Abstract
In order to determine the role of ferrous iron transport in Legionella pathogenesis, we identified and mutated the feoB gene in virulent Legionella pneumophila strain 130b. As it is in Escherichia coli, the L. pneumophila feoB gene was contained within a putative feoAB operon. L. pneumophila feoB insertion mutants exhibited decreased ferrous but not ferric iron uptake compared to the wild type. Growth on standard buffered charcoal yeast extract agar or buffered yeast extract broth was unaffected by the loss of L. pneumophila FeoB. However, the L. pneumophila feoB mutant had a reduced ability to grow on buffered charcoal yeast extract agar with a reduced amount of its usual iron supplementation, a phenotype that could be complemented by the addition of feoB in trans. In unsupplemented buffered yeast extract broth, the feoB mutant also had a growth defect, which was further exacerbated by the addition of the ferrous iron chelator, 2,2′-dipyridyl. The feoB mutant was also 2.5 logs more resistant to streptonigrin than wild-type 130b, confirming its decreased ability to acquire iron during extracellular growth. Decreased replication of the feoB mutant was noted within iron-depleted Hartmannella vermiformis amoebae and human U937 cell macrophages. The reduced intracellular infectivity of the feoB mutant was complemented by the introduction of a plasmid containing feoAB. The L. pneumophila feoB gene conferred a modest growth advantage for the wild type over the mutant in a competition assay within the lungs of A/J mice. Taken together, these results indicate that L. pneumophila FeoB is a ferrous iron transporter that is important for extracellular and intracellular growth, especially in iron-limited environments. These data represent the first evidence for the importance of ferrous iron transport for intracellular replication by a human pathogen.
Iron is essential for the growth of essentially all organisms, and its limitation plays an important role in host defense against infection by restricting bacterial replication (46, 60). L. pneumophila, the causative agent of Legionnaires' disease, is a facultative intracellular parasite of human macrophages and freshwater amoebae (31, 41, 50). The ability of L. pneumophila to acquire host cell iron is pivotal for it to establish successful intracellular infection (8-10, 25, 26, 29, 30, 42, 45, 58). For example, gamma interferon-activated macrophages are nonpermissive for L. pneumophila growth due to decreased intracellular iron (40). Also, sequestration of intracellular iron by treatment with chelators inhibits the growth of L. pneumophila in human macrophages (24). Furthermore, a reduced ability of legionellae to establish successful intracellular infection is correlated with decreased levels of the receptor for transferrin, the key iron transport protein of the host cell (8).
In order to occupy its intracellular niche, L. pneumophila likely has developed multiple iron acquisition mechanisms. Factors that may be involved in L. pneumophila iron acquisition include heme utilization (42); the iraAB locus, which may encode a transporter of iron-loaded peptides (58); the cytochrome c maturation (ccm) genes (59); the iron-regulated frgA, whose product has homology to aerobactin synthetases (30); legiobactin, the first example of a Legionella siderophore (36); and two internal ferric reductases (44).
Iron exists in a dynamic equilibrium between the ferrous (Fe2+) and ferric (Fe3+) forms, depending on its environment (1). In aerobic conditions at neutral pH, the majority of iron is generally in the insoluble ferric form (1). However, at acid pH or in anaerobic conditions, the equilibrium shifts to the soluble ferrous form (1). Within host cells, the majority of iron is sequestered by the eukaryotic storage proteins ferritin and hemosiderin (46, 62). Reports have indicated that Fe2+ is the predominant form of iron in the cytosolic pools in mammalian cells (5, 19).
Within its host cells, L. pneumophila resides in a specialized phagosome, where its precise source of iron is unclear but presumably involves active access to the iron pool (8). Although the L. pneumophila phagosome does not fuse with endosomes and lysosomes and is at nearly neutral pH during the early stages of the intracellular life cycle, it appears to fuse with low-pH cellular compartments during later stages of the infection (61). If there were a concomitant decrease in phagosomal pH, then the amount of soluble ferrous iron available to the bacteria may increase (54). Taken together, these data suggest that L. pneumophila may access significant amounts of ferrous iron for its intracellular replication. Furthermore, previous reports suggested that L. pneumophila can use ferrous iron during replication in bacteriological media, as reduced growth by wild-type bacteria was observed in the presence of the specific ferrous iron chelator 2,2′-dipyridyl (DIP) (47). Because ferrous iron may be important to the bacterium during infection of host cells, we began to consider the means by which L. pneumophila acquires and/or utilizes this form of iron.
Ferrous iron transport has been an important area of research in both eukaryotic and prokaryotic systems. For example, several determinants involved in ferrous iron transport have been identified in eukaryotic cells, such as Nramp-2 in mammalian cells and the FET3 gene in Saccharomyces cerevisiae (3, 4). In bacteria, the feoAB operon of Escherichia coli is the most-characterized ferrous iron transport system (33). FeoB is an inner membrane protein, believed to comprise an ATP-driven transporter (33). Genes homologous to feoB have been detected in the genomes of many bacterial species. In contrast, the function of FeoA remains unclear, and the feoA gene is not always present alongside feoB (33).
In E. coli, loss of feoA or feoB results in decreased ferrous iron uptake (33). Furthermore, E. coli feoB mutants exhibit decreased gut colonization in mice, suggesting a role for feoB in pathogenesis (53). However, the importance of feoB and Fe2+ transport in intracellular parasitism remains to be demonstrated. Given our desire to understand both the specific mechanisms of Legionella pathogenesis and the general importance of ferrous iron transport in intracellular infection, we sought to identify and characterize the L. pneumophila genes involved in the acquisition and utilization of Fe2+.
This study describes the characterization and mutational analysis of the L. pneumophila feoB gene and suggests that the acquisition of ferrous iron is relevant for the growth of L. pneumophila both extracellularly in artificial media and intracellularly in macrophages and amoebae.
MATERIALS AND METHODS
Bacterial strains, media, and extracellular growth conditions.
L. pneumophila strain 130b (ATCC BAA-74, also known as Wadsworth or AA100) was used for mutagenesis of the Legionella feoB gene and subsequently served as the wild-type control (18). L. pneumophila strains were routinely grown on standard buffered charcoal yeast extract (BCYE) agar for 3 days at 37°C or in buffered yeast extract (BYE) broth at 37°C with shaking at 225 rpm (Lab-Line Instruments; Dubuque, Iowa; model 3525) (16, 35). For selection of allelic exchange mutations, BCYE agar was supplemented with 5% (wt/vol) sucrose (35). Standard BCYE agar and BYE broth contain an iron supplement consisting of 0.25 g of ferric pyrophosphate (Sigma, St. Louis, Mo.) per liter.
In order to monitor extracellular growth in liquid media, L. pneumophila grown on BCYE agar was used to inoculate 25 ml of BYE broth overnight. The culture was washed once with BYE broth without iron supplementation and then used to inoculate 3 ml of BYE broth, which was incubated at 37°C with shaking at 225 rpm. Bacterial growth was monitored by determining the optical density of the culture at 660 nm (OD660; Beckman, Fullerton, Calif.; spectrophotometer model DU500). In order to assess bacterial growth in iron-limiting BYE broth, iron supplementation was omitted and, where stated, DIP (Sigma) was added at various concentrations. To further control the amount of iron in BYE broth cultures, Milli-Q water and acid-washed glassware were used throughout (14).
To assess the growth and survival of L. pneumophila on iron-limited solid media, bacteria grown on BCYE agar were resuspended in distilled water to an OD660 of 0.3 to 0.4, and then 100 μl of 10-fold serial dilutions was plated on BCYE agar containing either 32-fold less (0.0078 g per liter) iron supplementation than standard media or no iron supplementation at all. To further investigate the iron physiology of L. pneumophila, strains were tested for their resistance to streptonigrin (Sigma) (45). Specifically, bacteria were grown on standard BCYE agar and resuspended in distilled water to an OD660 of 0.3 to 0.4, and then 100 μl of 10-fold serial dilutions was plated on BCYE agar that contained no iron supplementation and either 0, 0.75, or 1.5 μM streptonigrin.
Escherichia coli DH5α was used as a host for recombinant plasmids and was cultured in Luria-Bertani broth or agar (27). Where appropriate, media were supplemented with the following antibiotics at final concentrations suitable for L. pneumophila (E. coli): kanamycin, 25 μg/ml (50 μg/ml), and chloramphenicol, 6 μg/ml (30 μg/ml).
Cloning and DNA sequence analysis.
The double-stranded sequence of the Legionella feoAB locus was obtained by using plasmid pMRJ. This plasmid was identified by colony hybridization of a genomic library that contained Sau3AI-digested fragments of strain 130b cloned into pBR322 (30). Hybridization was performed with a DNA probe produced by PCR amplification of a 1.4-kb internal fragment of the L. pneumophila feoB gene with primers FEOB2-F (5′-CATAGTATGCCAGCGAACAC-3′) and FEOB2-R (5′-AAGCAGGCTCAGTGACATAG-3′). Sequencing of the L. pneumophila feoB, feoA, and upstream region was performed by the dideoxy chain termination method with BigDye (ABI Prism, Applied Biosystems; Foster City, Calif.) with a series of primers devised from the DNA sequence of the feoB region from the L. pneumophila strain Philadelphia-1 unfinished genome database (http://genome3.cpmc.columbia.edu/≈legion).
DNA sequencing was performed by the Northwestern University Biotechnology Laboratory, and primers were obtained from Integrated DNA Technologies (Coralville, Iowa). Nucleotide sequences were analyzed with Seqman (DNAStar, Madison, Wis.), and Blast homology searches were conducted through GenBank at the National Center for Biotechnology Information. DNA motifs and structural analyses were conducted with the Prosite prediction model (http://us.expasy.org/prosite) and the SOSUI program (http://sosui.proteome.bio.tuat.ac.jp).
Mutation and complementation analyses.
An internal fragment of the L. pneumophila strain 130b feoB gene was amplified by PCR with primers FEOB2-F and FEOB2-R. The resultant 1.4-kb DNA fragment was then cloned into pGEMTEasy (Promega, Madison, Wis.) to give plasmid pMR3. The cloned L. pneumophila feoB was mutated by insertion of a 1.1-kb kanamycin resistance (Kmr) gene at the Klenow-treated Kpn21 site of pMR3, resulting in pMR4. The StuI-EcoRV fragment containing the Kmr gene was obtained from plasmid pVK4, a derivative of pVK3 with the antibiotic resistance gene in the opposite orientation (58). The mutated feoB gene was then cloned on a 2.6-kb NotI fragment into the counterselectable pBOC20 to yield pMR5. The chloramphenicol-resistant pBOC20 is based on a ColE1 replicon and facilitates allelic exchange in L. pneumophila by virtue of its sacB gene (35).
Production of competent 130b cells and electroporation of pMR5 into L. pneumophila were achieved as previously described (35). Potential mutants were selected based on chloramphenicol sensitivity and kanamycin and sucrose resistance, indicative of introduction of the mutated gene into the 130b chromosome by homologous recombination. Verification of mutant genotypes was carried out by PCR and Southern hybridization with the same primers and DNA probe used to identify pMRJ. Genomic DNA was isolated from L. pneumophila as described previously (17).
To facilitate complementation, a 2.4-kb PCR fragment containing only the feoB gene was first amplified from pMRJ with primers FEOB5KpnI (5′-CGGGGTACCCCGGTGAGGATGGCTCCATTAGG-3′) and pBRREV (5′-CGCCAGCAACCGCACCTGTG-3′). PCR fragments containing feoB plus the upstream feoA and promoter region and just the feoA gene plus the upstream promoter were generated with primers FEOB6 (5′-TATCTGAGTCGTCATCCTCG-3′) and pBR-R or FEOB6KpnI and FEOB9 (5′ CCG ATG CTC GGC AAT ATC CA 3′), respectively. Next, the PCR fragments were cloned into pGEMT (Promega) to yield pMR17, pMR18, and pMR19, encoding feoB, feoAB, and feoA, respectively. Primers used during amplification were designed against the known sequence of the feoB region from strain 130b. Next, the cloned feoB, feoAB, and feoA genes were transferred on PstI-KpnI fragments into the chloramphenicol-resistant pMMB207, yielding pMR20, pMR21, and pMR22, respectively (39). Plasmid pMMB207 provides a promoter for the expression of cloned genes and has been used previously for trans complementation of L. pneumophila mutants, including those with infectivity defects (37, 51, 59). Plasmids were transformed into L. pneumophila by electroporation by standard methods (35).
Iron uptake assays.
To investigate the ability of L. pneumophila strains to take up iron, studies were performed with 55FeCl3 (Perkin Elmer, Boston, Mass.) (16.07 mCi/ml) based on the protocol of Kammler et al. (33). L. pneumophila was grown overnight in 25 ml of BYE broth without supplemental iron to the late logarithmic phase (i.e., corresponding to an OD660 of 2) at 37°C with shaking. Bacteria were then subcultured into 25 ml of BYE broth without supplemental iron and allowed to grow again to the late logarithmic phase. The final cultures were centrifuged at 3,500 rpm for 15 min at 30°C, and then the bacterial pellet was resuspended in 25 ml of MOPS transport buffer (40 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.0], 100 mM MgSO4, 0.5 mM CaCl2 and 0.2% glucose, warmed to 37°C) to 109 CFU/ml, a concentration that was based on the OD660 of the suspension and confirmed by plating aliquots on BCYE agar.
In order to assess ferrous iron uptake, 10 ml of bacterial suspension was adjusted to 3 μM 55Fe by the addition of 10 μl of a 3 mM stock solution of 55FeCl3 in 0.5 M HCl in the presence of 1 mM sodium ascorbate (10 μl of a 1 M freshly prepared stock solution; Sigma). For monitoring ferric iron uptake, the bacteria and radiolabeled iron samples were mixed in the presence of 1 mM sodium citrate (10 μl of a 1 M freshly prepared stock solution; Sigma). At various times, 1 ml of the bacterial suspension was removed to a 0.45-μm nitrocellulose filter (Millipore, Bedford, Mass.) and washed three times with 1 ml of 0.1 M LiCl (Sigma) at room temperature. The filters were then allowed to dry at room temperature for 30 min, and their radioactivity was measured in 10 ml of Ultima Gold liquid scintillation cocktail (Packard Bioscience, Boston, Mass.) in a Beckman counter (model LS 3801). Preliminary experiments showed that three washes with 1 ml of LiCl removed >90% of the iron from filters containing known amounts of iron. Filtered bacterial suspensions without added radioactivity had no detectable radioactivity above that of the liquid scintillation cocktail. L. pneumophila siderophore activity was assessed with the chrome azurol S (CAS) assay, as described previously (36).
Intracellular infection of macrophages and amoebae by L. pneumophila.
Assessment of the ability of L. pneumophila strains to establish intracellular infections was performed in both human U937 cells differentiated into macrophage-like cells by treatment with phorbol myristate acetate (12, 43, 55) and H. vermiformis amoebae (21, 52). Growth kinetics of L. pneumophila in U937 macrophages was performed as previously reported (48, 49, 58). Briefly, U937 cells were cultivated in RPMI 1640 medium with l-glutamine (Cellgro, Herndon, Va.) supplemented with 10% heat-inactivated fetal calf serum (Gibco Life Technologies, Rockville, Md.) and 2.5 ml of fungizone (Gibco Life Technologies) in a 5% CO2 incubator at 37°C. At a multiplicity of infection (MOI) of 1, 106 adherent U937 macrophages were infected with bacteria that had been grown for 3 days on standard BCYE agar. After a 2-h incubation period to allow bacterial internalization, extracellular bacteria were removed by repeated washing, and then infected monolayers were incubated at 37°C in a 5% CO2 incubator. At various times postinoculation, intracellular bacteria were released by lysis of the monolayer with 10 μl of 10% saponin (Sigma). For estimation of viable cell counts, serial 10-fold dilutions from triplicate wells for each strain were plated on BCYE agar.
Cocultures with H. vermiformis and L. pneumophila were performed as described previously (48, 49, 58). This species of amoeba is known to support the growth of L. pneumophila and has been associated with clinical cases of legionellosis (21). Briefly, in 24-well tissue culture trays, 105 H. vermiformis trophozoites were infected with 103 bacteria grown for 3 days on BCYE agar. At 24-h intervals, samples were taken from triplicate wells, and viable cell counts were estimated as described above. Infection of iron-depleted macrophages and amoebae was accomplished by the addition of various concentrations of DIP to the medium for 24 h prior to infection with L. pneumophila and during the incubation period (24, 45, 58). All stages of the infections for these experiments, including washes, were carried out in the appropriate concentrations of DIP. L. pneumophila is unable to grow in the media in which the infections of macrophages or H. vermiformis were conducted; thus, increases in bacterial numbers reflect growth within the host cells (12, 43).
Cytopathicity assays.
Macrophage and H. vermiformis viability was assessed in order to establish the concentrations of DIP that did not adversely affect cellular viability and to determine whether there were differences in cytopathicity between the wild-type and mutant bacteria. Cytopathicity assays were carried out as previously described, by assessing by the ability of cell monolayers to reduce the dye AlamarBlue (Biosource International, Camarillo, Calif.) (2, 32, 48, 49). Briefly, 105 U937 macrophages or H. vermiformis cells were infected at various MOIs or left uninfected to test the viability of macrophages in the presence or absence of DIP. After 2 h, the infected and uninfected monolayers were washed twice with RPMI and then incubated at 37°C. At various time points, AlamarBlue was added (1/11th volume in RPMI 1640) to monolayers that had been washed twice and incubated at 37°C for 3 h, and fluorescence (excitation at 540 and emission at 584) was assessed.
Pulmonary infection of A/J mice with L. pneumophila.
Intratracheal infection of A/J mice by legionellae is a model system for acute L. pneumophila-induced pneumonia (2, 6, 7, 13). To assess the virulence of bacteria in the mouse lung, standard competition assays were performed, as we have described previously (22, 34, 38, 48, 63). Six- to eight-week-old female mice (Jackson Laboratories, Bar Harbor, Maine) were anesthetized and then inoculated intratracheally with 106 CFU of a ca. 1:1 ratio of wild-type and mutant bacteria which had been grown for 3 days on BCYE agar. At various time points, infected mice were sacrificed, and lungs were disrupted by extrusion of the tissue through a fine-mesh grid into 10 ml of phosphate-buffered saline. Host cell lysis was achieved by incubation of the tissue sample with 100 μl of 10% saponin for 15 min at 37°C, followed by vigorous pipetting. The numbers of viable bacteria and the ratio of wild type to mutant were estimated by plating 10-fold serial dilutions on both standard and kanamycin-supplemented BCYE.
Nucleotide sequence accession number.
The GenBank accession number for the L. pneumophila feoAB genes is AF492466.
RESULTS
Identification, sequence analysis, and mutation of L. pneumophila feoB.
Because L. pneumophila can utilize ferrous iron, the incomplete genome database of L. pneumophila strain Philadelphia-1 (http://genome3.cpmc.columbia.edu/≈legion) was examined for genes that might be involved in Fe2+ acquisition. A gene whose protein product appeared to have homology to FeoB in E. coli was identified (accession number CAA50387). The feoB-like gene and its upstream region were cloned from our virulent, serogroup 1 strain 130b. Complete sequence analysis of the cloned gene confirmed the existence of an L. pneumophila open reading frame that was predicted to encode a FeoB homologous (44% identity, 61% similarity) and similar in size (751 versus 773 amino acids) to E. coli FeoB (33).
Like its E. coli counterpart, L. pneumophila FeoB was predicted to have 10 transmembrane domains, based on Kyte and Doolittle estimations, consistent with an inner membrane location of this protein (33). Similar to the situation in E. coli FeoB, ATP/GTP-binding site motif A regions were identified at amino acids 8 to 15 and 380 to 387 in L. pneumophila FeoB (33). Immediately upstream of the feoB gene was an open reading frame, the predicted protein product of which bore homology (52% identity, 70% similarity) and was identical in size (75 amino acids) to E. coli FeoA (accession number CAA50386) (33). The fact that the predicted stop codon of feoA overlaps the predicted start codon of feoB suggests that L. pneumophila feoA and feoB form an operon, similar to E. coli feoAB (33).
Upstream of the L. pneumophila feoA coding region was a putative Fur binding site, which contained 14 of the 19 residues conserved in E. coli Fur-regulated promoters (15). This suggests that the L. pneumophila feoAB locus, like its E. coli counterpart, is subject to regulation by bacterial intracellular Fe2+ levels (15). Further upstream of L. pneumophila feoA was an open reading frame whose deduced protein product was homologous (41% identity, 60% similarity) to a hypothetical protein in Pseudomonas aeruginosa (accession number NP_253128). Analysis of the L. pneumophila genomic database indicates that downstream of feoAB there is an open reading frame that is transcribed in the opposite orientation and whose predicted product does not bear homology to any proteins in the GenBank database.
In order to assess the role of FeoB in L. pneumophila physiology and pathogenesis, a feoB mutation was generated in strain 130b by allelic exchange of the wild-type gene for a cloned copy that had been inactivated by a Kmr insertion. Mutant NU269, which contained a Kmr insertion at nucleotide 1600 of the 2,256-bp gene, was verified by PCR and Southern hybridization (data not shown).
Iron uptake by L. pneumophila strains.
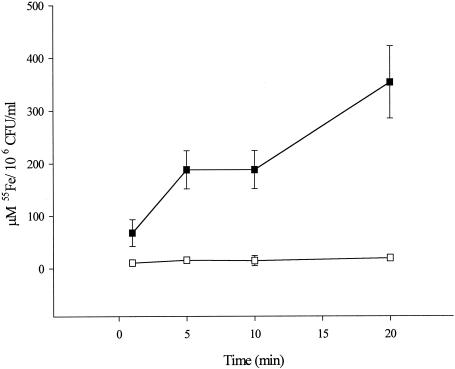
To investigate whether L. pneumophila FeoB was a functional homologue of E. coli FeoB, wild-type and mutant legionellae were compared in 55Fe iron uptake assays. To specifically monitor the uptake of ferrous iron, the procedure was performed in the presence of ascorbate, which is known to maintain iron in a reduced state over the necessarily short period of the assay (33). L. pneumophila 130b exhibited increasing ferrous iron accumulation over the assay period; i.e., there was 5.3-fold greater radiolabel incorporation at 20 min versus 1 min (Fig. 1). In comparison, the L. pneumophila feoB mutant did not exhibit significant ferrous iron uptake upon incubation (Fig. 1). This severe defect in NU269 is consistent with a role for FeoB as a ferrous iron transporter. Indeed, the mutant's defect in ferrous iron uptake could be complemented by the addition of feoB in trans (Table 1).
FIG. 1.
Kinetics of ferrous iron uptake by L. pneumophila strains. 55Fe2+ was supplied to wild-type 130b (solid squares) and feoB mutant NU269 (open squares) bacteria, and then, after various incubation times, the amounts of radioactive iron associated with 106 CFU were determined. Data show the means and standard deviations from triplicate samples. The mutant exhibited significantly less iron uptake at 1 min (Student's t test, P < 0.05) and at 5, 10, and 20 min (Student's t test, P < 0.01). These experiments were performed in duplicate with similar results.
TABLE 1.
Ferrous and ferric iron uptake by L. pneumophila strains
| Strain | Iron state | Mean μM 55Fe (μM/106 CFU/ml) ± SDa at:
|
|
|---|---|---|---|
| 1 min | 20 minb | ||
| 130b (pMMB207) | Fe2+ | 178 ± 86 | 346 ± 127 |
| NU269 (pMMB207) | 101 ± 70 | 62 ± 6 | |
| NU269 (pMR20) | 77 ± 8 | 168 ± 18 | |
| 130b | Fe3+ | 62 ± 32 | 104 ± 18 |
| NU269 | 82 ± 29 | 121 ± 34 | |
55Fe2+ or 55Fe3+ was supplied to wild type 130b and NU269 mutant bacteria, and 1-ml samples (n = 3) were tested for radioactivity. Complementation was assessed by introducing pMR20, which contains the intact feoB, into NU269 and then comparing NU269(pMR20) iron uptake to that of the wild type and mutant bacteria containing the empty vector pMMB207.
Significant differences were noted between NU269(pMMB207) and NU269(pMR20) (Student’s t test, P < 0.01) and 130b(pMMB207) and NU269(pMMB207) (Student’s t test, P < 0.05).
When the uptake assays were performed in the presence of citrate, which maintains the iron in the ferric form, the wild type and the feoB mutant exhibited similar levels of iron uptake (Table 1), indicating that L. pneumophila FeoB is not required for ferric iron uptake. In comparing the data in Fig. 1 and Table 1, less Fe3+ than Fe2+was associated with the wild type, suggesting that, in L. pneumophila, ferrous iron uptake may be more efficient than ferric iron uptake. The feoB mutant was not defective for legiobactin siderophore activity, based on the CAS assay (data not shown). Based on our sequence data and the defect in Fe2+ uptake exhibited by NU269, we conclude that L. pneumophila feoB encodes a factor that is required for ferrous iron transport and is the functional equivalent of E. coli FeoB.
Extracellular growth characteristics of L. pneumophila strains.
To investigate the role of feoB and ferrous iron transport in the extracellular growth of L. pneumophila, we compared wild-type and mutant bacteria for their ability to grow on both standard solid and liquid media and media that had been depleted of iron. Growth on BCYE agar was assessed by plating suspensions of bacteria and then noting both the number of recovered CFU and the time to colony formation. Strains 130b and NU269 grew similarly on standard BCYE agar, which is routinely supplemented with 0.25 g of ferric pyrophosphate per liter.
To start to investigate the role of iron in growth, BCYE agar was prepared with only 0.0078 g of the ferric pyrophosphate supplement per liter. This 32-fold decrease in iron supplementation did not affect the growth of 130b. However, the feoB mutant now took 10 as opposed to 3 days to form colonies, although similar numbers of CFU were eventually attained. When pMR20, containing an intact copy of feoB alone, was introduced into NU269, the mutant formed colonies in 4 days on the BCYE agar that had a diminished iron concentration. Further increasing the iron limitation, by omission of the ferric pyrophosphate, did not alter the growth of the wild type. However, the feoB mutant required 13 to 15 days for the generation of single colonies on unsupplemented BCYE agar. Taken together, these data, which were observed in at least three experiments, indicate that NU269 is defective for extracellular growth in iron-limited conditions and that this phenotype is specifically associated with the loss of feoB function. Interestingly, NU269 was able to form colonies in 4 to 5 days on unsupplemented BCYE agar when plated next to confluent lawns of either itself or wild-type bacteria.
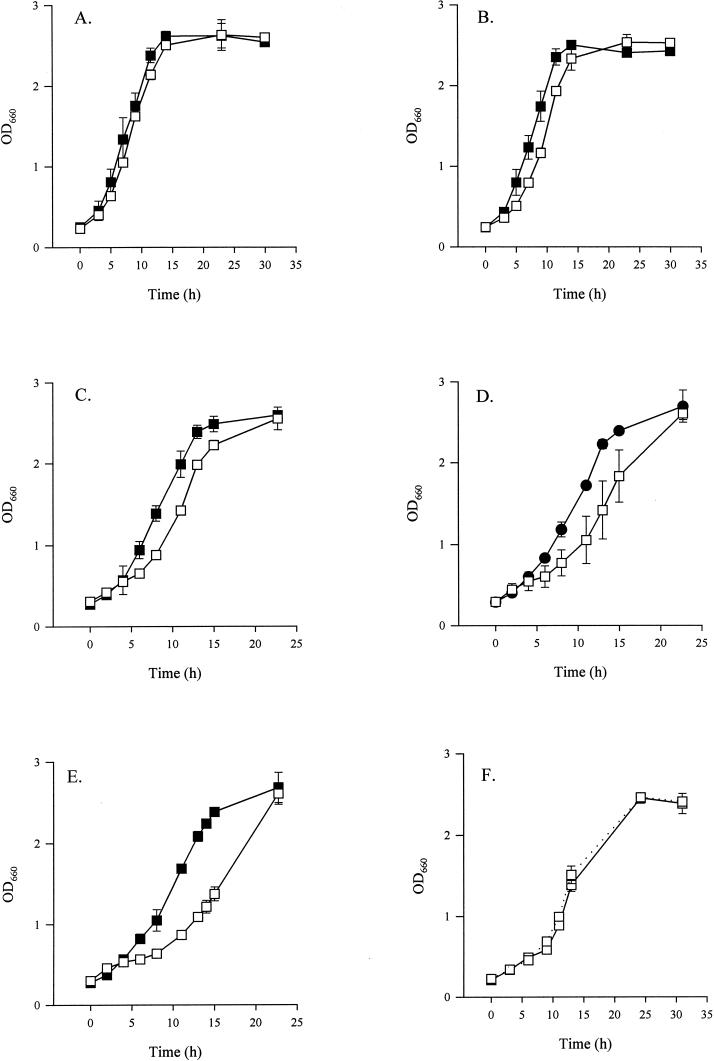
To more accurately compare the growth of the feoB mutant and its wild-type parent, we investigated the strains' behaviors in iron-replete and -depleted liquid media. NU269 grew similar to the wild type in BYE broth, which is routinely supplemented with 0.25g of ferric pyrophosphate per liter (Fig. 2A). In BYE broth with no iron supplementation, the wild type grew as it had in standard BYE broth, but the feoB mutant exhibited a small but reproducible defect in the early stages of growth compared to 130b (Fig. 2B). When available ferrous iron in unsupplemented BYE broth was reduced by the addition of 35, 40, or 45 μM of the Fe2+ chelator DIP, 130b grew similarly to how it had grown in BYE broth without iron supplementation (Fig. 2C and D and data not shown).
FIG. 2.
Growth of L. pneumophila in standard and iron-limited BYE broth. Wild-type 130b (solid squares) and feoB mutant NU269 (open squares) were grown in BYE broth (A) with standard iron supplementation, (B) without iron supplementation, (C) without iron supplementation in 35 μM DIP, (D) with 45 μM DIP, or (E) with 60 μM DIP. In panel F, NU269 was grown either in BYE broth that lacked its standard iron supplement (solid line) or in BYE broth that lacked its standard iron supplement but contained 60 μM DIP and 80 μM FeCl3 (dotted line). Data represent the means and standard deviations from triplicate samples from a representative experiment. In panels B to E, significant differences were noted between the wild-type and the feoB mutant between 5 and 11 h (Student's t test, P < 0.05). Experiments were repeated in triplicate with similar results.
Upon addition of 60 μM DIP, the wild-type bacteria entered the stationary phase slightly earlier than usual (Fig. 2E). In contrast, the feoB mutant had a decreased growth rate in unsupplemented BYE broth containing 35 μM DIP, which increased as the concentration of DIP increased to 45 μM and then to 60 μM (Fig. 2C to E). It is of note that even in the presence of 60 μM DIP, the feoB mutant eventually attained wild-type levels of growth (Fig. 2E). As confirmation that the inhibitory effect of DIP on the mutant was due to decreased iron in the media, maximal growth of NU269 in unsupplemented BYE broth was restored when 80 μM but not 10, 20, or 40 μM ferric chloride was added to the 60 μM DIP-containing cultures (Fig. 2F and data not shown). Taken together, these data indicate that L. pneumophila FeoB is not absolutely required for growth in bacteriological media but that, as iron becomes restricted, Fe2+ transport is relevant for extracellular replication.
Streptonigrin resistance of L. pneumophila.
To gain further support for the role of FeoB in L. pneumophila extracellular iron acquisition, we compared 130b and NU269 for their resistance to streptonigrin. Bacteria with low intracellular iron pools are more resistant to streptonigrin, as this antibiotic has a well-documented iron-dependent toxicity and has been used by others to actually select E. coli FeoB mutants (28). The survival of wild-type legionellae was diminished as the concentration of streptonigrin increased within the media from 0 to 1.5 μM, as in earlier studies (Table 2) (30, 45). Importantly, NU269 was 3-fold and 25-fold more resistant than 130b at 0.75 and 1.5 μM streptonigrin, respectively (Table 2). The increased resistance of NU269 suggests that L. pneumophila feoB mutants have less intracellular iron than the wild type, a situation that is compatible with an involvement of feoB in iron transport.
TABLE 2.
Streptonigrin resistance of L. pneumophila strainsa
| Streptonigrin (μM) | Mean growth (107 CFU/ml) ± SD
|
|
|---|---|---|
| 130b | NU269 | |
| 0 | 106.67 ± 12.58 | 105.33 ± 24.70 |
| 0.75 | 16.07 ± 0.72 | 48.67 ± 7.77 |
| 1.5 | 0.33 ± 0.21 | 8.33 ± 6.11 |
Bacteria were resuspended to 109 CFU/ml, and dilutions were plated on BCYE agar containing no iron supplementation and either 0, 0.75, or 1.5 μM streptonigrin. Data show the mean and standard deviation (n = 3). This experiment was performed in duplicate with similar results. Significant differences were noted between the wild type and the feoB mutant when plated on agar containing 0.75 μM streptonigrin (Student's t test, p < 0.01).
Intracellular growth of L. pneumophila in amoebae and macrophages.
The role of FeoB in L. pneumophila intracellular infection of protozoa was assessed by using H. vermiformis (21). Within the amoeba cocultures, both 130b and the feoB mutant were recovered in similar numbers at 0, 24, 48, and 72 h postinoculation (Fig. 3 and data not shown). These results could indicate that FeoB does not have an essential role in L. pneumophila growth and/or survival in H. vermiformis. However, as we observed in BYE broth cultures, it remained possible that the importance of feoB would be manifest in iron-depleted conditions. Hence, cocultures were performed in the presence of DIP, with a concentration of the iron chelator that did not adversely affect the viability of H. vermiformis, >90% of the amoebae reduced AlamarBlue at 0 to 50 μM DIP.
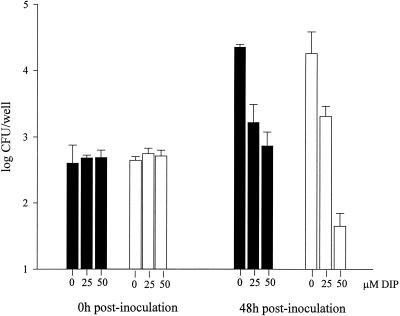
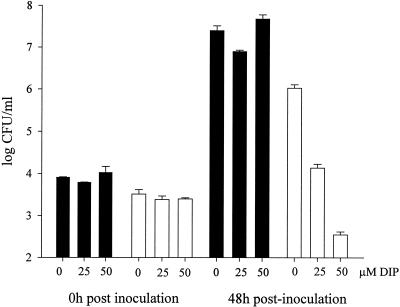
FIG. 3.
Coculture of L. pneumophila with H. vermiformis amoebae. Wild-type (solid bars) and NU269 (open bars) strains were mixed at an MOI of 0.1 to amoebae that were treated or not with 25 or 50 μM DIP, and then numbers of bacteria within the cocultures were sampled at 0 and 48 h postinoculation. Data represent the mean and standard deviation from triplicate samples. The experiment was repeated in triplicate with similar results. Significant differences between the wild-type and mutant strains were observed in the 48-h cultures containing 50 μM DIP (Student's t test, P < 0.005).
Upon sampling at 48 h postinoculation, a ca. 10-fold reduction in recovery of wild-type bacteria was noted at 25 and 50 μM DIP (Fig. 3). NU269 grew similarly to the wild type in cocultures containing 0 or 25 μM DIP (Fig. 3). However, in 50 μM DIP cocultures, the feoB mutant exhibited a 150-fold reduction in CFU recovered (Fig. 3). The numbers of NU269 recovered at 48 h postinoculation from cocultures containing 50 μM DIP were less than those originally inoculated, suggesting death of the mutant bacteria. Strains 130b and NU269 survived equally well in the medium used to culture the amoebae (data not shown). Together, these results indicate that feoB is important for growth and/or survival of L. pneumophila within iron-restricted amoebae.
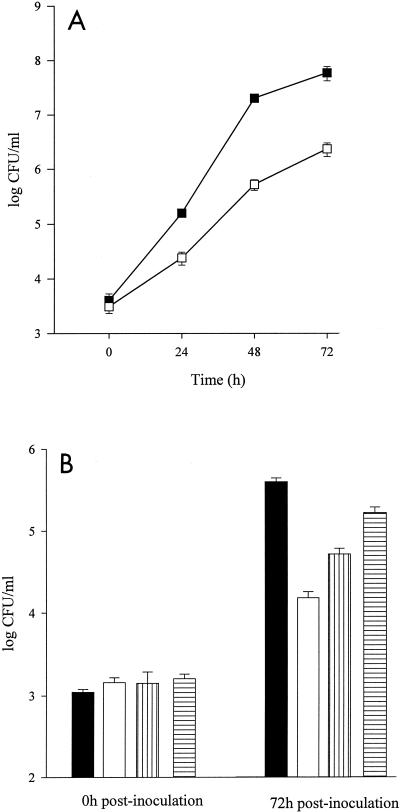
The role of the L. pneumophila FeoB in intracellular growth was next assessed in U937 cells, a widely used model for infection of human macrophages (43, 48, 49). Numbers of bacteria were monitored over time following infection of macrophages with either 130b or NU269. Equivalent numbers of bacteria were recovered from monolayers infected with 130b or NU269 at 0 h postinoculation (Fig. 4A), indicating that there was no obvious defect for NU269 in attachment and invasion of macrophages. However, the feoB mutant exhibited a 10-fold reduced recovery compared to the wild-type parent at 24 h, increasing to 15-fold at 48 and 72 h postinoculation (Fig. 4A). Strains 130b and NU269 survived equally well in the medium used to culture the macrophages (data not shown). Taken together, these data indicate that the feoB mutant has a reduced ability to replicate and/or survive in human macrophages.
FIG. 4.
Intracellular growth of L. pneumophila in U937 macrophages. (A) Wild-type 130b- (solid squares) and mutant NU269- (open squares) were used to infect U937 macrophages at an MOI of 1 and then, at various times postinoculation, the numbers of bacteria were assessed. Panel B shows complementation performed with NU269(pMR20) (vertical striped bars) or NU269(pMR21) (horizontal striped bars) containing plasmids encoding the entire feoB or feoAB genes, respectively. Complemented strains were compared to wild-type and mutant bacteria containing the empty vector pMMB207 (solid and open bars, respectively). Data represent the mean and standard deviation from triplicate samples. Experiments were performed in duplicate or triplicate with similar results. Significant differences were observed between the wild-type and mutant strains between 24 and 72 h postinoculation in panel A and between NU269(pMMB207) and NU269(pMR20) or NU269(pMR21) at 72 h postinoculation (Student's t test, P < 0.01).
Partial complementation of the growth defect of the feoB mutant was achieved with pMR20, which contains the feoB gene linked to a vector promoter (Fig. 4B). However, introduction of pMR21, which contains feoAB and its promoter region, produced greater complementation (Fig. 4B). The latter complementation event was due to the feoB gene, since introduction of pMR22, which contains only feoA and the promoter region, did not produce complementation in NU269 (data not shown). We strongly suspect that the increased complementation observed with pMR21 over pMR20 is associated with higher levels of expression of feoB from the native feoAB promoter. At the least, these data indicate that L. pneumophila feoAB is required for optimal intracellular infection of macrophages.
Further evidence of the role of L. pneumophila FeoB in infections of macrophages was provided by a cytopathicity assay, which monitors the reduction in host cell viability that is associated with Legionella infections (2). With the same MOI as during the kinetic studies (i.e., MOI = 1), the cytopathic effect of the wild type became apparent at 72 h postinoculation, when <5% of macrophages survived compared with uninfected monolayers (Fig. 5A). In contrast, the viability of macrophages infected with NU269 did not decrease significantly over the assay period (Fig. 5A). With an MOI of 10, a defect in cytopathicity was also observed; whereas 130b elicited 40% and <5% macrophage survival at 48 and 72 h, respectively, 50% of the NU269-infected cells remained viable after 72 h (Fig. 5B). Thus, loss of L. pneumophila feoB is associated with a decreased capacity for macrophage killing, substantiating the involvement of feoB in intracellular infection.
FIG. 5.
Cytopathicity of L. pneumophila for U937 macrophages. Wild-type (solid squares) and NU269 (open squares) strains were used to infect U937 macrophages at an MOI of (A) 1 or (B) 10, and host cell viability was assessed at various times postinoculation Cytopathicity data are presented as the percentage of surviving macrophages in infected monolayers compared to the numbers in uninfected monolayers and represent the mean and standard deviation from six wells. Experiments were performed in triplicate with similar results. Significant differences were observed between the wild-type and mutant strains at 48 and 72 h postinoculation (Student's t test, P < 0.05).
To provide a direct correlation between iron levels in macrophages and the growth defect observed in the feoB mutant, intracellular infection assays were repeated with U937 cells that had been depleted of iron by treatment with DIP. Preliminary experiments showed that the concentrations of DIP used did not cause any gross morphological changes in the macrophages and did not adversely affect their viability (data not shown). Wild-type bacteria were recovered in similar numbers independent of the iron-depleted state of the U937 cells (Fig. 6). In marked contrast, the 15-fold intracellular growth defect of the feoB mutant in untreated U937 cells at 48 h postinoculation was increased to 103-fold and 105-fold by the presence of 25 and 50 μM DIP, respectively (Fig. 6). In monolayers treated with 50 μM DIP, fewer mutant bacteria were recovered than were originally inoculated, suggesting death of the infecting bacteria (Fig. 6). In sum, these results indicate that the feoB mutant is hypersensitive to iron depletion within U937 cells and that ferrous iron transport is important during L. pneumophila intracellular infection of macrophages.
FIG. 6.
Intracellular growth of L. pneumophila in iron-depleted U937 cells. The numbers of wild-type (solid bars) and NU269 (open bars) cells were determined in U937 cell monolayers cultured in medium containing 0, 25, or 50 μM DIP. Data represent means and standard deviations recovered from triplicate wells. The experiment was performed in triplicate with similar results. Significant differences were observed between the wild-type and mutant strains at 25 and 50 μM DIP (Student's t test, P < 0.05).
Virulence of L. pneumophila in A/J mice.
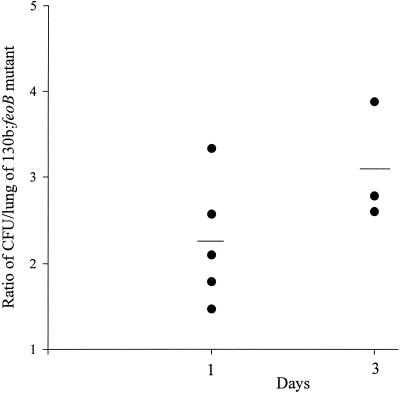
To investigate whether the L. pneumophila feoB mutant was defective for virulence, a competition assay was performed in the A/J mouse model as previously described (48). More specifically, bacteria were recovered from the lungs of mice following intratracheal inoculation of 130b and NU269 in a 1:1 ratio. Compared to NU269, increased numbers of 130b were recovered from infected mouse lungs at day 1; the average ratio of CFU of 130b to NU269 per lung was 2.25 (Fig. 7). By day 3, the average ratio of 130b to NU269 had increased to 3.09 (Fig. 7), suggesting the involvement of FeoB in L. pneumophila virulence in A/J mice.
FIG. 7.
Pulmonary infections of A/J mice with L. pneumophila. Wild-type 130b and mutant NU269 strains in a 1:1 ratio were introduced into the lungs of A/J mice by intratracheal inoculation. At days 1 and 3 postinoculation, the ratio of 130b to Kmr NU269 in infected lungs was determined. Data are representative of actual values obtained per mouse (n = 3 to 5), and a solid bar represents the mean value.
DISCUSSION
The requirement for iron by L. pneumophila for its replication both extracellularly and intracellularly in human macrophages and freshwater amoebae is well documented (8, 10, 11, 30, 59). Our laboratory has identified several determinants that may be involved in the acquisition and/or utilization of iron, likely in the ferric form (29, 30, 36, 42, 45, 58, 59). In this report, we addressed the importance of ferrous iron and its acquisition in L. pneumophila extracellular growth and intracellular infection. We identified and characterized L. pneumophila FeoAB, which bear homology to E. coli and Salmonella enterica serovar Typhimurium FeoAB (33, 56) and Helicobacter pylori FeoB, which lacks the accompanying FeoA (57). In those bacteria, FeoB has been shown to be a ferrous iron transporter. FeoA, although involved in ferrous iron uptake in some bacteria, is poorly understood, and hence our focus was the well-characterized FeoB-like protein in our studies (33, 56).
We strongly believe that L. pneumophila feoAB encodes a functional ferrous iron transporter for several reasons. First, L. pneumophila feoB mutants have decreased ferrous iron accumulation compared to the wild type, a phenotype that was complemented by a copy of the feoB gene supplied in trans. Second, L. pneumophila feoB mutants are streptonigrin resistant, a phenotype associated with iron acquisition mutants in general and feoB mutants in particular (28). Finally, there was strong similarity between the L. pneumophila feoAB predicted protein products and the E. coli and S. enterica FeoAB.
L. pneumophila feoB mutants grew similar to wild-type bacteria in standard BYE broth and BCYE agar, indicating that mutant phenotypes were not the result of a generalized growth defect. Growth in BYE broth without its usual iron supplementation resulted in a minor growth defect in NU269 compared to 130b, which was greatly exacerbated by the addition of the ferrous iron chelator DIP in a concentration-dependent manner. Growth of the feoB mutant could be restored by the addition of FeCl3 to the DIP-treated cultures. This is similar to the case with E. coli and H. pylori feoB mutants, which exhibit decreased extracellular growth in iron-depleted artificial media, which, in the case of the Helicobacter mutants, could be similarly restored by the presence of FeCl3 (53, 57). DIP-treated cultures of NU269 eventually attained wild-type levels of growth by stationary phase, which may indicate the ability of other iron acquisition systems to compensate for the defect in Fe2+ uptake due to the loss of FeoB.
Increased time to colony formation was noted in the feoB mutant compared to the wild type on BCYE agar containing 32-fold less of the usual iron supplementation or no supplement at all. Although this phenomenon has not been described for other bacterial feoB mutants, it was clearly due to the loss of FeoB, as it was complemented by the introduction of a cloned copy of feoB into NU269. Thus, FeoB plays a key role in extracellular L. pneumophila growth under iron-limited conditions. Although aerobic conditions typically result in a predominance of ferric iron, our data suggest that ferrous iron is present within Legionella cultures grown in the presence of oxygen.
Our in vitro infection data demonstrate, for the first time, the importance of a feoB gene in intracellular replication. The L. pneumophila feoB mutant exhibited a 15-fold decreased intracellular growth and/or survival in human U937 cell macrophages, which could be increased to 105-fold with iron-starved, DIP-treated macrophages. Importantly, the defect in intracellular replication in macrophages could be complemented by the addition of feoAB in trans. The only other feoB mutations in an intracellular parasite are in S. enterica serovar Typhimurium; however, the Salmonella mutants were not defective for growth in human epithelial Hep-2 or HeLa cells or in mouse J774 cell macrophages (56). However, S. enterica feoB mutants are defective for gut colonization, suggesting an inadequacy of cell models or a role for FeoB in extracellular growth in vivo.
In our own in vivo studies, the wild-type bacteria outcompeted the feoB mutant, indicating that ferrous iron is available during colonization of the lungs of mice by L. pneumophila and that feoB has a role in Fe2+ acquisition within a mammalian host. The role of L. pneumophila feoB in virulence in mice appears to be relatively modest, although it is of similar magnitude to the S. enterica feoB mutant at 3 days postinoculation; i.e., in S. enterica the ratio of CFU per milligram of feces of wild-type bacteria versus the feoB mutant was approximately 4, compared to the ratio of 3 in the CFU/lung of the L. pneumophila wild-type bacteria versus the feoB mutant (56).
A defect in recovery of the feoB mutant was also noted in iron-starved, DIP-treated H. vermiformis amoebae but not under standard iron-replete coculture conditions. The difference in the recovery of the feoB mutants in macrophages and amoebae under iron-replete conditions suggests that this gene is more important in macrophages than in amoebae. There are other examples of L. pneumophila mutants that are more defective in macrophages than in protozoa (23). Alternatively, it could indicate that more ferrous iron is present during amoebal cocultures or that L. pneumophila has greater access to Fe2+ during infection of H. vermiformis. That the feoB mutant is not completely defective for intracellular growth in macrophages or amoebae and exhibits only a modest virulence defect in mice implies that other uptake systems and/or iron sources can overcome the decreased ferrous iron uptake. The redundancy of determinants involved in using or acquiring iron underscores the key importance of this element for the growth of Legionella and other bacteria. For example, growth defects on iron-limiting agar are demonstrable in an E. coli feoB mutant only if that strain carries additional mutations in siderophore production and uptake determinants, i.e., an aroB tonB background (33, 56).
In summary, we have demonstrated the involvement of the L. pneumophila feoAB locus in ferrous iron transport, extracellular growth, and intracellular infection. The infectivity defect of the feoB mutant further suggests that ferrous iron is present and accessible in the L. pneumophila intracellular niche. Thus, we have shown, for the first time, the involvement of FeoB in bacterial growth in host cells and in the lungs of mice, findings that could have implications for other intracellular parasites and respiratory pathogens.
Acknowledgments
This work was funded by grant AI34937 from the National Institute of Health.
For performing CAS assays, we thank Kimberley Allard. For technical assistance with the mouse competition assays, we thank Antje Flieger, Joseph Garonski-Salerno, Ombeline Rossier, and Virginia Aragon. For helpful discussions and comments, we thank past and present members of the Cianciotto laboratory.
Editor: J. T. Barbieri
REFERENCES
- 1.Aisen, P. 1976. Iron metabolism. Ciba Found. Symp. 51: 1-17. [DOI] [PubMed] [Google Scholar]
- 2.Alli, O. A., L. Y. Gao, L. L. Pedersen, S. Zink, M. Radulic, M. Doric, and Y. Abu Kwaik. 2000. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect. Immun. 68:6431-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askwith, C., D. Eide, A. Van Ho, P. S. Bernard, L. Li, S. Davis-Kaplan, D. M. Sipe, and J. Kaplan. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bellamy, R. 1999. The natural resistance-associated macrophage protein and susceptibility to intracellular pathogens. Microbes Infect. 1:23-27. [DOI] [PubMed] [Google Scholar]
- 5.Breuer, W., S. Epsztejn, and Z. I. Cabantchnik. 1995. Iron acquired from transferrin by K562 cells is delivered into a cytoplasmic pool of chelatable iron(II). J. Biol. Chem. 270:24209-24215. [DOI] [PubMed] [Google Scholar]
- 6.Brieland, J., P. Freeman, R. Kunkel, C. Chrisp, M. Hurley, J. Fantone, and C. Engleberg. 1994. Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. A murine model of human Legionnaires' disease. Am. J. Pathol. 145:1537-1546. [PMC free article] [PubMed] [Google Scholar]
- 7.Brieland, J., M. McClain, L. Heath, C. Chrisp, G. Huffnagle, M. LeGendre, M. Hurley, J. Fantone, and C. Engleberg. 1996. Coinoculation with Hartmannella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires' disease. Infect. Immun. 64:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd, T. F., and M. A. Horwitz. 2000. Aberrantly low transferrin receptor expression on human monocytes is associated with nonpermissiveness for Legionella pneumophila growth. J. Infect. Dis. 181:1394-1400. [DOI] [PubMed] [Google Scholar]
- 9.Byrd, T. F., and M. A. Horwitz. 1991. Chloroquine inhibits the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. A potential new mechanism for the therapeutic effect of chloroquine against intracellular pathogens. J. Clin. Investig. 88:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd, T. F., and M. A. Horwitz. 1989. Interferon gamma-activated human monocytes downregulate transferrin receptors and inhibit the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. J. Clin. Investig. 83:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrd, T. F., and M. A. Horwitz. 1991. Lactoferrin inhibits or promotes Legionella pneumophila intracellular multiplication in nonactivated and interferon gamma-activated human monocytes depending upon its degree of iron saturation. Iron-lactoferrin and nonphysiologic iron chelates reverse monocyte activation against Legionella pneumophila. J. Clin. Investig. 88:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, J. D., S. L. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox, C. D. 1994. Deferration of laboratory media and assays for ferric and ferrous ions. Methods Enzymol. 235:315-329. [DOI] [PubMed] [Google Scholar]
- 15.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engleberg, N. C., C. Carter, D. R. Weber, N. P. Cianciotto, and B. I. Eisenstein. 1989. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect. Immun. 57:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epsztejn, S., O. Kakhlon, H. Glickstein, W. Breuer, and I. Cabantchik. 1997. Fluorescence analysis of the labile iron pool of mammalian cells. Anal. Biochem. 248:31-40. [DOI] [PubMed] [Google Scholar]
- 20.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 21.Fields, B. S., T. A. Nerad, T. K. Sawyer, C. H. King, J. M. Barbaree, W. T. Martin, W. E. Morrill, and G. N. Sanden. 1990. Characterization of an axenic strain of Hartmannella vermiformis obtained from an investigation of nosocomial legionellosis. J. Protozool. 37:581-583. [DOI] [PubMed] [Google Scholar]
- 22.Francis, M. S., and C. J. Thomas. 1997. Mutants in the CtpA copper transporting P-type ATPase reduce virulence of Listeria monocytogenes. Microb. Pathog. 22:67-78. [DOI] [PubMed] [Google Scholar]
- 23.Gao, L. Y., O. S. Harb, and Y. Abu Kwaik. 1998. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect. Immun. 66:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebran, S. J., C. Newton, Y. Yamamoto, R. Widen, T. W. Klein, and H. Friedman. 1994. Macrophage permissiveness for Legionella pneumophila growth modulated by iron. Infect. Immun. 62:564-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gebran, S. J., Y. Yamamoto, S. McHugh, C. Newton, T. W. Klein, and H. Friedman. 1994. Differences and similarities in permissive A/J versus non-permissive BALB/c murine macrophages infected with Legionella pneumophila: the role of iron. FEMS Immunol. Med. Microbiol. 9:7-14. [DOI] [PubMed] [Google Scholar]
- 26.Gebran, S. J., Y. Yamamoto, C. Newton, T. W. Klein, and H. Friedman. 1994. Inhibition of Legionella pneumophila growth by gamma interferon in permissive A/J mouse macrophages: role of reactive oxygen species, nitric oxide, tryptophan, and iron(III). Infect. Immun. 62:3197-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 28.Hantke, K. 1987. Ferrous iron transport mutants in Escherichia coli K12. FEMS Microbiology Lett. 44:53-57. [Google Scholar]
- 29.Hickey, E. K., and N. P. Cianciotto. 1994. Cloning and sequencing of the Legionella pneumophila fur gene. Gene 143:117-121. [DOI] [PubMed] [Google Scholar]
- 30.Hickey, E. K., and N. P. Cianciotto. 1997. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect. Immun. 65:133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi, A. D., and M. S. Swanson. 1999. Comparative analysis of Legionella pneumophila and Legionella micdadei virulence traits. Infect. Immun. 67:4134-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krogfelt, K. A., M. Hjulgaard, K. Sorensen, P. S. Cohen, and M. Givskov. 2000. rpoS gene function is a disadvantage for Escherichia coli BJ4 during competitive colonization of the mouse large intestine. Infect. Immun. 68:2518-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liles, M. R., P. H. Edelstein, and N. P. Cianciotto. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol. Microbiol. 31:959-970. [DOI] [PubMed] [Google Scholar]
- 36.Liles, M. R., T. A. Scheel, and N. P. Cianciotto. 2000. Discovery of a nonclassical siderophore, legiobactin, produced by strains of Legionella pneumophila. J. Bacteriol. 182:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthews, M., and C. R. Roy. 2000. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect. Immun. 68:3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34:836-849. [DOI] [PubMed] [Google Scholar]
- 39.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 40.Nash, T. W., D. M. Libby, and M. A. Horwitz. 1988. IFN-gamma-activated human alveolar macrophages inhibit the intracellular multiplication of Legionella pneumophila. J. Immunol. 140:3978-3981. [PubMed] [Google Scholar]
- 41.Nash, T. W., D. M. Libby, and M. A. Horwitz. 1984. Interaction between the legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J. Clin. Investig. 74:771-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Connell, W. A., E. K. Hickey, and N. P. Cianciotto. 1996. A Legionella pneumophila gene that promotes hemin binding. Infect. Immun. 64:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearlman, E., A. H. Jiwa, N. C. Engleberg, and B. I. Eisenstein. 1988. Growth of Legionella pneumophila in a human macrophage-like (U937) cell line. Microb. Pathog. 5:87-95. [DOI] [PubMed] [Google Scholar]
- 44.Poch, M. T., and W. Johnson. 1993. Ferric reductases of Legionella pneumophila. Biometals 6:107-114. [DOI] [PubMed] [Google Scholar]
- 45.Pope, C. D., W. O'Connell, and N. P. Cianciotto. 1996. Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect. Immun. 64:629-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 47.Reeves, M. W., L. Pine, S. H. Hutner, J. R. George, and W. K. Harrell. 1981. Metal requirements of Legionella pneumophila. J. Clin. Microbiol. 13:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robey, M., W. O'Connell, and N. P. Cianciotto. 2001. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossier, O., and N. P. Cianciotto. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 51.Segal, G., and H. A. Shuman. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol. Microbiol. 30:197-208. [DOI] [PubMed] [Google Scholar]
- 52.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stojiljkovic, I., M. Cobeljic, and K. Hantke. 1993. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol. Lett. 108:111-115. [DOI] [PubMed] [Google Scholar]
- 54.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sundstrom, C., and K. Nilsson. 1976. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 17:565-577. [DOI] [PubMed] [Google Scholar]
- 56.Tsolis, R. M., A. J. Baumler, F. Heffron, and I. Stojiljkovic. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64:4549-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 58.Viswanathan, V. K., P. H. Edelstein, C. D. Pope, and N. P. Cianciotto. 2000. The Legionella pneumophila iraAB locus is required for iron assimilation, intracellular infection, and virulence. Infect. Immun. 68:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Viswanathan, V. K., S. Kurtz, L. L. Pedersen, Y. Abu Kwaik, K. Krcmarik, S. Mody, and N. P. Cianciotto. 2002. The cytochrome c maturation locus of Legionella pneumophila promotes iron assimilation and intracellular infection and contains a strain-specific insertion sequence element. Infect. Immun. 70:1842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinberg, E. D. 2000. Microbial pathogens with impaired ability to acquire host iron. Biometals 13:85-89. [DOI] [PubMed] [Google Scholar]
- 61.Wiater, L. A., K. Dunn, F. R. Maxfield, and H. A. Shuman. 1998. Early events in phagosome establishment are required for intracellular survival of Legionella pneumophila. Infect. Immun. 66:4450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wooldridge, K. G., and P. H. Williams. 1993. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 12:325-348. [DOI] [PubMed] [Google Scholar]
- 63.Yildiz, F. H., and G. K. Schoolnik. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J. Bacteriol. 180:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]