Abstract
MicroRNAs (miRNAs) regulate cellular fate by controlling the stability or translation of mRNA transcripts. Although the spatial and temporal patterning of miRNA expression is tightly controlled, little is known about signals that induce their expression nor mechanisms of their transcriptional regulation. Furthermore, few miRNA targets have been validated experimentally. The miRNA, miR132, was identified through a genome-wide screen as a target of the transcription factor, cAMP-response element binding protein (CREB). miR132 is enriched in neurons and, like many neuronal CREB targets, is highly induced by neurotrophins. Expression of miR132 in cortical neurons induced neurite outgrowth. Conversely, inhibition of miR132 function attenuated neuronal outgrowth. We provide evidence that miR132 regulates neuronal morphogenesis by decreasing levels of the GTPase-activating protein, p250GAP. These data reveal that a CREB-regulated miRNA regulates neuronal morphogenesis by responding to extrinsic trophic cues.
Keywords: neurite, neurotrophin, plasticity, microRNA, transcription
Hormones, growth factors, and electrical activity regulate proliferation, differentiation, survival, and plasticity by triggering programs of gene expression. Although many transcription factors respond to environmental cues, the basic leucine zipper transcription factor, cAMP-response element binding protein (CREB), is considered prototypical because it was among the first identified, is expressed widely, and regulates many rapid-response genes (1). Phosphorylation of a conserved residue in its activation domain is required for CREB function (2). Although CREB was identified by virtue of its responsiveness to cAMP signaling, it is activated by an array of other cellular signals, including, but not limited to, neurotrophic factors, cytokines, and neuronal activity. The ability of CREB to recruit the coactivator, CREB binding protein (CBP) (3), in a phosphorylation-dependent manner underlies its capacity to mediate stimulus-induced transcription. CBP recruitment is believed to trigger transcriptional activation via intrinsic or associated acetylase activities and/or by interacting with general transcription factors (4). Recently, signal-dependent nuclear translocation of the coactivator transducer of regulated CREB has also been implicated in CREB activation (5).
The role of CREB in cellular adaptive responses has been studied most extensively in the nervous system. Early experiments revealed that stimuli known to effect neuronal maturation and plasticity, including cAMP signaling, membrane depolarization, and neurotrophins, were robust activators of CREB-dependent gene expression. These observations inspired other studies suggesting that CREB is a central regulator of memory formation and other forms of behavioral adaptation believed to require programs of de novo gene expression (6). Subsequent studies supported a role for CREB as a key regulator of developmental plasticity, addiction, and circadian rhythmicity (1).
A prominent role for CREB in neuronal survival has hampered analysis of its actions in regulating neuronal maturation and function. Nevertheless, a recent study provided strong evidence that CREB mediates neurotrophin-dependent morphogenesis of peripheral neurons (7). Additional studies have suggested a role for CREB in CNS morphogenesis (8, 9). The set of genes that directly mediate these effects has not been defined, however. To gain insight into the molecular mechanisms underlying CREB-regulated plasticity, we developed a genome-wide screen, termed Serial Analysis of Chromatin Occupancy that can profile transcription factor binding sites in an unbiased manner (10). This screen identified hundreds of CREB-binding sites that are tightly associated with noncoding transcripts and microRNAs (miRNAs) (S.I. and R.H.G., unpublished data). In this study, we characterize one of these unique transcripts.
miRNAs were first described in Caenorhabditis elegans (11, 12). Subsequent work demonstrated that these molecules encode 19- to 24-bp double-stranded RNAs that mediate gene silencing (13-15). miRNAs have since been identified in vertebrates and have been proposed to regulate a significant fraction of cellular mRNAs. miRNAs regulate a diverse set of biological functions, including regulation of developmental timing and neuronal asymmetry in C. elegans (11, 12, 16, 17); cell proliferation, suppression of apoptosis (18), and fat metabolism (19) in Drosophila; and hematopoetic and adipocyte differentiation (20, 21), insulin secretion (22), cardiomyocyte development (23), oncogenesis (24-27), and viral defense in mammals (28-30). Mature miRNAs silence gene expression by binding to the 3′UTRs of target mRNAs and promote translational repression or mRNA degradation (31).
Significant strides have been made in deciphering the stepwise processing of miRNAs from their larger precursors (32). The relatively few primary transcripts thus far studied suggest that miRNAs are derived from spliced coding or noncoding transcripts (33-35). Because many miRNA genes are near complex transcriptional loci, the regulatory elements responsible for miRNA biogenesis have been difficult to determine. Most studies indicate that miRNAs are derived from polyadenylated RNA polymerase II transcripts, but polymerase III-dependent mechanisms have also been suggested (36).
By analogy with protein-coding genes, identification of the pathways that control miRNA transcription should provide insight into the biological functions of these molecules. We focused on miRNA 132 (miR132) because it was tightly associated with a CREB-binding site and was highly responsive to neurotrophin signaling, suggesting a role in neuronal differentiation. Overexpression of this miRNA in cortical neurons dramatically increased the sprouting of neuronal processes. Conversely, inactivation of this miRNA reversed this process. We used bioinformatic databases to identify putative targets of miR132 and found that a highly conserved putative target, p250 GTPase-activating protein (GAP), had previously been linked to neuronal differentiation. We confirmed that p250GAP protein levels were controlled by miR132 in cortical neurons and showed that the ability of miR132 to regulate neuronal morphogenesis depends, at least in part, on its ability to target p250GAP.
Materials and Methods
Cell Culture and Stimulation. PC12 cells and neonatal rat cortical neurons were cultured as described (37). Brain-derived neurotrophic factor (BDNF; Chemicon) was dissolved in dH2O and used at 50 ng/ml (final concentration).
Plasmids. The premiR1-1 and premiR132 hairpins were amplified from rat genomic DNA by using the following primers: miR1-1 forward, TGGCGAGAGAGTTCCTAGCCTG; miR1-1 reverse, GTGTGCACAACTTCAGCCCATA; miR132 forward, CTAGCCCCG CAGACACTAGC; miR132 reverse, CCCCGCCTCCTCTTGCTCTGTA. premiR1-1, premiR132, GFP, LacZ, and ACREB were cloned in pCAG. The p250GAP (CTAGAAAAAATTCAACAACCACGGACTGACTCTTGATCAGTCCGTGGTTGTTGAA and TTTGTTCAACAACCACGGACTGATCAAGAGTCAGTCCGTGGTTGTTGA-ATTTTTT) duplex, short hairpin RNA (shRNA) was cloned into mU6pro (38). An shRNA vector that targets luciferase was used as a control (38). The NFκB-luc reporter was from Clontech. GFP-p250GAP is described in ref. 39. Details on the subcloning and purification of plasmids will be provided on request.
Transfection and Reporter Assays. Electroporation of rat cortical neurons was conducted by using the rat neuron nucleofection kit according to the manufacturer's instructions (Amaxa). The 2′O-methyl oligos (sense, UCCAUUGUCAGAUGUCGGUACCAGCGGGGCG; antisense, GGGCAACCGUGGCUUUCGAUUGUUACUGUGG; IDT, Coralville, IA) were electroporated by using the Amaxa (Gaithersburg, MD) nucleofector at 1, 10, and 100 nM. Luciferase and β-galactosidase activity were measured by using Luciferase and Galacoto-Light assay kits (PerkinElmer).
Western Blotting and Chromatin Immunoprecipitation. Western blotting was conducted as described (37). The following primary Abs were used overnight at 4°C in Tris-buffered saline containing 0.1% Triton X-100, 10 mM NaF, and 5% BSA: polyclonal anti-p250GAP (1:1,000) (39), monoclonal anti-GFP (Clontech), polyclonal anti-CBP (Santa Cruz Biotechnology, A22), and polyclonal anti-LacZ (5′ 3′). Then, 1-2 × 106 neonatal day 5 rat cortical neurons were subjected to chromatin immunoprecipitation as described (10).
Immunocytochemistry and Quantitation of Neuronal Morphology. Cortical neurons were fixed in 4% formaldehyde in PBS, pH 7.4, with 75 mM Hepes for 5 min. Neurons were blocked (1-2 h at 22°C) in 5% BSA in PBS with 0.05% Triton X-100 and 10 mM NaF. The rabbit anti-p250 Ab was used at 1:1,000, and anti-MAP2 mAb (Sigma) was used at 1:2,000. The Abs were visualized with 1 μg/ml Alexa 594 anti-rabbit IgG or 1 μg/ml Alexa 488 anti-mouse IgG (Molecular Probes). Neurites from GFP fluorescent neurons were traced and quantified by using the neuronj program (50) that allows semiautomatic tracings of neurites. Neurites were traced and quantified in a blinded manner.
Northern Blotting. RNA was purified by using the mirVana miRNA isolation kit (Ambion) and separated on a 15% denaturing acryl-amide gel. RNAs were transferred onto GeneScreen Plus membranes (PerkinElmer), UV crosslinked, and baked for 1 h at 80°C. A locked nucleic acid probe (40) antisense to the mature miR132 was hybridized at 40°C as described (13). Levels of tRNA were stained with EtBr. Locked nucleic acid oligonucleotide (IDT) probes used were: miR132 antisense, CGTACCATG*GCTGT*AG*AC*TGT*TA and mir132 sense, CG*ACCATG*GCTGT*AG*AC*TGT*TA. A modified nucleotide is preceded by an asterisk.
Statistics. Data with homogenous variances were analyzed by using the Student t test or analysis of variance and Tukey's post test. Nonparametric data were analyzed by using the Kruskal-Wallis multiple comparisons test.
In Vivo Genomic Footprinting. Genomic footprints were conducted as described (41) by using DNA isolated from dimethyl sulfate-treated PC12 cells. Control DNA was isolated from cells and treated with dimethyl sulfate in vitro. The DNA was subsequently cleaved, and the footprints were revealed by ligation-mediated PCR by using the following nested primer sets: miR132 (CCTCAGTAACAGTCTCCA, TCTCCAGTCACGGCCACC, and GCACGCCCGTCCA-AGGTC) and miR212 (CGTGACTGGAGACTGTTACT, CAGTAAGCAGTCTAGA-GCCAAGGT, and CGTCGCCCGCAGTTGAGAGTG-ATG).
Reverse Transcription and Real-Time PCR. Total RNA was isolated by using TRIzol (Invitrogen), and 100 ng of RNA was reverse-transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and 100 ng of random primers (Invitrogen). Primers were designed by using MIT's primer3 software (51) with default parameters except for the following settings: “rodent and simple repeat” was selected, amplicon size was 50-150 bp, primer size was 18-27 bases, Tm was 68°C, maximum self-complementarity was 4, and maximum 3′ complementarity was 2. Primer sequences are available on request. PCRs (10 μl) contained 1 μl of 10× PCR buffer, 2.5 mM MgCl2, 200 μM dNTP (Roche), 0.125 μM primer, 1× SYBR green I (Invitrogen), and 1 unit of Platinum Taq (Invitrogen). PCR was performed on an Opticon OP346 (MJ Research) for 40-50 cycles at 94°C for 15 s and 68°C for 40 s. All experimental points were within the linear range of a standard curve with an R2 of at least 0.99. PCR amplicons were confirmed by electrophoresis on 4% NuSeive GTG agarose gels (Cambrex, East Rutherford, NJ) and by automated sequencing (Applied Biosystems).
RNA Ligase-Mediated Rapid Amplification of cDNA Ends. Rat cortical neuron cDNA (2 μg) was subjected to 5′ oligo-capping RACE and conventional 3′ RACE according to the GeneRacer protocol (Invitrogen). The following primers were used for 3′ RACE: GACGCAACATCCGATGTATCTGCGG and CACCACTCCCGAGTTCTGCCAGCCTG. The reverse complements of the above primers were used for 5′ RACE. The miR132 cDNA sequence was deposited in the GenBank database.
Results
CREB Regulates miR132 Transcription. The CREB Serial Analysis of Chromatin Occupancy screen identified 13 genomic signature tags that fell in the vicinity of the miR212/132 locus (Fig. 1A). The largest cluster of genomic signature tags was located within the miR132 transcript. Two consensus CREs (TGACGTCA) are located immediately 5′ to the predicted miR212 transcript and another is located closer to miR132. The entire miR212/132 locus falls within a CpG island. CREB binding was confirmed at the consensus CRE sequences but not within a region between the two miRNAs (Fig. 1B), and these results were confirmed by using in vivo DNase I footprinting assays (Fig. 1C). Because miR212 has not yet been experimentally verified, we focused our analysis on miR132. To characterize its expression, we used real-time RT-PCR analysis of RNAs isolated from selected brain regions. These studies showed that premiR132 is enriched in hippocampus, cortex, and brainstem (Fig. 1D).
Fig. 1.
The neuronal miRNA miR132 is regulated by CREB. (A) A diagram indicating the relative positions of genomic signature tags (GSTs, red), CRE motifs (blue), and predicted premiRNA sequences for the miR212 and miR132 cluster. (B) Neocortical neurons were subjected to chromatin immunoprecipitation by using a CREB Ab or IgG control. Real-time PCR was conducted by using primers that interrogate the regions indicated in A.(C) In vivo genomic footprinting was performed in PC12 cells. (Left) Footprint surrounding two consensus CREs upstream of miR212. (Right) Footprint surrounding the single CRE upstream of miR132. The left lanes of each panel indicate dimethyl sulfate-cleaved naked genomic DNA and the right lanes show cleaved DNA from cells treated with dimethyl sulfate in vivo. Arrows denote footprinted bands and asterisks denote hypersensitive sites. (D) Real-time PCR for premiR132 was performed on cDNA from the indicated E18 mouse tissues (hip, hippocampus; cx, neocortex; bs, brainstem; skm, skeletal muscle). The data were normalized to GAPDH cDNA levels.
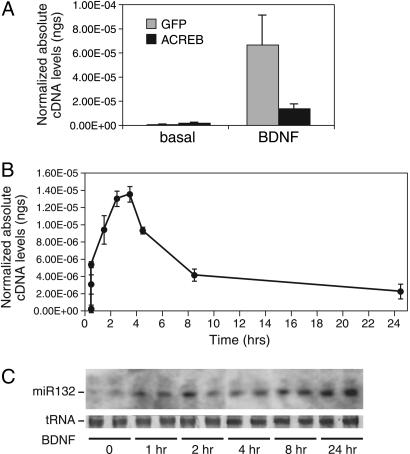
Response of the miR132 Gene Is Rapid and Long-Lasting. CREB has been shown to mediate rapid responses to stimuli, such as neurotrophins, by inducing the expression of immediate early genes. We used real-time RT-PCR to determine whether CREB similarly regulated miR132 transcription. BDNF markedly increased the expression of the miR132 precursor (Fig. 2A). Cotransfection of ACREB, a mutant that inhibits DNA binding of endogenous CREB (42), reduced miR132 transcription in response to BDNF. ACREB expression had no effect on BDNF-stimulated transcription of an NFκB-responsive reporter gene (data not shown). BDNF stimulation of premiR132 persists for at least 24 h, with a maximal level of stimulation at 2-3 h (Fig. 2B). Northern blot analysis confirmed that BDNF triggers the rapid induction and persistent expression of mature miR132 (Fig. 2C).
Fig. 2.
miR132 is induced by neurotrophins. (A) Cortical neurons were transfected with dominant negative CREB (ACREB) or vector control (GFP) and treated 36 h later with BDNF for 60 min. RNA was reverse-transcribed and analyzed by real-time PCR with premiR132 primers. The data were normalized to GAPDH cDNA levels. Error is SEM of five to six replicates. (B) Cortical neurons were treated with BDNF for the indicated times. RNA was reverse-transcribed and subjected to real-time PCR with primers specific to premiR132. The data were normalized to GAPDH cDNA levels. Error is SEM of three replicates. (C) Cortical neurons were treated with BDNF for the indicated times. Small molecular weight RNA was isolated and subjected to Northern analysis by using a probe to mature miR132. tRNA levels are shown as a loading control.
miR132 Is Processed from the Stable Intron of an Ephemeral Noncoding RNA. miRNAs are frequently processed from the introns of canonical mRNAs that ultimately are transported to the cytoplasm. In an effort to clone the miR132 primary transcript, we examined rat mRNA and EST annotation surrounding miR132 and found only a few low-confidence unspliced transcripts. Using RT-PCR, we mapped the 5′-splice site of a unique transcript whose first exon was located upstream of miR212 and whose second exon began downstream of miR132 (Fig. 3A). 5′ cap-trapping RACE and 3′ RACE produced single products that confirmed a two exon transcript without a significant ORF. We next determined whether the parent transcript was activated by neurotrophins. Real-time RT-PCR revealed neurotrophin-induced transcription only in areas that overlapped the cloned rat cDNA (Fig. 3B). Surprisingly, the exonic sequences were present at ≈40-fold lower levels than the products of the intron (Fig. 3B). These experiments suggest that the intron represents the biologically important transcript. These results are reminiscent of the generation of guide small nucleolar RNAs, which are often processed from the stabilized introns of noncoding transcripts (43, 44).
Fig. 3.
miR132 is transcribed from the stable intron of a cryptic noncoding RNA. (A) A diagram indicating the position of premiR132 and premiR212 and a unique rat noncoding transcript on chromosome 10 (UCSC Genome Center Rn3 assembly). A CpG island and mouse, rat, human, fugu, dog conservation track are indicated. All annotation is from the UCSC Genome Center. The red box in the RT-PCR track indicates a PCR product that interrogates premiR132. (B) Cortical neurons were treated with BDNF or vehicle for 60 min. RNA was reverse-transcribed and analyzed by real-time PCR by using primers interrogating the locations indicated in A. Error is SEM of four replicates.
Expression of miR132 Promotes Neurite Outgrowth. CREB is widely believed to regulate axonal and dendritic development. Genetic rescue experiments suggest that CREB is a critical regulator of NGF-induced axonal outgrowth in peripheral neurons (7). Likewise, attenuation of CREB function inhibits cAMP- and neurotrophin-induced neurite outgrowth in cortical neurons (8, 9). Because miR132 is enriched in brain tissue, we investigated whether miR132 might contribute to neuronal morphogenesis. Overexpression of the miR132 precursor in primary cortical neurons induced a marked increase in primary neurite outgrowth in comparison to cells transfected with GFP alone (Fig. 4 A and B). Northern blot analysis confirmed that expression of the miR132 precursor was associated with expression of the 22-bp mature miRNA (data not shown). This phenotype is specific for miR132, as overexpression of miR1-1, a heart-specific miRNA, does not effect neurite outgrowth. The average total neurite length for miR132-transfected neurons was 42% greater than that of miR1-1 transfected controls (Fig. 4B). Furthermore, transfection of an antisense 2′O-methyl RNA, but not a sense control, attenuated neurite outgrowth (Fig. 5A). Morphometric analysis revealed that transfection of the antisense 2′O-methyl RNA induced a significant decrease in both the number of primary neurites and the average total neurite length (Fig. 5B).
Fig. 4.
Expression of miR132 induces neurite sprouting. (A) Neonatal cortical neurons were transfected with a GFP reporter (green) and cotransfected with vector control, or expression constructs for premiR1-1 or premiR132. Cells were immunostained for the neuronal marker MAP2 (red). (B) Neurons were transfected as in A and analyzed morphometrically. The histogram depicts the distribution of neurons plotted as bins of neurites. The distributions were statistically distinct (P < 0.01; Kruskal-Wallis). (Inset) The average total neurite length (TNL) of miR1-1 (n = 109) and miR132 (n = 137) transfected neurons from four independent experiments. *, P < 0.01 for miR132 vs. miR1-1 (Student's t test).
Fig. 5.
Transfection of a 2′O-methyl inhibitor of miR132 markedly attenuated neurite outgrowth. (A) Cortical neurons were transfected with a GFP reporter (green) and cotransfected with empty vector or 2′O-methyl oligoribonucleotide directed against sense or antisense miR132. Cells were immunostained for the neuronal marker MAP2 (red). (B) Neurons were transfected as in A and analyzed morphometrically. The histogram depicts the distribution of neurons plotted as bins of neurites. The distributions were statistically distinct (P < 0.01; Kruskal-Wallis). (Inset) Average total neurite length (TNL) of vector (n = 94), sense (n = 64), and antisense (n = 69) transfected neurons from three independent experiments. *, P < 0.01 for antisense vs. vector or sense (ANOVA, Tukey's posttest).
miR132 Targets p250GAP. Because miRNAs generally inhibit expression of their target mRNAs, we hypothesized that the target of miR132 encodes a protein that represses neurite outgrowth. Analysis of miR132 (through the UCSC Genome Bioinformatics web site) reveals a high degree of conservation amongst vertebrates, suggesting that its targets may be similarly conserved. To predict possible miR132 targets, we used the miranda prediction algorithm (52) and looked for evolutionary conservation of the target 3′UTRs in neuronally expressed genes (45). Although many predicted targets for miR132 were identified, a region of the 3′UTR of p250GAP, a member of the Rac/Rho family of GAPs, was among the most highly conserved evolutionarily. A comparison of the 3′UTRs of p250GAP reveals that the putative target site for miR132 is conserved from humans to zebrafish (Fig. 6A). Interestingly, p250GAP is highly enriched in the CNS (39). To test whether miR132 could block expression of p250GAP, we transfected a p250GAP-GFP reporter and miR132 precursor into primary neurons. Cotransfection of miR132, but not miR1-1, selectively repressed the expression of a luciferase gene that contained a p250GAP miRNA response element in its 3′UTR (data not shown). Overexpression of miR132 reduced levels of p250GAP-GFP whereas miR1-1 did not (Fig. 6B). Furthermore, miR132 specifically reduced endogenous p250GAP levels in primary neurons (Fig. 6C), confirming that p250GAP is a target of miR132 in vivo. To examine the specificity of miR132 regulation of p250GAP protein expression, we blocked miR132 function in primary neurons by using an antisense 2′O-methyl oligoribonucleotide. We found that this sequence-specific miRNA inhibitor increased levels of the p250GAP-GFP and endogenous p250GAP, whereas a sense strand 2′O-methyl oligoribonucleotide did not (Fig. 6 D and E). As shown above, the antisense (but not the sense strand) 2′O-methyl oligoribonucleotide also blocked neurite outgrowth (Fig. 5B). Because we did not observe an miR132-induced reduction in p250GAP mRNA (data not shown), we suggest that miR132 regulates p250GAP by a translational block.
Fig. 6.
p250GAP is a target of miR132. (A) The miR132 target sequence in the p250GAP 3′UTR is conserved across vertebrate evolution. Yellow indicates perfect base pairing, green indicates wobble base pairing, and gray indicates no match. (B) Cortical neurons were cotransfected with GFP-tagged p250GAP and premiRNA132 or premiRNA1-1 expression constructs. LacZ was costransfected as a control. Cell lysates were immunoblotted for GFP and LacZ. -, cells not transfected with p250GAP. (C) Neonatal cortical neurons were transfected with premiR1-1 and premiR132 expression constructs. Cell lysates were immunoblotted for p250GAP or CBP. (D) Cortical neurons were cotransfected with GFP-tagged p250GAP and sense or antisense 2′O-methyl oligos. LacZ was costransfected as a control. Cell lysates were immunoblotted for GFP and LacZ. -, cells not transfected with p250GAP. (E) Neonatal cortical neurons were transfected with sense or antisense 2′O-methyl oligos. Cell lysates were immunoblotted for p250GAP or CBP.
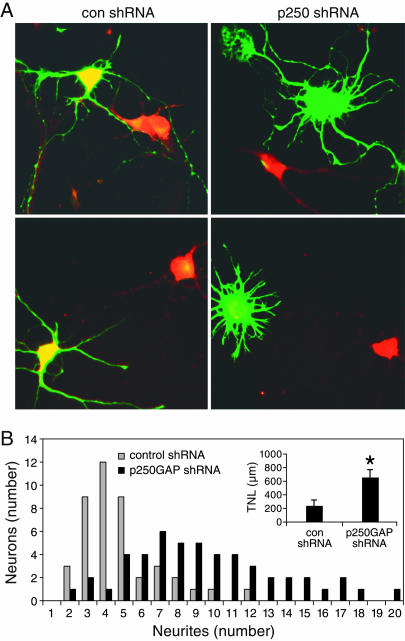
Inhibition of p250GAP Expression Promotes Neurite Outgrowth. Because miR132 stimulates neurite outgrowth, our data suggests that this phenotype is negatively regulated by p250GAP. To test this hypothesis, we generated a shRNA that represses expression of endogenous p250GAP in cortical neurons (Fig. 7A). We observed a striking increase in neurite outgrowth in cells transfected with the p250GAP shRNA construct as compared to a control shRNA construct (Fig. 7A). Morphometric analyses revealed that reduction of p250GAP levels induced a marked increase in primary neurite number and average total neurite length (Fig. 7B). These results support the hypothesis that miR132 mediates neuronal outgrowth by suppressing expression of p250GAP.
Fig. 7.
Down-regulation of p250GAP phenocopies miR132 expression. (A) Cortical neurons were transfected with a GFP reporter (green) and a p250GAP or control shRNA expression construct. Cells were immunostained for p250GAP (red). (B) Neurons were transfected as in A and analyzed morphometrically. The histogram depicts the distribution of neurons plotted as bins of neurites. The distributions were statistically distinct (P < 0.01; Kruskal-Wallis). (Inset) The average total neurite length (TNL) for control shRNA (n = 43) and p250GAP shRNA (n = 50) transfected neurons from two independent experiments. *, P < 0.05 (Student's t test).
Discussion
Experiments more than a decade ago in the sea snail, Aplysia, suggested a role for CREB in neuronal plasticity (6). Subsequently, a large body of work has implicated CREB in a variety of adaptive neuronal responses. Among the possible CREB targets, rapid response genes have received the most attention. Nevertheless, few CREB targets capable of directly regulating neuronal plasticity have been identified. Our unbiased genome-wide screen allowed us to identify many, if not all, potential CREB targets (10). Surprisingly, this screen revealed that a significant fraction of the rapid response genes under CREB control had little coding potential (S.I. and R.H.G., unpublished data). In the current study, we focused on miR132 because it was one of the most highly inducible genes characterized. We used overexpression and 2′O-methyl RNA blockers to show that miR132 is a key regulator of cortical neuron morphogenesis. Moreover, we demonstrate that miR132 downregulates the expression of p250GAP, a protein proposed to regulate neuronal differentiation (39, 46). Consistent with the hypothesis that p250GAP is a major target of miR132, knock-down of p250GAP phenocopies the regulation of neuronal morphogenesis by miR132.
Studies showing that p250GAP regulates neurite outgrowth via Rac/cdc42 signaling are consistent with a role for miR132 in early neuronal morphogenesis (39, 46). Of note, p250GAP also interacts with the glutamate receptor NR2A/B subunits, PSD-95, and β-catenin (39, 47). Interestingly, several additional predicted targets of miR132 are known to regulate the development of postsynaptic densities and spines. It is conceivable, therefore, that miR132 targets additional regulators of neuronal differentiation. The observation that miR132 is selectively expressed in zebrafish brain is consistent with the idea that it plays a role in neuronal development. The miRNA recognition element in p250GAP is one of the few predicted targets that, like miR132 itself, is conserved from fish to humans, suggesting a role for miR132 in vertebrate neurogenesis. miR132 can be considered to be a rapid response gene because its induction phase parallels that of c-fos (data not shown). However, mature miR132 shows a persistence not observed with other CREB-regulated immediate early genes. The pairing of rapid inducibility with persistent expression suggests that miR132 may function as a signal-dependent switch that regulates neuronal homeostasis over the long-term. Moreover, the ability of miRNAs to down-regulate the synthesis of a population of proteins provides a powerful mechanism for the coordinate control of neuronal function. miRNAs and the RNA-induced silencing complex machinery are associated with polyribosomes and dendritic RNA-binding proteins, raising the possibility that miRNAs are localized to discrete dendritic compartments (48, 49). Because some proteins have half-lives on the order of minutes, it is conceivable that induction of miR132 triggers a rapid and persistent downregulation of protein levels. Loss of function and over expression experiments should help reveal the precise role of miR132 in neuronal development and maturation. Conditional mouse mutants would also help determine whether miR132 contributes to memory consolidation and long-term neuronal plasticity.
Acknowledgments
We thank Gail Mandel and Karl Obrietan for their comments on the manuscript. This work was supported by National Institutes of Health Grants NS047176 (to S.I.) and DK45423 (to R.H.G.) and a grant from the Rett Syndrome Research Foundation (to R.H.G.).
Author contributions: R.H.G. and S.I. designed research; N.V., M.E.K., O.V., D.M.K., and S.I. performed research; T.Y. contributed new reagents/analytical tools; N.V., M.E.K., O.V., D.M.K., and S.I. analyzed data; and N.V., M.E.K., R.H.G., and S.I. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: CREB, cAMP-response element binding protein; miRNA, microRNA; GAP, GTPase-activating protein; BDNF, brain-derived neurotrophic factor; shRNA, short hairpin RNA; CBP, CREB binding protein; premiR, pre-miRNA.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ223059).
References
- 1.Lonze, B. E. & Ginty, D. D. (2002) Neuron 35, 605-623. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez, G. A. & Montminy, M. R. (1989) Cell 59, 675-680. [DOI] [PubMed] [Google Scholar]
- 3.Chrivia, J. C., Kwok, R. P., Lamb, N., Hagiwara, M., Montminy, M. R. & Goodman, R. H. (1993) Nature 365, 855-859. [DOI] [PubMed] [Google Scholar]
- 4.Vo, N. & Goodman, R. H. (2001) J. Biol. Chem. 276, 13505-13508. [DOI] [PubMed] [Google Scholar]
- 5.Conkright, M. D., Canettieri, G., Screaton, R., Guzman, E., Miraglia, L., Hogenesch, J. B. & Montminy, M. (2003) Mol. Cell 12, 413-423. [DOI] [PubMed] [Google Scholar]
- 6.Pittenger, C. & Kandel, E. R. (2003) Philos. Trans. R. Soc. London B. 358, 757-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lonze, B. E., Riccio, A., Cohen, S. & Ginty, D. D. (2002) Neuron 34, 371-385. [DOI] [PubMed] [Google Scholar]
- 8.Redmond, L., Kashani, A. H. & Ghosh, A. (2002) Neuron 34, 999-1010. [DOI] [PubMed] [Google Scholar]
- 9.Fujioka, T., Fujioka, A. & Duman, R. S. (2004) J. Neurosci. 24, 319-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Impey, S., McCorkle, S. R., Cha-Molstad, H., Dwyer, J. M., Yochum, G. S., Boss, J. M., McWeeney, S., Dunn, J. J., Mandel, G. & Goodman, R. H. (2004) Cell 119, 1041-1054. [DOI] [PubMed] [Google Scholar]
- 11.Lee, R. C., Feinbaum, R. L. & Ambros, V. (1993) Cell 75, 843-854. [DOI] [PubMed] [Google Scholar]
- 12.Wightman, B., Ha, I. & Ruvkun, G. (1993) Cell 75, 855-862. [DOI] [PubMed] [Google Scholar]
- 13.Lau, N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. (2001) Science 294, 858-862. [DOI] [PubMed] [Google Scholar]
- 14.Lee, R. C. & Ambros, V. (2001) Science 294, 862-864. [DOI] [PubMed] [Google Scholar]
- 15.Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. (2001) Science 294, 853-858. [DOI] [PubMed] [Google Scholar]
- 16.Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., Horvitz, H. R. & Ruvkun, G. (2000) Nature 403, 901-906. [DOI] [PubMed] [Google Scholar]
- 17.Johnston, R. J. & Hobert, O. (2003) Nature 426, 845-849. [DOI] [PubMed] [Google Scholar]
- 18.Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. (2003) Cell 113, 25-36. [DOI] [PubMed] [Google Scholar]
- 19.Xu, P., Vernooy, S. Y., Guo, M. & Hay, B. A. (2003) Curr. Biol. 13, 790-795. [DOI] [PubMed] [Google Scholar]
- 20.Esau, C., Kang, X., Peralta, E., Hanson, E., Marcusson, E. G., Ravichandran, L. V., Sun, Y., Koo, S., Perera, R. J., Jain, R., et al. (2004) J. Biol. Chem. 279, 52361-52365. [DOI] [PubMed] [Google Scholar]
- 21.Chen, C. Z., Li, L., Lodish, H. F. & Bartel, D. P. (2004) Science 303, 83-86. [DOI] [PubMed] [Google Scholar]
- 22.Poy, M. N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P. E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., et al. (2004) Nature 432, 226-230. [DOI] [PubMed] [Google Scholar]
- 23.Zhao, Y., Samal, E. & Srivastava, D. (2005) Nature 436, 214-220. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., Labourier, E., Reinert, K. L., Brown, D. & Slack, F. J. (2005) Cell 120, 635-647. [DOI] [PubMed] [Google Scholar]
- 25.He, L., Thomson, J. M., Hemann, M. T., Hernando-Monge, E., Mu, D., Goodson, S., Powers, S., Cordon-Cardo, C., Lowe, S. W., Hannon, G. J., et al. (2005) Nature 435, 828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, J., Getz, G., Miska, E. A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B. L., Mak, R. H., Ferrando, A. A., et al. (2005) Nature 435, 834-838. [DOI] [PubMed] [Google Scholar]
- 27.O'Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V. & Mendell, J. T. (2005) Nature 435, 839-843. [DOI] [PubMed] [Google Scholar]
- 28.Bennasser, Y., Le, S. Y., Yeung, M. L. & Jeang, K. T. (2004) Retrovirology 1, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lecellier, C. H., Dunoyer, P., Arar, K., Lehmann-Che, J., Eyquem, S., Himber, C., Saib, A. & Voinnet, O. (2005) Science 308, 557-560. [DOI] [PubMed] [Google Scholar]
- 30.Pfeffer, S., Zavolan, M., Grasser, F. A., Chien, M., Russo, J. J., Ju, J., John, B., Enright, A. J., Marks, D., Sander, C., et al. (2004) Science 304, 734-736. [DOI] [PubMed] [Google Scholar]
- 31.Bartel, D. P. (2004) Cell 116, 281-297. [DOI] [PubMed] [Google Scholar]
- 32.Kim, V. N. (2005) Nat. Rev. Mol. Cell. Biol. 6, 376-385. [DOI] [PubMed] [Google Scholar]
- 33.Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H. & Kim, V. N. (2004) EMBO J. 23, 4051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai, X., Hagedorn, C. H. & Cullen, B. R. (2004) RNA 10, 1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seitz, H., Youngson, N., Lin, S. P., Dalbert, S., Paulsen, M., Bachellerie, J. P., Ferguson-Smith, A. C. & Cavaille, J. (2003) Nat. Genet. 34, 261-262. [DOI] [PubMed] [Google Scholar]
- 36.Pfeffer, S., Sewer, A., Lagos-Quintana, M., Sheridan, R., Sander, C., Grasser, F. A., van Dyk, L. F., Ho, C. K., Shuman, S., Chien, M., et al. (2005) Nat. Methods 2, 269-276. [DOI] [PubMed] [Google Scholar]
- 37.Arthur, J. S., Fong, A. L., Dwyer, J. M., Davare, M., Reese, E., Obrietan, K. & Impey, S. (2004) J. Neurosci. 24, 4324-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, J. Y., DeRuiter, S. L. & Turner, D. L. (2002) Proc. Natl. Acad. Sci. USA 99, 6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakazawa, T., Watabe, A. M., Tezuka, T., Yoshida, Y., Yokoyama, K., Umemori, H., Inoue, A., Okabe, S., Manabe, T. & Yamamoto, T. (2003) Mol. Biol. Cell 14, 2921-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valoczi, A., Hornyik, C., Varga, N., Burgyan, J., Kauppinen, S. & Havelda, Z. (2004) Nucleic Acids Res. 32, e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riggs, A. D., Singer-Sam, J. & Pfeifer, G. P. (1998) in Chromatin: A Practical Approach, ed. Gould, H. (Oxford Univ. Press, Oxford), pp. 79-109.
- 42.Ahn, S., Olive, M., Aggarwal, S., Krylov, D., Ginty, D. D. & Vinson, C. (1998) Mol. Cell. Biol. 18, 967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tycowski, K. T., Shu, M. D. & Steitz, J. A. (1996) Nature 379, 464-466. [DOI] [PubMed] [Google Scholar]
- 44.Huttenhofer, A., Brosius, J. & Bachellerie, J. P. (2002) Curr. Opin. Chem. Biol. 6, 835-843. [DOI] [PubMed] [Google Scholar]
- 45.John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C. & Marks, D. S. (2004) PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura, T., Komiya, M., Sone, K., Hirose, E., Gotoh, N., Morii, H., Ohta, Y. & Mori, N. (2002) Mol. Cell. Biol. 22, 8721-8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okabe, T., Nakamura, T., Nishimura, Y. N., Kohu, K., Ohwada, S., Morishita, Y. & Akiyama, T. (2003) J. Biol. Chem. 278, 9920-9927. [DOI] [PubMed] [Google Scholar]
- 48.Kim, J., Krichevsky, A., Grad, Y., Hayes, G. D., Kosik, K. S., Church, G. M. & Ruvkun, G. (2004) Proc. Natl. Acad. Sci. USA 101, 360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson, P. T., Hatzigeorgiou, A. G. & Mourelatos, Z. (2004) RNA 10, 387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meijering, E., Jacob, M., Sarria, J.-C. F., Steiner, P., Hirling, H. & Unser, M. (2004) Cytometry 58A, 167-176. [DOI] [PubMed] [Google Scholar]
- 51.Rozen, S. & Skaletsky, H. J. (2000) in Bioinformatics Methods and Protocols: Methods in Molecular Biology, eds. Krawetz, S. & Misener, S. (Humana, Totowa, NJ), pp. 365-386.
- 52.Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C. & Marks, D. S. (2003) Genome Biol. 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]