathogen.
Chlamydia pneumoniae is an obligatory intracellular prokaryotic pathogen. It is a major cause of human respiratory infections, responsible for approximately 10% of all community-acquired pneumoniae. It is also an important cause of bronchitis and sinusitis (1, 5; K. Hashigucci, H. Ogawa, T. Suzuki, and Y. Kazuyama, Letter, Clin. Infect. Dis. 15:570-571, 1992). C. pneumoniae has also been associated with several important chronic human diseases, such as asthma (2, 19), sarcoidosis (13, 17), and atherosclerosis (6, 9). Despite this broad spectrum of clinical manifestations C. pneumoniae strains differing in pathogenic or virulence characteristics have not been described. Three C. pneumoniae genomes have been sequenced (AR-39 [14], CWL-029 [7], and J183 [15]). Comparisons between strains indicate a high level of sequence homology (e.g., AR-39 versus CWL-029 differences consisted of ca. 300 single-nucleotide polymorphisms and single-base frameshift mutations in addition to seven insertions and deletions [14]). The largest polymorphism involves the duplication of a 1,649-bp fragment containing a tyrP gene (encoding an aromatic amino acid transporter) and a truncated yccA gene (encoding a transmembrane protein, possibly a permease). Although Read et al. (14) do not speculate on the source of the internal polymorphisms in the AR-39 sequence, the data indicate that the DNA used for sequencing came from multiple variants within the AR-39 isolate. Alternatively, C. pneumoniae may be inherently polymorphic due to an elevated spontaneous mutation frequency. In either case, genetic studies as well as investigations on biological characteristics associated with genetic polymorphisms require genetically homogeneous, clonal populations, a concept widely accepted in the field of bacteriology but neglected in C. pneumoniae research so far due to a lack of an appropriate method for cloning.
Here we describe the isolation of C. pneumoniae genomic variants from a polymorphic parent strain. While the wild-type strain showed the presence of both a single and duplicate copy of the tyrP gene, clones obtained from this strain possessed either a single or duplicate copy of the gene. We used a novel focus- and plaque-forming assay for cloning C. pneumoniae. The assay allows the identification of different morphotypes of the prototype strains AR-39, TW-183, and CWL-029 that are not detectable by a similar assay described for the plaque cloning of C. trachomatis (12). Clonal C. pneumoniae isolated, obtained by this new method, can be utilized for future comparative genomic and pathogenetic studies.
MATERIALS AND METHODS
Strains.
C. pneumoniae strains AR-39, TW-183, CWL-029, and MUL-250 and C. trachomatis serovar D were grown in HeLa 229 cells, and elementary bodies (EB) were purified on density gradients of Renocal-76 (Bracco Diagnostics, Princeton, N.J.) by methods previously described (3). Inclusion-forming units (IFU) of EB preparations were determined following titrations on monolayers of HeLa 229 in 24-well cluster plates and indirect fluorescent antibody (IFA) assays (16). Strain MUL-250 was isolated from a patient with lower respiratory tract disease and subsequent reactive arthritis. The strain was identified as C. pneumoniae by staining with a species-specific antibody (C. pneumoniae fluorescein-5-isothiocyanate [FITC] research reagent; Dako, Hamburg, Germany) as well as PCR amplification using a species-specific primer set followed by detection with a species-specific probe (10).
Focal assay media.
The following stock solutions were prepared aseptically: (i) 1.1% agarose (SeaKem ME agarose; FMC BioProducts, Rockland, Maine) in distilled water; (ii) 2× Dulbecco's Modified Eagle Medium (2× DMEM), without phenol red, with high glucose (Invitrogen, Frederick, Md.) supplemented with 4 mM l-glutamine, 20 mM HEPES, 2 mM sodium pyruvate, 0.11 mM β-mercaptoethanol (Invitrogen), and 88 mM sodium bicarbonate (J. T. Baker, Inc., Philipsburg, N.J.) and (iii) cycloheximide (1 mg/ml) in phosphate-buffered saline (PBS), and (iv) gentamicin (1 mg/ml; Invitrogen). Agarose medium was made by adding an equal volume of 1.1% agarose equilibrated to 45°C to 2× DMEM supplemented with 20% (vol/vol) heat-inactivated fetal bovine serum (FBS) (HyClone, Logan, Utah), cycloheximide (16 μg/ml), and gentamicin (20 μg/ml) that had also been equilibrated to 45°C. Liquid overlay medium was made by mixing an equal volume of 2× DMEM, supplemented with 20% (vol/vol) FBS and gentamicin (20 μg/ml), with an equal volume of sterile distilled water. The overlay was prewarmed to 37°C prior to use.
Plaque assay.
The plaque assay for cloning C. trachomatis trachoma biovars using McCoy cells was performed as described by Matsumoto et al. (12). Plaque formation of C. trachomatis serovar D and C. pneumoniae strain AR-39 were compared using this assay.
Focus-forming assay. (i) Cell culture and infection.
The focus-forming assay for cloning C. pneumoniae was done as follows. Monolayers of HeLa 229 cells were grown in six-well cluster plates at a density of 1.2 × 106 per well in 3 ml of DMEM with high glucose (Mediatech Cellgro; Fisher Scientific, Pittsburgh, Pa.), supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 0.055 mM β-mercaptoethanol, and gentamicin (10 μg/ml). Duplicate monolayers were infected with 100, 50, and 25 IFU per well, respectively, in 2 ml of SPG (250 mM sucrose-10 mM sodium phosphate-5 mM l-glutamic acid, pH 7.2) by centrifugation at 543 × g for 1 h at 32°C followed by rocking in an ambient-air incubator for 30 min at 35°C. The inoculum was aspirated from the wells, and 2 ml of agarose medium was gently added to the surface of the monolayer in each well. The plates were allowed to stand for 10 min at room temperature to allow the agarose to solidify, and then 2 ml of liquid overlay medium was added to the surface of the solidified agarose medium. The plates were incubated at 35°C in 95% air-5% CO2 for 10 to 12 days. During the incubation period, liquid overlay medium was gently aspirated from each well and replaced with fresh liquid medium every 4 days.
(ii) IFA staining of foci.
Overlay medium was aspirated from wells, and the agarose plugs were gently removed with a sterile spatula taking care not to disturb the underlying monolayer of cells. The viable cells of the monolayers were then stained by IFA in the following manner. Monolayers were washed once with 2 ml of prewarmed Hanks' balanced salt solution (HBSS) containing 10 mM HEPES. Wash and antibody solutions were prewarmed to 37°C prior to use. A 2-ml volume of monoclonal antibody (MAb) GZD1E8 (20) diluted 1:500 in HBSS was then added to the monolayers, and the plates were further incubated for 1 h at 37°C with rocking. MAb GZD1E8 is C. pneumoniae species specific and recognizes the major outer membrane protein (MOMP) (20). The antibody was removed, and the monolayers were gently washed three times with 2 ml of HBSS. A 2-ml volume of HBSS containing a 1:200 dilution of FITC-conjugated goat anti-mouse immunoglobulin G (ICN, Cappel Research Reagents, Costa Mesa, Calif.) was added to each monolayer, the plates were incubated for 30 min at 37°C with rocking and washed three times with 2 ml of HBSS, and then 2 ml of HBSS was added to each well. Monolayers were examined for C. pneumoniae foci using a Leica DM IRB inverted epifluorescence microscope (Bartels and Stout, Inc., Bellevue, Wash.) at a magnification of ×25. To detect C. pneumoniae inclusions in infected cells in IFA-stained monolayers, cells were fixed with methanol at room temperature for 5 min, washed once with PBS, and restained with a 1:500 dilution of MAb GZD1E8. The fixed cells were washed with PBS and stained with 2 ml of a 1:20 dilution of tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse immunoglobulin G (ICN). The twice-stained monolayers were observed for green-staining foci and red-staining inclusions by epifluorescence, and images were recorded on Ektachrome 400 35-mm color film (Kodak, Rochester, N.Y.).
(iii) Isolation of chlamydiae from foci.
Monolayers infected with C. pneumoniae were stained at 10 days postinfection by IFA as described above. Plates were viewed for immunoreactive foci by epifluorescence at a magnification of ×25. Individual foci were identified, and their location on the monolayer was marked on the underside of the plate using a fine-tip marker (Sarstedt, Inc., Newton, N.C.). This was accomplished by placing the pen tip between the objective lens and the bottom of the plate. After marking foci in this fashion the HBSS was aspirated from each well and individual foci were picked using a sterile Calgiswab (Spectrum Laboratories, Inc., Houston, Tex.). The tip of the swab was gently positioned onto the monolayer over the marked area on the underside of plate identifying the stained foci. Swabs containing cells were placed into 1.5-ml microtubes that contained three 4-mm-diameter sterile glass beads and 350 μl of SPG, disturbed for 2 min with an Eppendorf 5432 mixer (Eppendorf AG, Hamburg, Germany) and frozen at −80°C. Samples were assayed for recoverable IFU on monolayers of HeLa 229 cells grown in 96-well cluster plates. The plates were centrifuged as described above, rocked for 30 min, aspirated, and then fed with 100 μl of DMEM containing cycloheximide (1 μg/ml). After incubation for 50 h at 35°C, the monolayers were fixed with methanol, stained by IFA, and assayed for IFU.
Correlation between input IFU and foci formation.
HeLa 229 cell monolayers were infected with serial twofold dilutions (five wells for each dilution) of C. pneumoniae AR-39 ranging from 25 to 1.56 IFU, respectively. The plates were processed for focus staining, and the number of focus-forming units (FFU) produced was then compared to input IFU.
Cloning of strain MUL-250 and duplication PCR.
Strain MUL-250 was cloned as described for C. pneumoniae AR-39. Individual clones were propagated for one passage in one well of a 96-well plate, followed by two further passages in a 48-well plate and a final, fourth passage in a 24-well plate. Infected cells were harvested; one aliquot was frozen for later expansion and Southern blot analysis. The DNA of the second aliquot was extracted via the use of a DNase tissue kit (Qiagen, Valencia, Calif.) using the protocol for animal tissue. DNA was eluted in 300 μl of H2O, and 5 μl of the eluate was used per 50-μl PCR mixture. Each sample was screened by two different Taqman (Applied Biosystems) primer-probe pairs on an ABI Prism 7700 (Applied Biosystems, Foster City, Calif.). Standard PCR conditions were used as recommended by the manufacturer. In order to detect a duplication of the tyrP gene, a primer-probe set was designed to bind at the 3′ end of the yccA and the 5′ end of the tyrP gene (forward primer, TCTTTTTGGTGGGTGTGGTGT; reverse primer, CCATCACTTGACCAACTACCGA; probe, TACGCTAGGCGTGTCTTTCTTTATCAACTCTAA [synthesized by Applied Biosystems]). The quantity of C. pneumoniae DNA in the extracted sample was determined by a second quantitative PCR targeting the euo gene (forward primer, GGAATACCTGTGCAGAAGGTCTACT; reverse primer, CCCAAGCGGCTCCCTTAC; probe, CCTCGTATGGTTCCCGTACGAGTTGCA [synthesized by Applied Biosystems]). PCR products were visualized on a 5% MetaPhor agarose gel (FMC BioProducts). The sensitivity limit of the duplication PCR was determined by limiting dilutions of DNA from gradient-purified AR-39 EB. A 0.1-pg amount of DNA (ca. 80 genome equivalents) of AR-39 was detectable by the duplication PCR.
Southern blotting.
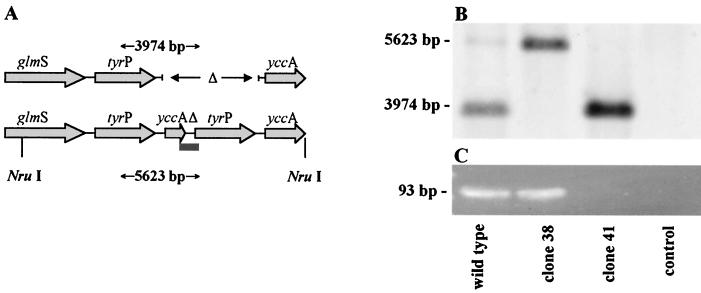
One clone (clone 38), identified by PCR as containing the duplicated tyrP gene, and one representative single tyrP gene clone (clone 41) were propagated for four further passages. Infected cells were disrupted by glass beads, and the supernatant separated from the cell debris (15 min of centrifugation at 380 × g) and further purified by 30 min of centrifugation at 24,000 × g over 30% Renocal-76 (Bracco Diagnostics). DNA was extracted by phenol-chloroform-isoamyl alcohol according to standard procedures. The concentration of DNA was measured by spectrophotometric analysis. Two hundred nanograms of DNA prepared from each clone and uninfected HeLa cells were digested with NruI (New England BioLabs, Beverly, Mass.), separated on a 1% agarose gel, and transferred to Hybond-N+ membrane (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, Great Britain) by capillary blotting using the neutral transfer protocol as described by the manufacturer. A probe was generated by amplification of a 1-kb fragment of the tyrP gene (forward primer, GCAGGTTCTGCCATCGGTGCTGG; reverse primer, GGGAATACCCCGATAATCACGGC) and purification of the product from an agarose gel using a QIAEX II kit (Qiagen). The probe was then labeled with alkaline phosphatase using the AlkPhos direct kit (Amersham Pharmacia Biotech). Hybridization was performed according to the manufacturer's instructions. The hybridized probe was detected via a chemiluminescent ECF substrate (Amersham Pharmacia Biotech) and scanned on a phosphorimager (Storm 860; Amersham Pharmacia Biotech). A 3,974-bp fragment was indicative for a single tyrP copy variant, and a 5,623-bp fragment was indicative for a tyrP duplication (see Fig. 4A).
FIG. 4.
(A) Genomic map of the tyrP region. Sequencing data revealed a 1,649-bp polymorphism containing a duplication of the tyrP and yccAΔ genes (14). Lines indicate the cutting sites of NruI resulting in 3,974- and 5,623-bp fragments, respectively. The bar indicates the region amplified by the duplication-PCR. (B) Southern blot analysis after Nru I digestion. The MUL-250 wild-type strain shows two bands of 3,974 and 5,623 bp, respectively, indicating the presence of both a single tyrP copy population and a double tyrP copy population. Clones obtained by the focus assay showed either a double copy (clone 38) or a single copy (clone 41). HeLa DNA was used as a negative control (line 4). (C) Agarose gel analysis of duplication-PCR products (93 bp). Screening of 60 C. pneumoniae strain MUL-250 clones revealed a single clone (clone 38) containing a duplicated tyrP gene as indicated by a positive PCR product. A negative duplication-PCR for clone 41 indicates the presence of a single tyrP copy only. The MUL-250 wild-type strain was used as a positive control, and H2O was used as a negative control (lane 4).
RESULTS
C. pneumoniae plaque and foci formation.
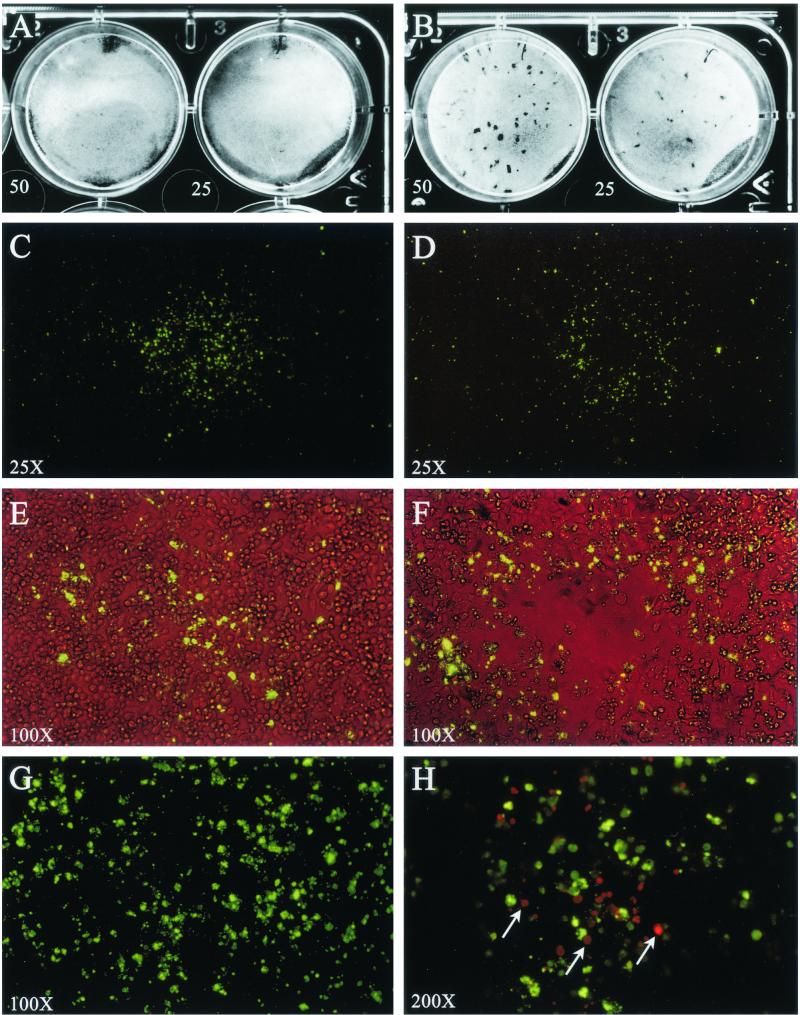
We initially attempted to plaque C. pneumoniae using the McCoy cell plaque assay described by Matsumoto et al. for plaque cloning of C. trachomatis (12). Despite our ability to reproducibly generate plaques of C. trachomatis strains, we were unable to detect plaques with C. pneumoniae strain AR-39 by this method (Fig. 1). We therefore substituted HeLa 229 cells, commonly used for the in vitro propagation of C. pneumoniae, for McCoy cells with the rationale that they would be more permissive for C. pneumoniae infection (11). In brief, we observed that the HeLa cells had a propensity to overgrow in the assay despite the presence of cycloheximide (1 μg/ml) in the agarose. We therefore performed experiments varying the concentration of cycloheximide added to the agarose to determine conditions that would sustain HeLa cell viability over an extended incubation period but prevent overgrowth of the monolayer. We found that the addition of cycloheximide at 8 μg/ml to the agarose medium on day 0 maintained a confluent monolayer of HeLa cells with minimal overgrowth over a 14-day culture period. We then attempted to plaque C. pneumoniae AR-39 using these culture conditions. Monolayers infected with strain AR-39 did not show plaque formation, not visible on a native monolayer (Fig. 2A) or after neutral red stain (data not shown), but IFA staining of the fixed monolayer showed the presence of C. pneumoniae-infected cells. We concluded from these results that although C. pneumoniae AR-39 failed to form plaques it might form foci, similar to some nonlytic viruses (4), that would be detectable following the staining of the viable monolayer with C. pneumoniae-specific antibody. To test this hypothesis HeLa cell monolayers were infected with various amounts of C. pneumoniae IFU, the agarose overlay was carefully removed at 10 to 14 days postinfection, and the unfixed monolayer was stained with a MAb specific to C. pneumoniae MOMP. Distinct focal areas of immunoreactivity were visualized on the surface of cells following IFA staining (Fig. 2C). Combined phase and fluorescence microscopy showed that the foci were localized to the surface of viable cells of the confluent monolayer (Fig. 2E). Foci ranged in size from 0.5 to 2.0 mm and displayed a noncontiguous staining pattern dispersed across the monolayer surface. When observed at higher magnification, the staining pattern appeared unstructured, consisting of immunoreactive material of heterogenous size (Fig. 2G). To determine if the foci were associated with C. pneumoniae-infected cells, viably stained cells were fixed with methanol, strained with anti-MOMP antibody, and then stained with a TRITC-labeled secondary antibody. The green-stained foci were distinctly positioned over C. pneumoniae-infected cells that contained red-stained chlamydial inclusions (Fig. 2H). We found that MAb specific to chlamydial lipopolysaccharide (LPS) stained foci in a manner similar to that observed for anti-MOMP staining. In contrast, chlamydial HSP60 MAb did not react with the foci (data not shown).
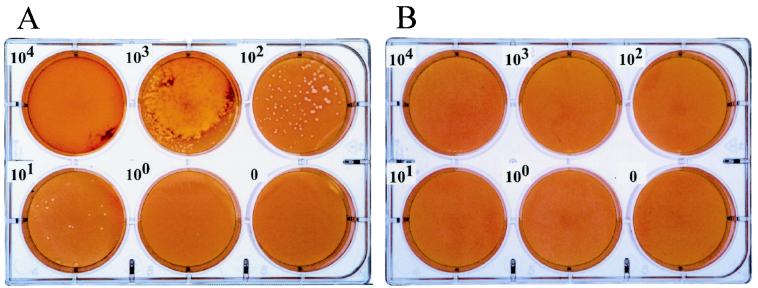
FIG. 1.
Comparative plaquing efficiency of C. trachomatis serovar D and C. pneumoniae strain AR-39 on murine McCoy cells. Chlamydiae were inoculated onto monolayers of McCoy cells at serial 10-fold dilutions ranging from 105 to 101 IFU per well and processed for plaque formation as described by Matsumoto et al. (12). C. trachomatis was incubated at 37°C, and C. pneumoniae was incubated at 35°C, for 10 days, respectively. The monolayers were then stained with neutral red to detect plaque formation. C. trachomatis formed distinct plaques in a dose-dependent manner by this assay, whereas no plaques were observed for C. pneumoniae strain AR-39 at any of the input IFU.
FIG. 2.
Focus and plaque morphology of C. pneumoniae AR-39 (A, C, E, G, and H) and TW-183 (B, D, and F) by immunostaining of viable HeLa 229 cell monolayers. Monolayers of HeLa 229 cells were infected with C. pneumoniae AR-39 or TW-183 at a multiplicity of infection of 50 and 25 IFU/well. After 10 days of incubation at 35°C, the infected monolayers were stained by IFA as either viable or fixed cells and examined for fluorescence. (A and B) Native monolayer after removal of the agarose overlay. AR-39 infection does not affect the monolayer (A), whileTW-183 forms small plaques with diameters of about 0.5 to 1.5 mm (B). (C and D) Monolayer of viable cells incubated with anti-MOMP MAb GZD1E8 followed by staining with FITC-labeled goat anti-mouse antibody (total magnification, ×25). Note positive-staining focus on the monolayer's surface exhibiting a galaxy-like fluorescence pattern. The staining of the foci is noncontiguous and radiates in a circumferential pattern away from its center. Foci ranged in size from 0.5 to 3.0 mm. (C) The focus shown (strain AR-39) is approximately 1.8 mm in diameter. (D) Some foci of TW-183 show a central clearing in the fluorescence pattern. (E and F) Similar foci to those shown in panels C and D but observed by simultaneous phase and fluorescent microscopy (magnification, ×100). While AR-39 does not affect the confluency of the monolayer (E), most foci of TW-183 show a central cell lysis (F) that is correlated to the central clearing of fluorescence shown in panel D. (G) Same focus as that shown in panel C but viewed at a magnification of ×200. The immunostaining with MOMP MAb produced a nonstructural nonuniform punctate staining pattern on the surface of the monolayer. (H) A monolayer stained for foci (AR-39), fixed with methanol, and secondarily stained with anti-MOMP and a TRITC-labeled goat anti-mouse antibody (magnification, ×200). C. pneumoniae inclusions (red) were found in HeLa cells underlying the infection focus (green). Inclusions were only observed in association with foci, demonstrating that the source of antigen detected on the viable cell surface was derived from the underlying infected cells of the monolayer.
We then investigated whether two other prototype C. pneumoniae strains, TW-183 and CWL-029, formed identifiable foci in this assay. CWL-029 formed foci of the same size as AR-39 and behaved similarly in all aspects. In contrast, TW-183 was very heterogeneous in the foci morphology. In addition to foci similar to those in AR-39 and CWL-029, it formed small plaques with diameters that ranged from 0.5 to 1.5 mm. The plaques were observable in the native monolayer after removal of the agarose overlay (Fig. 2B). Neutral red staining allowed the identification of plaques also, but the borders were poorly defined in comparison to C. trachomatis plaques in McCoy cells, and staining had no advantage over native viewing (data not shown). IFA staining showed that these plaques were associated with C. pneumoniae antigen (Fig. 2D). The staining pattern was similar to that in AR-39, except that cell lysis occurred in a small area in the center of the foci (Fig. 2F). Together, these findings suggest that C. pneumoniae infection spreads from cell to cell producing a progressively larger circumference of infected cells with extended incubation time. AR-39 and CWL-029 infection do not result in cell lysis, while TW-183 is unique in its ability to cause, in addition to nonlytic foci, plaques with a central area of cell lysis that can differ in size. Because only antibodies specific to antigens contained in the organism's outer membrane (e.g., MOMP and LPS) were reactive, it is likely that the focal antigen material represents chlamydial organisms released from cells that have either reattached or nonspecifically associated with the surface of uninfected or previously infected cells.
Concordance of input IFU and formation of foci.
To determine if the foci were the result of infection by a single organism, monolayers were infected with a known number of IFU of AR-39 and then assayed for foci, and the concordance of the two numbers was evaluated. There was a nearly 1:1 relationship between input IFU and foci detected at each of the input IFU dilutions tested (Table 1). Thus, based on this analysis, we conclude that a focus is generated from a single infectious chlamydial particle.
TABLE 1.
Mean observable C. pneumoniae AR-39 foci from two separate focal assays compared to serially diluted input IFU
| Assay no.b | No. of observable foci with input IFUa
|
||||
|---|---|---|---|---|---|
| 25.0 | 12.5 | 6.25 | 3.12 | 1.56 | |
| 1 | 26.0 | 11.0 | 5.0 | 3.0 | 1.0 |
| 22.0 | 13.0 | 7.0 | 1.0 | 3.0 | |
| 24.0 | 15.0 | 10.0 | 3.0 | 0.0 | |
| 25.0 | 15.0 | 6.0 | 4.0 | 2.0 | |
| 25.0 | 9.0 | 6.0 | 4.0 | 1.0 | |
| Mean ± SEc | 24.4 ± 0.7 | 12.6 ± 1.2 | 6.8 ± 0.9 | 3.0 ± 0.5 | 1.4 ± 0.5 |
| 2 | 18.0 | 12.0 | 6.0 | 2.0 | 1.0 |
| 25.0 | 11.0 | 2.0 | 3.0 | 0.0 | |
| 25.0 | 15.0 | 6.0 | 3.0 | 4.0 | |
| 24.0 | 11.0 | 8.0 | 2.0 | 1.0 | |
| 28.0 | 13.0 | 4.0 | 3.0 | 4.0 | |
| Mean ± SEc | 24.0 ± 1.6 | 12.4 ± 0.7 | 5.2 ± 1.0 | 2.6 ± 0.2 | 2.0 ± 0.8 |
Input IFU were calculated as 25 IFU per well, and four twofold serial dilutions were performed to obtain additional concentrations of 12.5, 6.25, 3.12, and 1.56 IFU per well.
Focal assays 1 and 2 consisted of five wells of each final concentration of input IFU.
Mean values (n = 5) ± standard error.
Recovery of C. pneumoniae from foci.
We next determined the percentage of foci that yielded recoverable C. pneumoniae organisms (Fig. 3). We screened 75 foci of AR-39 and found that 74 of 75 (98.7%) of the foci produced recoverable organisms. Recoverable IFU ranged from 7 to 5,600, with a median of 147. Chlamydiae were not recovered when samples were collected for areas of the monolayer that did not contain foci. Based on these results we conclude that the focus-forming assay is capable of cloning C. pneumoniae and that clones can be recovered from individual foci in a highly efficient manner.
FIG. 3.
Quantitation of infectious C. pneumoniae AR-39 recovered from individual foci. A total of 75 foci were assayed for recoverable chlamydial IFU as described in the text. Chlamydiae were recovered from 74 of the75 foci assayed (98.7%). The number of IFU recovered from individual foci ranged from 7 to 5,600, with a median of 147. No infectious organisms were recovered from areas of the monolayer that were negative for focus staining (data not shown).
Cloning and genetic analysis of C. pneumoniae strain MUL-250.
The focus-forming assay was used to prove the concept of intrastrain polymorphism and to isolate clonal variants from a polymorphic parent strain. The presence of at least two genotypes within the strain MUL-250 was shown by two tyrP-encoding loci in the Southern blot analysis (single tyrP indicated by a 3,974-bp band and double tyrP indicated by a 5,623-bp band; Fig. 4B, lane 1). A total of 60 foci were picked and expanded, and the cloned organisms were screened for the duplicated tyrP region by PCR. One clone (clone 38) was positive by PCR, indicating the presence of a tyrP duplication. The other clones (clone 41 shown representatively) were PCR negative, consistent with clonal organisms possessing a single copy of the tyrP gene (Fig. 4C). To definitively demonstrate that these clones represented distinct genovars, DNA of clones 38 and 41 was digested by Nru I. Southern blot analysis showed that the genotypic tyrP variants present in the parent C. pneumoniae strain MUL-250 material (single and double tyrP) were isolated as clonal bacterial lines with either a single or double tyrP.
DISCUSSION
In this study we describe the isolation of clonal variants of C. pneumoniae using a novel focus-forming assay with HeLa 229 cells. Foci were easily detected by IFA staining using MOMP-specific antibody on the surface of the HeLa monolayer. Following infection and an approximate period of three to four growth cycles, foci that are associated with underlying C. pneumoniae-infected cells form a galaxy-like fluorescence pattern at the monolayer's surface (Fig. 2). The foci are thought to originate from a single EB based on our findings that the input IFU/FFU ratio approached unity (Table 1). Most importantly, we proved the concept of intrastrain polymorphism by cloning a clinical isolate that consisted of a mixed population of genovars possessing either a single or duplicated copy of the tyrP gene. Using the focus-forming assay, we separated the polymorphic C. pneumoniae strain into genotypically distinct clonal variants. This method will enable future studies using clonal isolates to determine whether genotypic variants have different biological or pathogenic properties.
A method for plaque cloning C. trachomatis has been described previously using murine McCoy cells (12). This method utilized the ability of this species to lyse the host cells and to cause an identifiable isolated plaque following staining with neutral red. However, we observed that none of the C. pneumoniae prototype strains formed similar plaques using this assay (Fig. 1). Thus, we adapted this assay but incorporated human epithelial cells (HeLa 229) as host cells instead of McCoy cells, since the former are known to be a more permissive cell line for the growth of C. pneumoniae (11). Furthermore, we found that it was necessary to use a higher concentration of cycloheximide in the agarose overlay to prevent overgrowth of HeLa cells during extended culture periods required for foci formation. Despite these changes to optimize the assay's sensitivity for C. pneumoniae growth, we found that all three prototype strains formed nonlytic foci and only TW-183 formed both foci and small plaques of different size. Therefore, IFA staining of the viable HeLa cell monolayers is necessary to detect foci of AR-39 and CWL-029 as well as nonplaquing clones of TW-183.
We hypothesize from our observations that focus formation with AR-39 and CWL-029 strains resulted from a cell-to-cell spread of the chlamydiae, producing a progressively larger circumference of infection with extended incubation time. This conclusion is supported by the observation that a, FFU originates from a single organism (Table 1). It is unclear why only some cells in these loci contain inclusions (Fig. 2H). Previously infected nonlysed cells may become refractory to reinfection. Regardless of the reasons, it is evident that (i) the organism forms a clearly identifiable focus on the cell surface, (ii) there is a near-unity relationship between the input IFU inoculum size and resultant FFU, and (iii) C. pneumoniae can be readily isolated from individual foci. The nature of the IFA-reactive, chlamydial material on the monolayer surface is unknown. Our findings that foci could be detected with antibodies specific to surface-exposed outer membrane molecules of C. pneumoniae (MOMP and LPS), but not non-surface-exposed cytoplasmic antigen (HSP60), argues that the focus material deposited on the monolayer surface is composed of structurally intact organisms. It is possible that these particles have been released from infected cells but remain associated with the focal area by reattaching or associating nonspecifically with neighboring cells.
The C. pneumoniae in vitro growth cycle has been reported to result in host cell lysis following the burst of the chlamydial inclusion (8). The absence of cell lysis in AR-39, CWL-029, and some foci of TW-183 contradicts this. Our results show that release of C. pneumoniae can occur without destruction of the host cell. Nonlytic spread has been shown before for C. trachomatis, and electron microscopic studies suggested exocytosis as a mechanism (18). The relationship of this growth phenotype and pathogenesis is not known, but it is interesting to make the analogy that C. pneumoniae infections are commonly persistent and associated with chronic inflammatory disease, a correlation that might reflect this nonlytic in vitro growth phenotype.
We observed differences among C. pneumoniae strains in their ability to form foci (AR-39, CWL-029) or small plaques and foci (TW-183). It is intriguing to speculate that these different morphotypes might be inherently connected to genotypic differences. The two focus-forming strains AR-39 and CWL-029 have been sequenced and exhibit >99.9% homology (14). The sequence of the plaque-forming strain TW-183 is unknown and therefore might differ significantly from either AR-39 or CWL-029. Moreover, the presence of foci and plaques of different sizes within TW-183 might reflect intrastrain polymorphisms. It is possible that a more careful genetic and biological analysis of the small-plaque- and focus-forming TW-183 phenotypes will lead to the discovery of differences in infectivity or pathogenesis. With the ability to clone these phenotypes, it is now possible to address these questions experimentally.
The concept of polymorphism within a C. pneumoniae strain was first introduced by the sequencing of strain AR-39 (14). Read et al. identified 304 intrastrain polymorphisms, most of which were single-nucleotide polymorphisms, and a deletion of one unit of a tandem 1,649-bp repeat containing a tyrP gene. We were unable to demonstrate a similar tyrP polymorphism with the AR-39 strain used in our laboratory. The reasons for these differences in findings are not understood, but extensive in vitro propagation may have led to the selection of a homogenous tyrP population. The polymorphic nature of the tyrP locus was, however, confirmed in three clinical isolates, of which MUL-250 was chosen for separation of the tyrP variants (Fig. 4). Using the focus-forming assay, we were able to clone organisms that contained either a single or duplicate copy of this gene. The origin of the tyrP polymorphism is unclear, but given the nature of the gene product (aromatic amino acid transport into bacterial cell) and the dependence of C. pneumoniae on host cell sources of aromatic amino acids, the duplication may well be an important pathogenic adaptation. It will be interesting to determine if these genovars have different pathogenic propensities in infection models, experiments that can only be done following clonal isolation of the genotypic variants. In summary, the polymorphic nature of C. pneumoniae suggests that differences in pathogenic potential exists. The ability to work with genetically homogenous clonal populations should be a major factor in future studies aimed at understanding the pathogenesis of C. pneumoniae.
Acknowledgments
We appreciate the technical assistance of John Carlson and secretarial assistance of Kelly Matteson. We thank Gary Hettrick and Anita Mora for their help in preparation of the figures.
This work was partially supported by Public Health Service grant AI 42790 (to G.I.B.). J.G. was supported by a fellowship of the Deutsche Forschungsgemeinschaft, Bonn, Germany (GI 344/1-1).
Editor: A. D. O'Brien
REFERENCES
- 1.Blasi, F. 1996. Clinical features of Chlamydia pneumoniae acute respiratory infection. Clin. Microbiol. Infect. 1:S14-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasi, F., P. Tarsia, C. Arosio, L. Fagetti, and L. Allegra. 1998. Epidemiology of Chlamydia pneumoniae. Clin. Microbiol. Infect. 4:S1-S6. [PubMed] [Google Scholar]
- 3.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czub, M., S. Czub, F. J. McAtee, and J. L. Portis. 1991. Age-dependent resistance to murine retrovirus-induced spongiform neurodegeneration results from central nervous system-specific restriction of virus replication. J. Virol. 65:2539-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grayston, J. T. 1992. Infections caused by Chlamydia pneumoniae strain TWAR. Clin. Infect. Dis. 15:757-761. [DOI] [PubMed] [Google Scholar]
- 6.Grayston, J. T., and L. A. Campbell. 1999. The role of Chlamydia pneumoniae in atherosclerosis. Clin. Infect. Dis. 28:993-994. [DOI] [PubMed] [Google Scholar]
- 7.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 8.Kuo, C. C., and J. T. Grayston. 1990. A sensitive cell line, HL cells, for isolation and propagation of Chlamydia pneumoniae strain TWAR. J. Infect. Dis. 162:755-758. [DOI] [PubMed] [Google Scholar]
- 9.Maass, M., C. Bartels, P. M. Engel, U. Mamat, and H. H. Sievers. 1998. Endovascular presence of viable Chlamydia pneumoniae is a common phenomenon in coronary artery disease. J. Am. Coll. Cardiol. 31:827-832. [DOI] [PubMed] [Google Scholar]
- 10.Maass, M., and K. Dalhoff. 1994. Comparison of sample preparation methods for detection of Chlamydia pneumoniae in bronchoalveolar lavage fluid by PCR. J. Clin. Microbiol. 32:2616-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maass, M., and U. Harig. 1995. Evaluation of culture conditions used for isolation of Chlamydia pneumoniae. Am. J. Clin. Pathol. 103:141-148. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto, A., H. Izutsu, N. Miyashita, and M. Ohuchi. 1998. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J. Clin. Microbiol. 36:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puolakkainen, M., L. A. Campbell, C. C. Kuo, M. Leinonen, C. Gronhagen-Riska, and P. Saikku. 1996. Serological response to Chlamydia pneumoniae in patients with sarcoidosis. J. Infect. 33:199-205. [DOI] [PubMed] [Google Scholar]
- 14.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirai, M., H. Hirakawa, M. Kimoto, M. Tabuchi, F. Kishi, K. Ouchi, T. Shiba, K. Ishii, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28:2311-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su, H., and H. D. Caldwell. 1995. CD4+ T cells play a significant role in adoptive immunity to Chlamydia trachomatis infection of the mouse genital tract. Infect. Immun. 63:3302-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tauber, E., C. Wojnarowski, E. Horcher, G. Dekan, and T. Frischer. 1999. Necrotizing sarcoid granulomatosis in a 14-yr-old female. Eur. Respir. J. 13:703-705. [DOI] [PubMed] [Google Scholar]
- 18.Todd, W. J., and H. D. Caldwell. 1985. The interaction of Chlamydia trachomatis with host cells: ultrastructural studies of the mechanism of release of a biovar II strain from HeLa 229 cells. J. Infect. Dis. 151:1037-1044. [DOI] [PubMed] [Google Scholar]
- 19.Von Hertzen, L., T. Vasankari, K. Liippo, E. Wahlstrom, and M. Puolakkainen. 2002. Chlamydia pneumoniae and severity of asthma. Scand. J. Infect. Dis. 34:22-27. [DOI] [PubMed] [Google Scholar]
- 20.Wolf, K., E. Fischer, D. Mead, G. Zhong, R. Peeling, B. Whitmire, and H. D. Caldwell. 2001. Chlamydia pneumoniae major outer membrane protein is a surface-exposed antigen that elicits antibodies primarily directed against conformation-dependent determinants. Infect. Immun. 69:3082-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]