Abstract
A bovine-specific cDNA microarray system containing 721 unique leukocyte expressed sequence tags (ESTs) and amplicons representing known genes was used to compare gene expression profiles of peripheral blood mononuclear cells (PBMCs) from clinical and subclinical Johne's disease-positive Holstein cows (n = 2 per group). Stimulation of PBMCs from clinically infected cows with Mycobacterium paratuberculosis tended to decrease expression of 83 genes (fold change, >1.5). Of these 83 genes, 16 displayed significant down regulation across both clinical cows (P < 0.1), including genes encoding microspherule protein 1, fibroblast growth factor, and the Lyn B protein kinase. Only eight genes from PBMCs of clinically infected cows exhibited a modest up regulation following stimulation with M. paratuberculosis, including those encoding bovine CD40L, gamma interferon, interleukin-10 (IL-10), and tissue inhibitor of matrix metalloproteinases (TIMP) 4. In contrast, stimulation of PBMCs from subclinically infected cows with M. paratuberculosis tended to up regulate expression of 71 genes representing 68 unique transcripts. Of these, 11 genes showed significant up regulation (fold change, >1.5; P < 0.1) across both animals, including those encoding bovine CD40L, several matrix metalloproteinases, and SPARC (secreted protein, acidic and rich in cystine). Repression of gene expression was also observed in PBMCs from the subclinical cows, with 16 genes being significantly down regulated (fold change, >1.5; P < 0.1) across both animals, including those encoding the bovine orthologs of cytochrome oxidase subunit III, IL-1 receptor type I, and fibrinogen-like 2 protein. Only one clone, representing an unknown bovine EST, was similarly down regulated in PBMCs from both the clinical and subclinical cows. Thus, the most prominent change induced by exposure of PBMCs from clinical cows to M. paratuberculosis in vitro tended to be repression of gene expression, while changes in similarly treated PBMCs from subclinical cows was balanced between gene activation and repression. Comparison of gene expression profiles between PBMCs from clinical and uninfected (control) cows stimulated with the general mitogen concanavalin A were highly similar (overall r = 0.84), suggesting that M. paratuberculosis-induced gene repression in clinically infected cow PBMCs was not due to a general failure of the immune response in these animals.
Johne's disease is a chronic infectious disease of ruminants caused by Mycobacterium avium subspecies paratuberculosis (M. paratuberculosis). Many ruminant infections with M. paratuberculosis lead to granulomatous enteritis, persistent diarrhea, progressive wasting, and finally death. Associated losses due to reduced milk production and premature culling combine to make Johne's disease one of the most costly infectious diseases of dairy cattle. Overall estimates suggest infections with M. paratuberculosis cost the U.S. dairy industry more than $200 million per year (16). As sobering as these estimates are, many cattle with subclinical M. paratuberculosis infections go undiagnosed for several years, and thus the national infection rate and actual cost to dairy and livestock producers may be much higher. A controversial but developing link between M. paratuberculosis and human Crohn's disease suggests that this pathogen may also be an important food safety concern (4, 9). Prevalence of M. paratuberculosis infection in the national dairy herd, the grave economic and welfare consequences of Johne's disease, and the potential for linkage to human disease combine to make a powerful case for learning more about how M. paratuberculosis infections progress and about the host immune response to this fastidious pathogen.
Cattle and other ruminants are usually infected with M. paratuberculosis in utero or in the first few months of life, via the fecal-oral route or by ingestion of infected colostrum (19, 25, 27, 30). Shortly after initial infection, most animals begin to develop a T-cell response, characterized by release of the proinflammatory cytokines gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α), as well as production of interleukin-2 (IL-2) (for a review, see reference 10). This proinflammatory cytokine cascade leads to recruitment and activation of cytotoxic CD8+ T cells and other immune components, including neutrophils and additional activated macrophages, to sites of M. paratuberculosis infection (1, 2, 7, 15, 17, 18). Activation of CD4+ T cells, production of proinflammatory cytokines, and recruitment of cytolytic CD8+ T cells to the site of infection comprise development of an acquired Th1-like immune response against M. paratuberculosis. Rapid development and continued maintenance of this response is a critical factor in controlling mycobacterial infections in a variety of species (14, 15, 21, 22).
During the long subclinical phase of infection (2 to 5 years) with M. paratuberculosis, the ruminant host immune system shifts away from a beneficial cytotoxic response in favor of a Th2-like response that elicits nonprotective antibody production from B cells (17, 18, 24). Late-stage subclinical and clinical infections with M. paratuberculosis are often associated with a strong immunoglobulin G1 antibody response (for a review, see reference 10). During the clinical stage of Johne's disease, circulating CD4+ T-cell responses to M. paratuberculosis may be severely depressed, as assessed by pathogen-stimulated proliferation assays and measurement of IFN-γ production in peripheral blood mononuclear cells (PBMCs) isolated from sick animals (5-7, 23, 24). This antigen-specific anergy may also be accompanied by a general reduction in response of T cells to nonspecific mitogens (24), indicating significant changes in general T-cell responses of M. paratuberculosis-infected animals. Very late clinical stage animals also exhibit a reduction in B-cell response to M. paratuberculosis antigens, although B-cell responses to mitogens remain intact and overall circulating B-cell numbers are not reduced relative to levels in control uninfected animals (29). The mechanisms responsible for these profound and ultimately disastrous changes in the immune response to M. paratuberculosis and other mycobacteria remain unknown.
In the present report, we describe experiments using a novel bovine total leukocyte (BOTL-2) cDNA microarray resource to evaluate the response of PBMCs from cattle naturally infected with M. paratuberculosis and in various stages of Johne's disease to antigen stimulation. Prior application of the microarrays employed in this report has clearly demonstrated the utility of gene expression profiling in monitoring immune responses of cattle to general mitogens (31) and to immune system stress (8). We reasoned that application of gene expression profiling would help define the complex interactions between M. paratuberculosis and the bovine immune system.
MATERIALS AND METHODS
Experimental animals and preparation of PBMCs.
The infected and control cattle used in this study were all multiparous Holstein cows ranging in age from 24 to 48 months and were all housed on the same commercial dairy operation. The immune status of all animals with regard to M. paratuberculosis infection had been monitored by a serum enzyme-linked immunosorbent assay (ELISA) on a bimonthly basis for over 20 months prior to the initiation of experiments. Additional tests for M. paratuberculosis infection included IFN-γ ELISA following ex vivo stimulation of PBMCs using a commercial system (BioCor, Inc., Des Moines, Iowa) and periodic fecal culture testing using a U.S. Department of Agriculture-approved testing laboratory (Michigan State University Animal Health Diagnostic Laboratory, East Lansing, Mich.). The control uninfected animals (n = 3) had shown negative responses in over 2 years of testing by all assays used. The clinical M. paratuberculosis-infected animals (n = 2) were strongly positive by serum ELISA (index value, >100) over the entire testing period, exhibited occasional diarrhea, and were slightly emaciated by the conclusion of these studies. The subclinically M. paratuberculosis-infected cows (n = 2) displayed low to moderate positive results on serum ELISA testing (index value between 50 and 100) and shed very low (less than 5 colonies per g) levels of M. paratuberculosis in feces. Tests for IFN-γ in the clinical and subclinical animals were strongly positive when results were analyzed as recommended by the manufacturer (BioCor, Inc.).
Blood samples were obtained from all animals via the coccygeal (tail) vein by using 2.5-cm 21-gauge multiple sample needles and a series of four 8-ml Vacutainer tubes containing heparin as an anticoagulant. Peripheral blood monocytes were prepared as previously described (28, 31). Briefly, blood samples were centrifuged at 4°C for 20 min at 1,000 × g, and the resulting buffy coat (approximately 2 ml) was transferred to a new 50-ml conical tube containing 34 ml of ice-cold sterile phosphate-buffered saline (PBS) overlaid on a 10-ml cushion of Percoll (1.084 g/ml; Sigma Chemical Co., St. Louis, Mo.). Cells were centrifuged at 1,000 × g for 40 min at room temperature to separate erythrocytes and polymorphonuclear leukocytes from mononuclear cells. Following careful aspiration of the PBS, PBMCs lying at the PBS-Percoll interface were transferred to new 50-ml conical tubes, rinsed once with 20 ml of sterile PBS, and finally resuspended in maintenance medium (RPMI 1640) containing 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (complete RPMI 1640).
Stimulation of PBMCs was conducted using 108 live M. paratuberculosis bacteria per ml of media or concanavalin A (ConA) at a final concentration of 10 μg per ml of media, each added in 200 μl of PBS. Control stimulations (nil) were conducted by adding 200 μl of PBS only. Separate aliquots of PBMCs from each cow were stimulated with the various treatments (nil, M. paratuberculosis, or ConA) and were incubated at 37°C for 16 to 18 h in complete RPMI 1640 without antibiotics in a humidified atmosphere of 95% air, 5% CO2. For RNA extraction and microarray analysis, each treatment typically consisted of two to three 75-cm2 flasks, with each flask containing approximately 109 PBMCs. In previous microarray analyses using ConA stimulation of PBMCs, a dramatic enhancement of immune cell gene expression was observed when PBMCs from different animals were pooled prior to stimulation, versus stimulation in individual aliquots (31). Therefore, all treatments used in experiments reported here were performed using individual animal PBMCs rather than pooled PBMCs.
BOTL-2 cDNA microarrays.
The cDNA microarrays (BOTL-2) used in these experiments have been described previously (8, 31). Briefly, each microarray contained 3,072 total spots consisting of 721 unique bovine expressed sequence tag (EST) clone inserts or amplicons representing known genes derived from the bovine sequence. Known genes represented on the BOTL-2 cDNA microarrays included those encoding the cytokines IL-1, IL-4, IL-5, IL-6, IL-10, IL-12, TNF-α, IFN-γ, and transforming growth factor β, several matrix metalloproteinases (MMPs), CD40L, tissue inhibitors of matrix metalloproteinases (TIMPs), and various growth factor and hormone receptors. All ESTs and known genes were spotted in triplicate. Control genes included 144 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control gene spots (3 spots in each of 48 patches), 96 synthetic lambda Q control gene spots (2 per patch), 48 negative control spots (1 per patch), and numerous blank spots (13 per patch). The entire array was organized in a 4-by-12 pattern of patches, with each patch containing 64 spots in an 8-by-8 pattern. Use of these microarrays to monitor the response of bovine PBMCs to ConA demonstrated that the arrays could readily detect immune cell activation in cattle (31). A web-accessible resource has been established (http://www.nbfgc.msu.edu) to allow scanning of microarray contents as well as to provide a readily accessible resource for searching by cDNA clone name or gene name. This resource may be used to obtain additional information on EST clones discussed in this report.
RNA extraction, preparation of labeled cDNA, and microarray analysis.
RNA was extracted from the nil, M. paratuberculosis-, and ConA-stimulated PBMCs by using Trizol reagent (Invitrogen Life Technologies, Inc., Gaithersburg, Md.) as previously described (8, 31). The quantity and quality of extracted total RNA were estimated by UV spectrophotometry and electrophoresis on 1.2% native agarose gels. To evaluate the gene expression profiles of PBMCs following various treatments, total RNA (20 μg) from nil, M. paratuberculosis-, or ConA-stimulated PBMCs of each animal were used as templates in reverse transcription reactions, using a commercial cDNA microarray labeling kit (Atlas Glass labeling system; Clontech Inc., Alameda, Calif.) and oligo(dT)15 as primer. Synthesis of cDNA in this system incorporates an amino-modified dUTP into the cDNA. To provide a control for cDNA synthesis and labeling efficiency, as well as for subsequent cDNA microarray hybridization, 1 to 2 ng of synthetic lambda Q gene RNA containing an engineered poly(A) tail was spiked into each reaction mixture. Following first-strand cDNA synthesis, cDNAs from nil and M. paratuberculosis-stimulated or nil and ConA-stimulated PBMCs of each animal were differentially labeled using N-hydroxysuccinimide-derivatized Cy3 and Cy5 dyes, respectively (Amersham Pharmacia, Ltd.). Labeled cDNAs were extensively purified to remove unincorporated dyes, combined, and concentrated to 4 to 10 μl using Microcon 30 spin concentrators (Millipore Corp.). Microarray hybridizations for each animal were performed by addition of concentrated Cy3 (nil) and Cy5 (ConA or M. paratuberculosis) labeled probe cDNAs to 45 μl of GlassHyb (Clontech, Inc.) supplemented with 10 μg of bovine serum albumin/ml and 10 μg of denatured salmon sperm DNA/ml. Hybridizations were conducted for 18 to 20 h at 50°C in sealed and humidified chambers (Arrayit; TeleChem International, Inc., Sunnyvale, Calif.). Following hybridizations, the microarrays were washed twice at room temperature in 1× SSC-0.05% sodium dodecyl sulfate (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and once at 50°C in the same solution. Washed microarrays were rinsed in double-distilled H2O and dried by centrifugation in a cushioned 50-ml conical centrifuge tube. This process yielded microarrays allowing direct comparison of PBMC gene expression profiles following stimulation with M. paratuberculosis or ConA versus nil stimulation for each animal.
Final microarrays were scanned immediately using a GeneTAC LS IV microarray scanner and GeneTAC LS software (Genomic Solutions, Inc., Ann Arbor, Mich.). GeneTAC analyzer software was then used to process microarray images, find spots, integrate robot-spotting files with the microarray image and, finally, to create reports of spot intensity ratios and total spot reports.
Microarray data analysis.
Total spot report data from each microarray experiment were imported into a Microsoft Excel worksheet for analysis as described previously (8, 31). Briefly, spot intensity data for the GAPDH, negative, blank, and lambda Q gene controls were separated from the sample spot report to allow assessment of any potential RNA loading, background, and cDNA synthesis efficiency effects. GAPDH Cy3 intensity was regressed on GAPDH Cy5 intensity for each of the 144 GAPDH gene spots to determine if significant dye or RNA loading effects were present. Typical slopes for the GAPDH regression line ranged from 1 to 3 for experiments reported here. If necessary, the GAPDH regression line equation was used to globally normalize sample spot Cy3 or Cy5 intensity values, both within each microarray and across multiple microarrays. Total uncorrected and GAPDH-corrected sample spot intensities were also plotted to assess the efficiency of GAPDH normalization within an experiment. In most cases, plots of all uncorrected sample intensities yielded a regression line with a slope in good agreement with that derived by using only the GAPDH control spots, supporting the use of GAPDH as a control gene for normalization in experiments with total bovine PBMCs.
For analysis of test samples, corrected spot intensity data were sorted by clone name and microarray address to isolate the triplicate data for each of the Cy3- and Cy5-labeled BOTL cDNAs and known genes within each microarray. Expression ratios (M. paratuberculosis/PBS or ConA/PBS) were independently calculated for each of the three gene replicates for an animal (one microarray per animal). Expression ratios were log transformed and used to calculate a mean log expression ratio for each gene for an animal. The mean expression ratios for each animal within our groups were combined to yield an overall average log gene expression ratio, standard error, t statistic, and P value for each gene represented on the microarray. Thus, the presented P values represent the confidence that the combined log expression ratio (n = 2 for the clinical and subclinical groups and n = 3 for the control group) is different from 0. For comparisons across groups, a Student's two-tailed t test was performed to assess the confidence that mean expression ratios between groups were different.
RESULTS
Differences in gene expression profiles of PBMCs from subclinical and clinical Johne's disease-positive cows following stimulation with M. paratuberculosis in vitro.
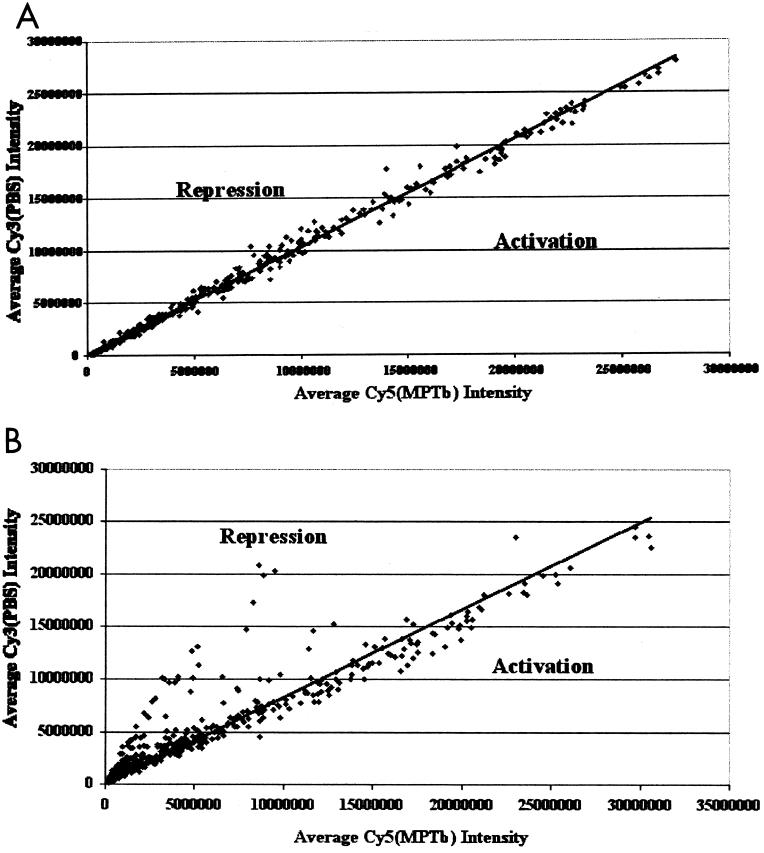
As a first step in analyzing the effect of M. paratuberculosis on PBMCs from infected cattle, we hypothesized that stimulation of PBMCs from Johne's disease-positive cows in various stages of disease progression would result in vastly different immune cell gene expression profiles. As a first step in these studies, we compared gene expression profiles of PBMCs from three uninfected control cows following stimulation with M. paratuberculosis or PBS (nil). When the average relative fluorescence intensities for each gene represented on the BOTL-2 microarray were plotted for the uninfected cows, most transcripts represented on the BOTL-2 microarray were similarly expressed following nil and M. paratuberculosis stimulation (Fig. 1). Following complete analysis of the data, only six genes (0.8%) exhibited significantly activated expression (fold change, >1.5; P < 0.1) following stimulation of the uninfected control animal PBMCs with M. paratuberculosis. This is as expected, since PBMCs from cows not previously exposed to M. paratuberculosis would not be expected to respond strongly to antigens from this pathogen. Importantly, this study suggested that most gene expression changes observed in PBMCs from infected animals would be due to infection and not to an inherent response of bovine PBMCs to M. paratuberculosis.
FIG. 1.
Scatter plots of total sample Cy3 (PBS) and Cy5 (M. paratuberculosis) relative fluorescence intensities corrected for GAPDH. (A) Average gene expression intensities for control uninfected cows, with plot of average normalized intensity values for uninfected control animal (n = 3) PBMCs stimulated with PBS (Cy3; y axis) or M. paratuberculosis (Cy5; x axis). RNA isolation, probe labeling, and microarray hybridizations were conducted as described in Materials and Methods. Raw microarray intensity data for the Cy3 and Cy5 channels across three microarrays (one per animal) were normalized using GAPDH control genes, also as described in Materials and Methods. Resulting normalized intensity values were used to derive a mean relative fluorescence intensity value for each gene on the BOTL-2 microarray across all animals. The resulting mean normalized Cy3 (PBS) and Cy5 (M. paratuberculosis) intensity values were plotted, with Cy3 values on the y axis and Cy5 values on the x axis. The correlation (r2) of all spots to the resulting regression line is shown (0.9956). (B) Plot of average gene expression intensities comparing PBS versus M. paratuberculosis stimulation of PBMCs derived from two clinical-stage Johne's disease-positive cows. In this analysis, average gene expression intensities for each gene on the BOTL-2 cDNA microarray were derived as described for panel A using the normalized data from two separate microarrays (one per animal). In this case, the correlation coefficient (r2) derived by regression of average PBS intensities against average M. paratuberculosis intensities was 0.88, and the line was skewed from unity (y = 0.8394x), likely due to the number of genes showing repressed expression following stimulation with M. paratuberculosis relative to simulation with PBS.
The next phase of this investigation was to analyze responses of PBMCs from clinical M. paratuberculosis-infected cows to stimulation with M. paratuberculosis, relative to nil stimulation. This process produced two separate microarrays, with each one comparing nil versus M. paratuberculosis stimulation of PBMCs from one clinical Johne's disease-positive cow. In contrast to the uninfected control animals, a total gene average fluorescence intensity plot for the clinically infected cows revealed an obvious suppression of gene expression (Cy3 [PBS] fluorescence intensity > Cy5 [M. paratuberculosis] fluorescence intensity) as one major outcome of PBMC stimulation with M. paratuberculosis (Fig. 1B). Following complete data analysis, expression of 83 genes tended to be repressed (mean fold change, >1.5) by stimulation of PBMCs from the clinical cows with M. paratuberculosis relative to nil stimulation. Of these genes, 16 showed significantly repressed gene expression in both animals, based on our criteria of fold repression of >1.5 and a P value of <0.1 (Table 1). Repressed genes included those encoding the Lyn B tyrosine kinase, a cell cycle-regulated microspherule protein, and a fibroblast growth factor binding protein, as well as several genes of unknown function.
TABLE 1.
Genes and EST clones corresponding to transcripts significantly repressed across both clinical M. paratuberculosis-infected cows
| Gene or EST name | Description | Avg fold repression | P valuea |
|---|---|---|---|
| BOTL0100004XG07R | Unknown bovine EST sequence matching human chromosome 19 gene | 1.861 | 0.01133 |
| BOTL0100006XB04R | Unknown bovine EST | 1.957 | 0.00334 |
| BOTL0100006XG04R | Unknown bovine EST | 1.819 | 0.06342 |
| BOTL0100006XH06R | Bovine EST ortholog of human Lyn B protein kinase gene (v-ves-related oncogene) | 1.738 | 0.04571 |
| BOTL0100008_C08 | Bovine EST similar to human DAZ-associated protein 2 mRNA | 1.709 | 0.09946 |
| BOTL0100009_B07 | Bovine EST similar to murine midnolin mRNA | 2.094 | 0.07973 |
| BOTL0100009_E09 | Bovine EST similar to Bos taurus microsatellite MNB-134 | 2.717 | 0.08134 |
| BOTL0100009_E10 | Bovine EST similar to human diacylglycerol O-acyltransferase mRNA | 1.843 | 0.07205 |
| BOTL0100009_H10 | Bovine EST similar to human sequence encoding hypothetical 116-kDa protein | 2.068 | 0.09816 |
| BOTL0100010_E07 | Bovine EST similar to human BTG gene family and viral receptor | 1.698 | 0.01641 |
| BOTL0100011_E04 | Bovine EST similar to human cell cyclo-regulated microspherule protein 1 mRNA | 2.631 | 0.00466 |
| BOTL0100011_G08 | Bovine EST similar to human mRNA encoding protein of unknown function | 1.822 | 0.02551 |
| BOTL0100012_C04 | Bovine EST similar to sequence encoding human hypothetical protein MGC5442 | 2.216 | 0.09546 |
| BOTL0100012_C10 | Bovine EST similar to human fibroblast growth factor binding protein mRNA | 1.768 | 0.06311 |
| BOTL0100012_G08 | Bovine EST similar to human PX domain containing protein kinase mRNA | 1.718 | 0.08744 |
| BOTL0100013_B10 | Bovine EST similar to region of human chromosome 19 sequence | 1.840 | 0.06908 |
Paired t test (n = 2).
In these same animals, the expression of eight genes tended to be up regulated, but the up regulation was not significant (P > 0.1) across both animals. The observed variability in expression ratios of these genes was primarily due to a failure to activate expression in PBMCs from one clinical infected animal (Table 2). When examined within an animal, using the three gene replicates on each microarray, expression levels of IL-10 and IFN-γ were significantly (P < 0.1) greater in PBMCs stimulated with M. paratuberculosis than in the nil control stimulated PBMCs for each of our clinical infected cows, although the absolute degree of activation was lower in PBMCs from clinical cow number 2 (Table 2). Expression levels of the remaining genes in Table 2 were significantly greater in M. paratuberculosis-stimulated PBMCs relative to nil stimulated PBMCs from clinical animal number 1 only. Taken together, our results suggest that despite variability between animals, the major common response of PBMCs from both clinical M. paratuberculosis-infected cows used in this study to M. paratuberculosis was down regulation of gene expression.
TABLE 2.
Genes and EST clones tending to be up regulated following stimulation of PBMCs from clinical M. paratuberculosis-infected cows with M. paratuberculosis
| Gene or EST name | Description | Level of activation (fold)
|
||
|---|---|---|---|---|
| Clinical animal 1 | Clinical animal 2 | Avg | ||
| BOTL0100003XB05R | Bovine EST similar to human guanylate binding protein 1, interferon-inducible mRNA | 2.878 | 1.150 | 2.014 |
| BOTL0100004XD12R | Bovine EST similar to human selenophosphate synthetase 2 mRNA | 3.147 | 1.064 | 2.105 |
| BOTL0100009_C01 | Bovine EST similar to human chromosome 10 sequence | 3.447 | 0.944 | 2.196 |
| BOTL0100013_C10 | Bovine EST similar to rodent Fos-related antigen (Fra) mRNA | 3.164 | 1.177 | 2.170 |
| CD40L | Amplicon representing bovine CD40 ligand mRNA | 2.138 | 1.059 | 1.599 |
| IL-10 | Amplicon representing bovine IL-10 mRNA | 2.843 | 1.216 | 2.029 |
| IFN-γ | Amplicon representing bovine IFN-γ mRNA | 3.342 | 1.309 | 2.325 |
| TIMP 4 | Amplicon representing bovine TIMP 4 mRNA | 2.989 | 0.825 | 1.907 |
The effects of stimulating PBMCs from subclinical M. paratuberculosis-infected cows with M. paratuberculosis were examined next, allowing a comparison of the response of cells from these animals to that of the clinical infected cows. A plot of normalized average Cy3 (PBS) and average Cy5 (M. paratuberculosis) signal intensities for all genes represented on the BOTL-2 cDNA microarray suggested that the response of PBMCs from subclinical cows to stimulation with M. paratuberculosis was not nearly as pronounced as that of the clinical infected animals observed previously (data not shown). However, when the data were subsequently analyzed and selected to reveal genes that tended to be up regulated or down regulated (fold change, >1.5) by exposure of PBMCs to M. paratuberculosis, expression of 71 known genes and EST clones representing 68 unique transcripts tended to be enhanced relative to nil controls. Of these 71 known genes and EST clones, 11 were significantly (P < 0.1) activated across PBMCs from both animals (Table 3). As with PBMCs from the clinical cows, 16 genes were significantly repressed in PBMCs from the subclinical cows following stimulation with M. paratuberculosis (Table 4). Of these genes, only one transcript, corresponding to an EST clone (BOTL010006XG04R) of unknown function, was similarly down regulated in PBMCs from the clinical M. paratuberculosis-infected cows.
TABLE 3.
Genes and EST clones representing transcripts significantly activated across both subclinical M. paratuberculosis-infected cows
| Gene or clone name | Description | Mean fold activation | P valueb |
|---|---|---|---|
| BOTL0100001XF05R | Bovine EST similar to human genomic sequence of unknown function | 2.122 | 0.086 |
| BOTL0100002XB07R | Bovine EST similar to human glioma tumor suppressor candidate region gene | 1.748 | 0.055 |
| BOTL0100003XD05R | Bovine EST similar to human genomic sequence of unknown function | 2.123 | 0.055 |
| BOTL0100003XH08R | Bovine EST similar to pig and human 17-β-estradiol dehydrogenase 4 | 1.751 | 0.039 |
| BOTL0100006XB01R | Bovine EST similar to human mRNA for geminin | 1.857 | 0.040 |
| CD40La | Amplicon representing bovine CD40 ligand | 2.299 | 0.081 |
| MMP 15 | Amplicon representing bovine MMP 15 | 2.177 | 0.046 |
| MMP 16 | Amplicon representing bovine MMP 16 | 2.357 | 0.082 |
| MMP 9 | Amplicon representing bovine MMP 9 | 1.836 | 0.044 |
| SPARC | Amplicon representing bovine SPARC | 1.968 | 0.078 |
| TIMP 4a | Amplicon representing bovine TIMP 4 | 1.551 | 0.023 |
TIMP 4 and CD40L expression also tended to be activated in PBMCs from clinical M. paratuberculosis-infected cows.
Paired t test (n = 2).
TABLE 4.
Genes and EST clones representing transcripts significantly repressed across both subclinical M. paratuberculosis-infected cows
| Gene or clone name | Description | Mean fold repression | P valueb |
|---|---|---|---|
| BOTL0100001XC01R | Bovine EST similar to human fibrinogen-like 2 mRNA | 1.680 | 0.038 |
| BOTL0100002XE10R | Bovine EST similar to human acidic ribosomal phosphoprotein mRNA | 1.763 | 0.076 |
| BOTL0100002XF06R | Bovine EST representing mitochondrial cytochrome oxidase subunit III (COX III) gene | 2.053 | 0.074 |
| BOTL0100003XA07R | Bovine EST similar to mouse integral membrane protein TAPA-1 mRNA | 1.695 | 0.089 |
| BOTL0100005XH03R | Bovine EST similar to human CD164 mRNA | 1.802 | 0.096 |
| BOTL0100006XD05R | Bovine EST similar to human genomic sequence from chromosome 1 of unknown function | 1.704 | 0.084 |
| BOTL0100006XG02R | Bovine EST similar to murine β-chimaerin splicing factor | 1.924 | 0.055 |
| BOTL0100006XG04Ra | Bovine EST of unknown function | 2.021 | 0.071 |
| BOTL0100009_F03 | Bovine EST similar to human cDNA FLJ12695 | 1.730 | 0.098 |
| BOTL0100010_D01 | Bovine EST similar to human IL-1 receptor type 1 | 1.862 | 0.017 |
| BOTL0100010_E08 | Bovine EST representing adenylyl cyclase type VII | 1.839 | 0.052 |
| BOTL0100010_G01 | Bovine EST similar to human glutaryl-coenzyme A dehydrogenase mRNA | 1.748 | 0.056 |
| BOTL0100010_G03 | Bovine EST similar to human CA150 transcription factor mRNA | 1.770 | 0.098 |
| BOTL0100010_H09 | Bovine EST similar to human mRNA of unknown function | 1.950 | 0.007 |
| BOTL0100012_B01 | Bovine EST similar to murine Mdes mRNA and human fatty acid desaturase MLD mRNA | 1.965 | 0.049 |
| THR | Amplicon representing bovine thyroid hormone receptor | 1.672 | 0.069 |
BOTL010006XG04R expression was also significantly repressed in PBMCs from clinical M. paratuberculosis-infected cows.
Paired t test (n = 2).
Mean expression ratios of the 71 known genes and EST clones that tended to be up regulated by M. paratuberculosis in PBMCs from subclinical cows were compared with those from the clinical cows (Table 5). Although several genes, including those for many cytokines, appeared to be similarly activated by M. paratuberculosis in PBMCs from both clinical and subclinical M. paratuberculosis-infected cows, there were many clear differences. In most cases (>50), the expression levels of genes that were activated by M. paratuberculosis in PBMCs from subclinical cows were not similarly affected in PBMCs from the clinical cows. Perhaps more importantly, there were 13 genes where differences between the subclinical and clinical groups were highly significant (P < 0.1, paired t test with n = 2 per group) (Table 5). This list of genes includes IL-1, several MMPs (MMP 15, MMP 19, and MT1-MMP), SPARC (secreted protein, acidic and rich in cystine), and uPAR (urokinase plasminogen activator receptor).
TABLE 5.
Expression ratios for known genes and EST clones activated in PBMCs from subclinical M. paratuberculosis-infected cows following stimulation with M. paratuberculosis
| Gene or clone name | Mean expression ratio (M. paratuberculosis/PBS)
|
P value | |
|---|---|---|---|
| Subclinical | Clinical | ||
| 17a-hydroxylase | 2.160 | 0.976 | 0.39767 |
| 3BHSD | 1.520 | 1.300 | 0.81912 |
| a-2mac | 1.516 | 1.006 | 0.31539 |
| BOTL0100001XE01R | 1.509 | 0.935 | 0.53066 |
| BOTL0100001XF05R | 1.870 | 1.281 | 0.40416 |
| BOTL0100002XB07R | 1.748 | 0.850 | 0.30087 |
| BOTL0100002XE08R | 1.642 | 0.861 | 0.21130 |
| BOTL0100002XE12R | 1.743 | 1.205 | 0.46002 |
| BOTL0100003XA08R | 1.847 | 1.382 | 0.68034 |
| BOTL0100003XB05R | 2.243 | 1.737 | 0.79676 |
| BOTL0100003XD05R | 2.123 | 0.721 | 0.26725 |
| BOTL0100003XD06R | 2.204 | 1.083 | 0.45855 |
| BOTL0100003XG03R | 1.796 | 1.038 | 0.37513 |
| BOTL0100003XH08R | 1.751 | 1.272 | 0.52597 |
| BOTL0100006XB01R | 1.857 | 1.448 | 0.64477 |
| BOTL0100006XC07R | 1.690 | 1.083 | 0.64300 |
| BOTL0100006XC08R | 1.588 | 0.765 | 0.02881 |
| BOTL0100006XF09R | 2.315 | 0.957 | 0.49482 |
| BOTL0100007XE07R | 1.964 | 1.025 | 0.52231 |
| BOTL0100008_E02 | 3.127 | 0.782 | 0.25304 |
| BOTL0100009_C01 | 1.772 | 1.656 | 0.92095 |
| BOTL0100009_D10 | 1.666 | 0.711 | 0.02036 |
| BOTL0100009_H05 | 1.763 | 1.048 | 0.45466 |
| BOTL0100009_H07 | 1.523 | 0.529 | 0.13340 |
| BOTL0100010_D06 | 1.577 | 1.142 | 0.42540 |
| BOTL0100010_D09 | 2.536 | 1.236 | 0.37886 |
| BOTL0100011_A10 | 1.694 | 0.593 | 0.24960 |
| BOTL0100011_C03 | 1.575 | 0.981 | 0.47555 |
| BOTL0100012_C11 | 1.908 | 0.969 | 0.49981 |
| BOTL0100012_D12 | 1.519 | 0.470 | 0.01391 |
| BOTL0100012_H03 | 1.565 | 0.697 | 0.09590 |
| CD40L | 2.299 | 1.578 | 0.47264 |
| CRF | 1.929 | 1.102 | 0.58050 |
| CRFR1 | 1.638 | 1.050 | 0.45890 |
| EP2 | 1.600 | 0.983 | 0.11912 |
| FSH-R | 1.785 | 1.056 | 0.67017 |
| IL-1 | 2.033 | 0.926 | 0.03202 |
| IL-10 | 2.052 | 1.587 | 0.73982 |
| IL-12 | 1.977 | 1.386 | 0.73632 |
| IL-2 | 2.279 | 1.417 | 0.50863 |
| IL-4 | 1.895 | 1.453 | 0.72178 |
| IL-5 | 1.699 | 1.405 | 0.84261 |
| IFN-γ | 2.208 | 1.761 | 0.77541 |
| MMP 1 | 2.330 | 0.562 | 0.17830 |
| MMP 1 up | 1.997 | 0.584 | 0.12136 |
| MMP 15 | 2.177 | 1.047 | 0.08862 |
| MMP 16 | 2.357 | 1.180 | 0.22152 |
| MMP 19 | 1.850 | 0.859 | 0.07824 |
| MMP 23 | 2.939 | 0.770 | 0.25812 |
| MMP 2 short | 2.211 | 1.275 | 0.39144 |
| MMP 3 | 2.050 | 1.070 | 0.11977 |
| MMP 3B | 1.814 | 0.991 | 0.25395 |
| MMP 7 | 1.692 | 1.071 | 0.67467 |
| MMP 9 | 1.836 | 0.519 | 0.07064 |
| MT1-MMP | 1.743 | 0.668 | 0.05681 |
| MT1-MMP short | 2.347 | 1.020 | 0.19849 |
| PAI-1 | 2.035 | 0.974 | 0.06131 |
| PAI-2 | 2.099 | 0.979 | 0.01474 |
| PR | 1.817 | 1.243 | 0.41354 |
| SCC | 1.718 | 0.973 | 0.28871 |
| SPARC | 1.968 | 0.705 | 0.06443 |
| TGF-β | 2.231 | 1.290 | 0.56225 |
| TIMP 1 | 1.646 | 0.916 | 0.24659 |
| TIMP 2 | 2.427 | 1.047 | 0.20320 |
| TIMP 4 | 1.551 | 1.637 | 0.93343 |
| TNF-α | 1.836 | 1.191 | 0.47802 |
| tPA | 1.666 | 1.356 | 0.79660 |
| Transcobalamine | 2.317 | 0.776 | 0.14221 |
| uPA | 2.011 | 1.234 | 0.42537 |
| uPAR | 1.682 | 1.087 | 0.08025 |
| V3 | 1.571 | 0.968 | 0.34576 |
Most of the known cytokine genes represented on the BOTL-2 cDNA microarray tended to be activated following M. paratuberculosis stimulation of PBMCs from the subclinical and clinical infected animals (Table 5). However, variation in the absolute value of the ratio (M. paratuberculosis stimulation/nil stimulation) between animals within each group reduced the overall significance of these changes, despite the fact that the direction of change (positive or negative) was the same in both animals within a group.
Repression of immune cell gene expression in PBMCs from clinical M. paratuberculosis-infected animals is not due to a general loss of immune cell function.
Failure of M. paratuberculosis to activate many immune cell genes in PBMCs from the clinical cows, as well as the observed significant repression of numerous genes in these cells, could have resulted from an antigen-specific anergy or from a general failure of the immune cells from clinical cows to respond to any stimulant. To distinguish between these possibilities, we compared gene expression profiles of PBMCs from clinical M. paratuberculosis-infected cows (n = 2) with those from control uninfected cows (n = 3) to ConA, a general mitogen.
Consistent with our recently published results (31), ConA stimulation of PBMCs from the control uninfected cows tended to activate expression (>1.5-fold increase, ConA versus nil) of numerous immune cell genes. However, selection of these genes for only those where activation was greater than 1.5-fold and the P value was <0.1 reduced the number of significantly activated genes across all three control animals to 29 (data not shown). The list of significantly activated genes includes many that encode proteins commonly associated with immune cell function, including cytokines, MMPs, and genes or EST clones representing cell signaling proteins. No genes were significantly repressed in PBMCs from the control animals by ConA treatment.
ConA stimulation of PBMCs from the two clinical M. paratuberculosis-infected animals also tended to stimulate genes encoding important immune cell proteins, including cytokines, MMPs and their inhibitors, and other EST clones and known genes (Table 6). Differences were observed in the response of PBMCs from the control and clinical M. paratuberculosis-infected animals to ConA for several MMPs, glucocorticoid receptor (GCR), follicle stimulating hormone receptor (FSH-R), tissue plasminogen activator (tPA), and several EST clones (Table 6). In each case, it appeared that expression of these genes was not activated in PBMCs from the clinical M. paratuberculosis animals by ConA. ConA stimulation of PBMCs from the clinical M. paratuberculosis-infected cows significantly repressed expression (fold change, >1.5; P < 0.1) of only one EST (BOTL0100010_C06). This EST clone represents a bovine ortholog of the Notch 2 cell fate-determining factor and was unaffected by ConA treatment of PBMCs from the control uninfected animals (mean expression ratio = 1.089).
TABLE 6.
Mean expression ratios of various genes and clones in PBMCs of control and clinical M. paratuberculosis-infected animals following stimulation with ConA
| Gene or clone group and name | Description | Mean expression ratio
|
|
|---|---|---|---|
| Control animals | Clinical animals | ||
| Known cytokine genes | |||
| IL-12 | Amplicon representing bovine IL-12 (p35) | 2.455 | 1.703 |
| IL-2 | Amplicon representing bovine IL-2 | 2.533 | 2.343 |
| IL-4 | Amplicon representing bovine IL-4 | 2.133 | 1.883 |
| IL-5 | Amplicon representing bovine IL-5 | 2.256 | 1.702 |
| IFN-γ | Amplicon representing bovine IFN-γ | 1.704 | 1.771 |
| TNF-α | Amplicon representing bovine TNF-α | 2.472 | 2.283 |
| Known MMP and inhibitor genes | |||
| MMP 1 | Amplicon representing bovine MMP 1 | 2.560 | 1.356 |
| MMP 1 up | Amplicon representing bovine MMP 1 | 2.437 | 1.304 |
| MMP 13 | Amplicon representing bovine MMP 13 | 2.172 | 1.319 |
| MMP 16 | Amplicon representing bovine MMP 16 | 1.940 | 1.169 |
| MMP 19 | Amplicon representing bovine MMP 19 | 2.208 | 1.078 |
| MMP 3 | Amplicon representing bovine MMP 3 | 2.315 | 1.686 |
| MMP 9 | Amplicon representing bovine MMP 9 | 2.258 | 1.036 |
| MMP 9B | Amplicon representing bovine MMP 9 | 2.150 | 0.992 |
| TIMP 2 | Amplicon representing bovine TIMP 2 | 2.575 | 1.711 |
| TIMP 4 | Amplicon representing bovine TIMP 4 | 2.489 | 1.151 |
| Bovine EST clones and other known genes | |||
| BOTL0100001XF05R | Bovine EST similar to human sequence from clone RP5-852M4, chromosome 20; contains the XAP4 gene encoding the HBV-associated factor | 2.078 | 2.017 |
| BOTL0100002XB07R | Bovine EST similar to human glioma tumor suppressor candidate region gene 2 (GLTSCR2) | 2.437 | 2.473 |
| BOTL0100002XD02R | Bovine EST similar to bovine mRNA for cyclophilin | 1.677 | 1.270 |
| BOTL0100003XA08R | Bovine EST similar to Homo sapiens pituitary tumor-transforming 1 interacting protein mRNA | 1.946 | 1.785 |
| BOTL0100003XA09R | Bovine EST not similar to known genes | 2.183 | 2.434 |
| BOTL0100003XB05R | Bovine EST similar to Homo sapiens guanylate binding protein 1, interferon-inducible, 67 kD (GBP1). | 2.153 | 1.912 |
| BOTL0100006XB01R | Bovine EST similar to Homo sapiens geminin mRNA | 1.636 | 1.390 |
| BOTL0100006XC07R | Bovine EST similar to human G protein-coupled receptor (EBI 1) gene | 2.159 | 1.427 |
| BOTL0100008_A05 | Bovine EST similar to Macaca fascicularis brain cDNA, clone QccE-20770 | 1.941 | 1.142 |
| FSH-R | Amplicon representing bovine follicle stimulating hormone receptor | 2.213 | 1.335 |
| GCR | Amplicon representing bovine glucocorticoid receptor | 2.135 | 0.874 |
| SCC | Amplicon representing bovine ortholog of P450scc | 2.173 | 1.189 |
| tPA | Amplicon representing bovine tPA | 2.357 | 0.840 |
Despite individual animal variability and exceptions as noted above, the overall correlation of mean expression ratios (ConA/nil) for all genes on the BOTL-2 microarray between the clinical and control cows used in this study was high (r = 0.84). From these data, we concluded that the responses of PBMCs from clinical M. paratuberculosis-infected cows and uninfected cows to stimulation by a general mitogen (ConA) were similar and that there was little general immune suppression evident by analysis of gene expression profiles in the clinical M. paratuberculosis-infected cows.
DISCUSSION
The host immune response to mycobacteria tends to be quite dynamic in animals that progress to clinical disease states (10, 24). In M. paratuberculosis infection, the ruminant immune response begins with an appropriate cytotoxic and proinflammatory activity but gradually changes to an ineffective Th2-like response, characterized by immunoglobulin G1 antibody production (for a review, see reference 10). This is curious, since maintenance of a strong cytotoxic immune activity is essential for control of intracellular pathogens such as mycobacteria (14, 15, 21, 22). The underlying molecular mechanisms involved in shifting immune responses to mycobacterial pathogens are unknown but likely involve a complex interplay of antigen presentation, tolerance, and specific immune cell suppression (10). While there is no doubt that the commonly known cytokines, especially IFN-γ and IL-10, play a role in determining the outcome of the immune response to mycobacteria, there are likely many other, as yet unknown, factors involved.
In this report, we used gene expression profiling on glass slide cDNA microarrays containing 721 unique bovine genes to evaluate and compare the effect of M. paratuberculosis on PBMCs from naturally infected cows in various stages of Johne's disease. Our conclusions must be subject to the caveats that animal numbers used in these studies were low and that it is possible that differences in gene expression profiles observed between groups were due to changes in specific immune cell populations. Nevertheless, our results provide an intriguing insight into the response of immune cells from Johne's disease-afflicted cattle to M. paratuberculosis, suggesting targets for further studies by our group and by others.
A significant finding in the present study was the observation that expression levels of many genes (83 total) in PBMCs from clinical Johne's disease cows tended to be repressed following stimulation with M. paratuberculosis relative to nil stimulation. Conversely, only eight genes tended to be up regulated in the same cells, including those for IL-10 and IFN-γ. Thus, the major response of PBMCs from clinical Johne's disease cows used in this study was repression of gene expression. Of the 83 genes whose expression tended to be repressed in PBMCs from the clinical cows, only 16 were significantly repressed (P < 0.1) across both animals. Lack of significance for the remaining genes was due to differences in the absolute degree of repression, not to genes being affected in opposite directions in the two clinical cows. Of the 16 significantly repressed genes, 7 represent sequences encoding orthologs of proteins with unknown function or novel bovine ESTs, providing excellent candidates for further study. One of the more interesting known genes repressed in PBMCs from the clinical cows was the Lyn B protein tyrosine kinase. Lyn B is of critical importance in B-cell signaling through the B-cell antigen receptor (11, 26), forming one member of a cascade leading to cell proliferation, differentiation, or death (11). Lyn B deficiency has been associated with enhanced clonal expansion and reduced B-cell terminal differentiation (20). In addition, Lyn B is rapidly activated following Fc-gamma receptor-mediated phagocytosis and may be a critical factor in the cellular response to pathogen entry (26). Thus, repression of Lyn B in PBMCs from clinical M. paratuberculosis-infected cows could reduce both B-cell differentiation and macrophage signaling following uptake of opsonized bacteria.
Stimulation of PBMCs from clinical M. paratuberculosis-infected cows with the general mitogen ConA produced a pattern of gene expression very similar to that observed in PBMCs from uninfected cows. These results suggest that the M. paratuberculosis-induced gene repression in PBMCs from clinical Johne's disease cows employed in this study was not due to a general failure of the immune response. Notable exceptions to this included several MMPs, including MMP 9, as well asTIMP 4, GCR, FSH-R, tPA, and several EST clones representing unknown transcripts. The significance of these differences is not clear at present, but their role in general immune responses of M. paratuberculosis-infected cattle is worthy of further study. Likewise, significant ConA-induced repression of a bovine EST clone representing the Notch 2 gene ortholog is intriguing. Notch 2 is a receptor protein that has been associated with hematopoietic cell differentiation, proliferation, and death (3, 12, 13).
In sharp contrast to the situation with clinical Johne's disease cows, M. paratuberculosis stimulation of PBMCs from the two subclinical Johne's disease cows resulted in a predominate tendency to activate immune cell gene expression. In total, expression of 71 EST clones or known genes representing 68 unique transcripts showed a tendency to be up regulated in PBMCs from subclinical cows following exposure to M. paratuberculosis. When mean expression ratios of genes tending to be up regulated by M. paratuberculosis in PBMCs from subclinical cows were compared to expression ratios of the same genes in PBMCs from clinical cows, 13 genes showed significantly different (P < 0.1) expression profiles between the two groups of animals. This list of genes includes four bovine ESTs, IL-1, four different MMPs, plasminogen activator inhibitor 1 (PAI-1) and PAI-2, and uPAR. Many other potentially interesting differences are apparent in Table 5, but their significance is not discernible with the limited sample size employed in the present study. Nevertheless, our results lead us to conclude that gene expression profiles from PBMCs of clinical and subclinical Johne's disease cows are very different following exposure to M. paratuberculosis. Furthermore, our data have suggested that direct comparison of PBMC gene expression may help define the molecular basis for the antigen-specific anergy observed in clinical-stage M. paratuberculosis-infected cattle.
Acknowledgments
We acknowledge the outstanding support of Jianbo Yao, Associate Director of the MSU Center for Animal Functional Genomics, and Jeanne Burton for critical review of the manuscript and helpful suggestions during the course of this work. We also acknowledge the excellent assistance of Rob Tempelman, Department of Animal Science, Michigan State University, in statistical analysis of microarray data.
We also acknowledge the generous financial support of the College of Agriculture and Natural Science, the Michigan State University Agriculture Experiment Station, and the Office of the Vice President for Research and Graduate Studies at Michigan State University. Additional support for this project was provided by the Michigan Animal Industry Coalition and USDA IFAFS grant number 2001-52100-11211.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Adams, J. L., and C. J. Czuprynski. 1994. Mycobacterial cell wall components induce the production of TNF-alpha, IL-1, and IL-6 by bovine monocytes and the murine macrophage cell line RAW 264.7. Microb. Pathog. 16:401-411. [DOI] [PubMed] [Google Scholar]
- 2.Alzuherri, H. M., C. J. Woodall, and C. J. Clarke. 1996. Increased intestinal TNF-alpha, IL-1 beta and IL-6 expression in ovine paratuberculosis. Vet. Immunol. Immunopathol. 49:331-345. [DOI] [PubMed] [Google Scholar]
- 3.Aster, J. C., and W. S. Pear. 2001. Notch signaling in leukemia. Curr. Opin. Hematol. 8:237-244. [DOI] [PubMed] [Google Scholar]
- 4.Bakker, D., P. T. Willemsen, and F. G. van Zijderveld. 2000. Paratuberculosis recognized as a problem at last: a review. Vet. Q. 22:200-204. [DOI] [PubMed] [Google Scholar]
- 5.Bassey, E. O., and M. T. Collins. 1997. Study of T-lymphocyte subsets of healthy and Mycobacterium avium subsp. paratuberculosis-infected cattle. Infect. Immun 65:4869-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrells, C., C. J. Clarke, A. Colston, J. M. Kay, J. Porter, D. Little, and J. M. Sharp. 1999. Interferon-gamma and interleukin-2 release by lymphocytes derived from the blood, mesenteric lymph nodes and intestines of normal sheep and those affected with paratuberculosis (Johne's disease). Vet. Immunol. Immunopathol. 68:139-148. [DOI] [PubMed] [Google Scholar]
- 7.Burrells, C., C. J. Clarke, A. Colston, J. M. Kay, J. Porter, D. Little, and J. M. Sharp. 1998. A study of immunological responses of sheep clinically affected with paratuberculosis (Johne's disease). The relationship of blood, mesenteric lymph node and intestinal lymphocyte responses to gross and microscopic pathology. Vet. Immunol. Immunopathol. 66:343-358. [DOI] [PubMed] [Google Scholar]
- 8.Burton, J. L., S. A. Madsen, J. Yao, S. Sipkovsky, and P. M. Coussens. 2001. An immunogenomics approach to understanding periparturient immunosuppression and mastitis susceptibility in dairy cows. Acta Vet. Scand. 42:407-425. [PubMed] [Google Scholar]
- 9.Chiodini, R. J., and C. A. Rossiter. 1996. Paratuberculosis: a potential zoonosis? Vet. Clin. North Am. Food Anim. Pract. 12:457-467. [DOI] [PubMed] [Google Scholar]
- 10.Coussens, P. M. Mycobacterium paratuberculosis and the bovine immune system. Anim. Health Res. Rev. 2:141-162. [PubMed]
- 11.Hsueh, R., and R. Scheuermann. 2000. Tyrosine kinase activation in the decision between growth, differentiation, and death responses initiated from the B cell antigen. Adv. Immunol. 75:285-316. [DOI] [PubMed] [Google Scholar]
- 12.Hubmann, R., J. D. Schwarzmeier, M. Shehata, M. Hilgrath, M. Duechler, M. Dettke, and R. Berger. 2002. Notch 2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood 99:3742-3747. [DOI] [PubMed] [Google Scholar]
- 13.Jonsson, J. I., Z. Xiang, M. Pettersson, M. Lardelli, and G. Nilsson. 2001. Distinct and regulated expression of Notch receptors in hematopoietic lineages and during myeloid differentiation. Eur. J. Immunol. 31:3240-3247. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann, S. H. 1999. Cell-mediated immunity: dealing a direct blow to pathogens. Curr. Biol. 9:R97-R99. [DOI] [PubMed] [Google Scholar]
- 15.Munk, M. E., and M. Emoto. 1995. Functions of T-cell subsets and cytokines in mycobacterial infections. Eur. Respir. J. Suppl. 20:668S-675S. [PubMed] [Google Scholar]
- 16.National Animal Health Monitoring System. 1997. Johne's disease on U.S. dairy operations, no. N245.1097. Veterinary Services, Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Fort Collins, Colo.
- 17.Navarro, J. A., G. Ramis, J. Seva, F. J. Pallares, and J. Sanchez. 1998. Changes in lymphocyte subsets in the intestine and mesenteric lymph nodes in caprine paratuberculosis. J. Comp. Pathol. 118:109-121. [DOI] [PubMed] [Google Scholar]
- 18.Perez, V., J. Tellechea, J. M. Corpa, M. Gutierrez, and J. F. Garcia Marin. 1999. Relation between pathologic findings and cellular immune responses in sheep with naturally acquired paratuberculosis. Am. J. Vet. Res. 60:123-127. [PubMed] [Google Scholar]
- 19.Seitz, S. E., L. E. Heider, W. D. Heuston, S. Bech-Nielsen, D. M. Rings, and L. Spangler. 1989. Bovine fetal infection with Mycobacterium paratuberculosis. J. Am. Vet. Med. Assoc. 194:1423-1426. [PubMed] [Google Scholar]
- 20.Shih, T., M. Roederer, and M. Nussenzweig. 2002. Role of receptor affinity in T cell-independent antibody responses in vivo. Nat. Immunol. 3:399-406. [DOI] [PubMed] [Google Scholar]
- 21.Silva, C. L., M. F. Silva, R. C. Pietro, and D. B. Lowrie. 1996. Characterization of T cells that confer a high degree of protective immunity against tuberculosis in mice after vaccination with tumor cells expressing mycobacterial Hsp65. Infect. Immun. 64:2400-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sousa, A. O., R. J. Mazzaccaro, R. G. Russell, F. K. Lee, O. C. Turner, S. Hong, L. Van Kaer, and B. R. Bloom. 2000. Relative contributions of distinct MHC class I-dependent cell populations in protection to tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 97:4204-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stabel, J. R. 2000. Cytokine secretion by peripheral blood mononuclear cells from cows infected with Mycobacterium paratuberculosis. Am. J. Vet. Res. 61:754-760. [DOI] [PubMed] [Google Scholar]
- 24.Stabel, J. R. 2000. Transitions in immune responses to Mycobacterium paratuberculosis. Vet. Microbiol. 77:465-473. [DOI] [PubMed] [Google Scholar]
- 25.Streeter, R. N., G. F. Hoffsis, S. Bech-Nielsen, W. P. Shulaw, and D. M. Rings. 1995. Isolation of Mycobacterium paratuberculosis from colostrum and milk of subclinically infected cows. Am. J. Vet. Res. 56:1322-1324. [PubMed] [Google Scholar]
- 26.Strzelecka-Kiliszek, A., and A. Sobota. 2002. Sequential translocation of tyrosine kinases lyn and syk to the activated Fc-gamma receptors during phagocytosis. Folia Histochem. Cytobiol. 40:131-132. [PubMed] [Google Scholar]
- 27.Sweeney, R. W. 1996. Transmission of paratuberculosis. Vet. Clin. North Am. Food Anim. Pract. 12:305-312. [DOI] [PubMed] [Google Scholar]
- 28.Tooker, B. R., J. L. Burton, and P. M. Coussens. 2002. Survival tactics of M. paratuberculosis in bovine macrophage cells. Vet. Immunol. Immunopathol. 87:443-451. [DOI] [PubMed] [Google Scholar]
- 29.Waters, W. R., J. R. Stabel, R. E. Sacco, J. A. Harp, B. A. Pesch, and M. J. Wannemuehler. 1999. Antigen-specific B-cell unresponsiveness induced by chronic Mycobacterium avium subsp. paratuberculosis infection of cattle. Infect. Immun. 67:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells, S. J., and B. A. Wagner. 2000. Herd-level risk factors for infection with Mycobacterium paratuberculosis in US dairies and association between familiarity of the herd manager with the disease or prior diagnosis of the disease in that herd and use of preventive measures. J. Am. Vet. Med. Assoc. 216:1450-1457. [DOI] [PubMed] [Google Scholar]
- 31.Yao, J., J. L. Burton, P. Saama, S. Sipkovsky, and P. M. Coussens. 2001. Generation of EST and cDNA microarray resources for the study of bovine immunolobiology. Acta Vet. Scand. 42:391-406. [PubMed] [Google Scholar]