Abstract
A Salmonella enterica serovar Typhi gene that is selectively up-regulated upon bacterial invasion of eukaryotic cells was characterized. The open reading frame encodes a 298-amino-acid hydrophobic polypeptide (30.8 kDa), which is predicted to be an integral membrane protein with nine membrane-spanning domains. The protein is closely related (87 to 94% reliability) to different transport and permease systems. Gene expression under laboratory conditions was relatively weak; however, sevenfold induction was observed in a high-osmolarity medium (300 mM NaCl). The growth pattern in a laboratory medium of a serovar Typhi strain Ty2 derivative containing a 735-bp in-frame deletion in this gene, named gaiA (for gene activated intracellularly), was not affected. In contrast, the mutant was partially impaired in intracellular survival in murine peritoneal macrophages, as well as in human monocyte-derived macrophages. However, in the case of human macrophages, this survival defect was modest and evident only at late infection times (24 h). Despite the distinct intracellular survival kinetics displayed in macrophages of different species, the gaiA null mutant was significantly affected in its potential to trigger apoptosis in both murine and human macrophages. Provision of the gaiA gene in trans resulted in complementation of these phenotypes. Interestingly, the absence of a functional gaiA gene caused a marked attenuation in the mouse mucin model, as shown by the increase (3 orders of magnitude) in the 50% lethal dose of the mutant strain over that of the parental strain Ty2 (P ≤ 0.05). Altogether, these data indicate that the product encoded by the gaiA gene is required for triggering apoptosis and bacterial survival within murine macrophages, which is consistent with the in vivo results obtained in the mouse mucin model. However, gaiA was not required for initial intracellular survival in human cells, indicating that its role in the natural host might be more complex than is suggested by the studies performed in the murine system.
Infections caused by Salmonella enterica serovars constitute a major public health problem worldwide (1, 20). These pathogens can affect both humans and animals, causing food-borne diseases ranging from mild gastroenteritis to life-threatening systemic infections, such as those caused by S. enterica serotype Typhi in humans (19, 20, 41). The clinical management of patients infected with S. enterica serovars is rendered difficult due to the emergence of multidrug-resistant strains (47).
S. enterica serovar Typhi has a particularly complex infection cycle, in which the microorganism transits through different niches (41, 55). It has been demonstrated that during infection, the expression of bacterial products is tightly regulated according to environmental signals (10). This allows the pathogen to optimize the expression of the virulence factors required in each phase, avoiding the additional energetic cost associated with the production of unnecessary products. Better understanding of the molecular basis of Salmonella infections has led to identification of bacterial products which are essential for pathogenicity, such as virulence factors, regulatory proteins, and secretion systems (15, 19). The corresponding genes constitute potential targets for the development of attenuated strains, which can be used either as live vaccines against salmonellosis or as carriers for heterologous antigens (7, 32).
Serovar Typhi promoters that are activated mainly upon bacterial entry into eukaryotic cells have been identified previously (58). It is likely that the genes controlled by these promoters are involved in the infection process to some extent. Precedents exist in S. enterica serovar Typhimurium demonstrating that genes induced inside cultured macrophages or epithelial cells are essential for virulence. This is the case for the spv operon and the mig-14 gene (13, 61). In this work we describe the characterization of a novel serovar Typhi gene driven by one of those promoters. The results obtained suggest that this gene is involved in the interactions between bacteria and phagocytic cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are described in Table 1. Bacteria were grown in Luria-Bertani (LB) broth (48) or LB agar plates. Plasmids were maintained in the Escherichia coli strain XL1-Blue, and the INVαF′ strain was used as a recipient for cloning fragments amplified by PCR and cloned into the pCR2.1 vector. Media were supplemented with chloramphenicol (50 μg ml−1), ampicillin (200 μg ml−1), nalidixic acid (20 μg ml−1), or streptomycin (50 μg ml−1) when required.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant genotype or characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-Blue | Tcr; endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1 lac [F′ proAB lac1qZΔM15 Tn10] | Stratagene |
| INVαF′ | F′ endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1 φ80 lacZΔM15 Δ(lacZYA-argF)U169 deoR λ− | Invitrogen |
| SM10 (λpir) | Kmr; thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu-Km::Tn7 λpir | 34 |
| Serovar Typhi | ||
| Ty2 | Wild type | ATCC 19430 |
| Ty2 Na1r | Na1r; spontaneous derivative of Ty2 | This work |
| ΔgaiA | Na1r; Ty2 Na1r derivative containing an in-frame deletion of the gaiA gene | This work |
| Serovar Typhimurium | ||
| MT189 | recA1 galE496/F′ 100-12 λpir (gal+ bio+) | 24 |
| LT2 | Wild type | ATCC 15277 |
| Plasmids | ||
| PCR2.1 | Apr Kmr; high-copy-number vector for cloning PCR products | Invitrogen |
| pKNG101 | Smr; broad-host-range π-dependent suicide vector | 30 |
| pHOB35 | Smr; pKNG101 derivative containing a PCR fragment encompassing the gaiA gene with a 735-bp internal deletion | This work |
| pVDL8 | Cmr; derivative of pHSG575 with additional NotI sites flanking the polylinker | 60 |
| pHOB38 | Cmr; pVDL8 derivative containing a PCR fragment encompassing the gaiA ORF, generated with primers 5102XBA and SEQAPOR2 | This work |
| pUJ9TT | Apr; multicopy promoter probe vector to generate fusions with the lacZ gene | 29 |
| pHOB700 | Apr; pUJ9TT derivative containing a 431-bp EcoRI/BamHI PCR fragment generated with primers 5102ECO and 3LACZBAM | This work |
| pLS102 | Apr; pCB182 derivative containing an in vivo activated promoter from the Salmonella strain Ty2 | 58 |
DNA manipulations.
Plasmid DNA isolation, restriction endonuclease digestion, ligation, transformation, agarose gel electrophoresis, and other standard DNA techniques were carried out as described by Sambrook et al. (48). Oligonucleotides (Table 2) were synthesized by GIBCO. Colony PCR, extraction of PCR products, and cloning experiments were performed according to standard protocols (48). Inverse PCR was carried out as described by Ochman et al. (40). DNA sequencing was performed by using a Taq dye-deoxy terminator cycle sequencing kit and a model 373A automatic DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.), according to the manufacturer's instructions. Restriction and modification enzymes were purchased from New England Biolabs (Frankfurt, Germany). Electroporation was carried out with a gene pulser (Bio-Rad Laboratories, Richmond, Calif.) as described by O'Callaghan and Charbit (39). Searches in databases for nucleotide and amino acid sequence homologies were performed by using the BLASTP (2), BLASTP plus BEAUTY (2, 66), FASTA3 (42), and PSORT (36) algorithms.
TABLE 2.
Oligonucleotides used for PCR
| Name | Sequence (5′→3′) |
|---|---|
| 5102XBA | GGTCTAGAGATCACATTTTGTAATGTTACAC |
| 5102ECO | GGGAATTCGATCACATTTTGTAATGTTACAC |
| 5102B | GGAAGCTTTTTACGCTGCTATTGCTG |
| 5Δ1823 | GCGATGTCGCTTTCCGGCGGTCTGATG |
| 3Δ172 | GCCGGAAAGCGACATCGCCCAGAA |
| SEQAPOR2 | CGCGGAATTTACGACATCATCA |
| 3LACZBAM | CCGGATCCAAGACAGGCGCGGCC |
| REVSD | GGCTCGAGTCAGCAAAACGGCGAACAG |
Construction of a nonpolar mutation.
Overlap extension PCR (27) was used to generate an in-frame deletion in the gaiA gene. Two PCR fragments were obtained by using the primer pairs 5102XBA and 3Δ172 (447 bp) and 5Δ1823 and SEQAPOR2 (510 bp). The resulting products contain the first 87 bp and the last 72 bp of the gaiA open reading frame (ORF), respectively. An 18-bp overlap in their sequences allowed the amplification of a 934-bp fragment during a second PCR using the primer pair 5102XBA and SEQAPOR2. The resulting product, which encompasses a gaiA gene containing an internal 735-bp deletion, was digested with XbaI and cloned into XbaI-digested pKNG101 (30), generating pHOB35. This plasmid was transformed into strain SM10 (λpir) and then transferred by conjugation (25) into the recipient S. enterica serovar Typhi strain Ty2 (Nalr). Cointegration and excision of the suicide vector were performed as previously described (30). The in-frame deletion contained in the serovar Typhi gaiA mutant resulting from the allelic exchange was confirmed by PCR analysis using primers homologous to regions encompassed in the deleted fragments or to adjacent external sequences (data not shown). Primers 5102XBA and SEQAPOR2 were used to amplify the full-length gaiA gene and 360 bp of the region located upstream of the start codon, which was subsequently cloned into the low-copy-number vector pVDL8 (60), generating pHOB38, which was used for complementation studies.
Construction of a gaiA′-′lacZ fusion.
For construction of a gaiA′-′lacZ fusion, a 431-bp EcoRI/BamHI-fragment amplified by PCR using primers 5102ECO and 3LACZBAM, which contains 375 bp of the upstream sequence of gaiA and 56 bp of the gaiA gene, was cloned into pUJ9TT (29), generating pHOB700. β-Galactosidase activity was quantified by the method of Miller (33).
Tissue culture methods, invasion tests, and in vivo studies.
S. enterica serovar Typhi strains were tested for the ability to survive in Henle 407 cells (ATCC CCL-6) and the macrophage-like cell line J774A.1 (ATCC TIB 67). Primary macrophages were either obtained from the peritoneal cavities of BALB/c mice or derived from human peripheral blood mononuclear cells obtained from healthy volunteers as previously described (43). Henle 407 cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 25 mM HEPES, 10% (vol/vol) fetal calf serum (FCS), and 5 mM glutamine (GIBCO). Macrophages were maintained in DMEM (Sigma Chemie GmbH, Deisenhofen, Germany) supplemented with 4.5 g of glucose/liter, 10% FCS, 5 mM glutamine, and 1.5 g of NaHCO3/liter in an atmosphere containing 5% CO2 at 37°C. Cells seeded at a concentration of approximately 5 × 104 per well in 24-well tissue culture plates (Inter Med NUNC, Roskilde, Denmark) were infected with bacteria grown overnight in static LB broth cultures supplemented with 17.53 g of NaCl/liter during 90 min (Henle cells), 60 min (human macrophages), or 30 min (murine macrophages and J774A.1 cells), as previously described (22, 29). The number of apoptotic cells was determined by using an in situ cell death detection kit with fluorescein (Boehringer Mannheim GmbH) according to the manufacturer's instructions. The mouse mucin model described by Powell et al. (44) was used for determination of the 50% lethal dose (LD50) (45).
Statistical analysis.
The statistical significance of the results obtained was evaluated by the chi-square test.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in the EMBL database under accession number AJ006101.
RESULTS AND DISCUSSION
Identification of an S. enterica serovar Typhi gene that is activated upon infection of eukaryotic cells.
A previous study allowed the identification of serovar Typhi promoters that were activated upon invasion of eukaryotic cells (58). By use of the gene encoding chloramphenicol acetyltransferase (cat) as a reporter, it was estimated that the activity of the promoter contained in vector pLS102 was increased 93-fold when bacteria entered eukaryotic cells (58). This vector carries a 548-bp fragment of serovar Typhi DNA containing a promoter motif reminiscent of σ54-dependent promoters, a Shine-Dalgarno sequence, and the start of an ORF at position 372 (58).
Total DNA from the serovar Typhi strain Ty2 was digested with different restriction endonucleases recognizing sequences present in the insert from pLS102 and was subsequently analyzed by Southern blotting using the digoxigenin-labeled insert from pLS102 as a probe. A 1,600-bp fragment that reacted specifically with the probe was identified by using ApoI. To identify the sequences located downstream of the truncated ORF present in pLS102, this 1,600-bp fragment was amplified by inverse PCR using primers REVSD and 5102B, with ApoI-digested DNA as a template. Sequence analysis of the resulting fragment allowed us to design primer SEQAPOR2, corresponding to the 3′ end, which was used together with primer 5102XBA to PCR amplify the whole ORF by using chromosomal DNA from the serovar Typhi strain Ty2 as a template.
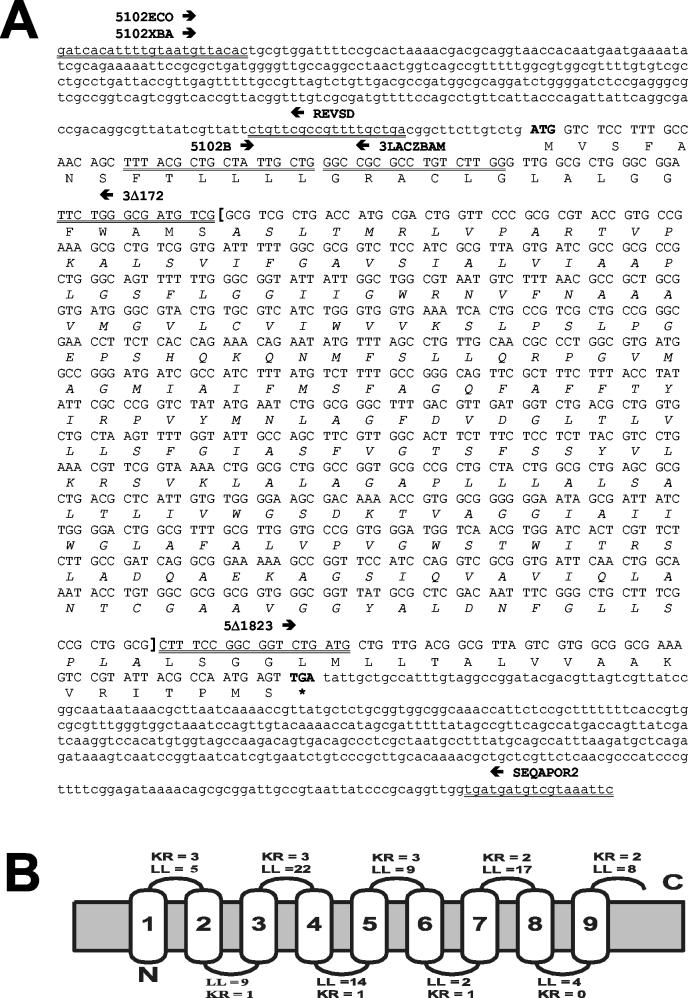
The gene present in the cloned fragment, which was named gaiA (for gene activated intracellularly), encodes a 298-amino-acid hydrophobic polypeptide with a predicted molecular mass of 30.8 kDa and a pI of 10.02. A potential leader peptide with a cleavage site at positions 18 and 19 (ACL-GL) was detected by using the SignalP algorithm (37). Use of the TopPred II algorithm (9) suggested with 95% reliability that the GaiA protein is an integral membrane protein with nine membrane-spanning domains (Fig. 1). Interestingly, nine predicted N-myristoylation sites were also detected; one of them was located at the predicted leader peptide cleavage site (GLALGG; positions 19 to 24). In gram-negative bacteria the myristoylation of lipid A is essential for its proinflammatory properties, which are essential for a virulent phenotype (23, 56, 57). However, the covalent attachment of a myristoyl group to an NH2-terminal glycine residue seems to occur exclusively in eukaryotic cells (3, 53). Nevertheless, recent studies have proved that proteins exported from prokaryotic cells can also be N myristoylated by the machinery of target cells (38, 54).
FIG. 1.
Identification of the product encoded by the gaiA gene from serovar Typhi. (A) Nucleotide sequence and predicted translation product of the gaiA locus. Amino acids are given in one-letter code, start and stop codons are boldfaced, sequences deleted in the ΔgaiA mutant are marked off by square brackets (with amino acids in italics), primer sequences are double underlined, and arrows indicate direction. (B) Predicted topology of the product encoded by the gaiA gene. The predicted leader sequence peptide has been removed for the prediction. The topology corresponding to the cytoplasmic location of the COOH terminus is shown. Abbreviations: LL, loop length; KR, lysine-plus-arginine profile.
Searches for homologies in databases showed that the product encoded by the gaiA gene exhibits 88% identity (293-amino-acid overlap) with the putative transport protein coded by the f451 ORF from E. coli (YICM_ECOLI; P31438) and 87% identity with the corresponding allelic variant of the E. coli strain EDL933 (AAG588858). The E. coli yicM gene is located in the intergenic fragment between the uhpT and nlpA genes (5). uhpT is involved in the uptake of phosphorylated sugars (14, 63, 64), whereas nlpA is a lipoprotein attached to the cytoplasmic membrane (68). The location of the gaiA gene was examined in the genomes of S. enterica serovars Typhi and Typhimurium by using the “Enteric” server (http://galapagos.cse.psu.edu/enterix/enteric/enteric.html). This analysis revealed that, unlike its E. coli homologue, gaiA/yicM is flanked by several ORFs of unknown function that are absent in the E. coli genome (10 and 9 ORFs in the upstream and downstream regions, respectively). Moreover, 12 ORFs upstream of gaiA/yicM is the mgtC gene, which has been shown to be the first gene of a 17-kb region encompassing Salmonella pathogenicity island 3 and required for intramacrophage survival (4). This evidence suggests that gaiA/yicM has been conserved between the two genera but that in the case of Salmonella, extensive chromosomal rearrangement of the genome has occurred in its flanking regions, including insertion of a pathogenicity island.
The product encoded by the gaiA gene was also analyzed by using the Propsearch algorithm (28), which detects functional or structural homologues belonging to putative protein families by using 144 properties (e.g., amino acid composition, molecular weight, content of bulky or small residues, average hydrophobicity, and charge). The Euclidian distance between the product encoded by gaiA and other database sequences suggested that this protein is related with a 87 to 94% reliability to different transport systems (e.g., sugars, cytosine, ABC transporters, arsenic pumps, branched amino acids, Na+/H+ antiporter). This suggests that the product encoded by this gene may be involved in the transport of substrates required for bacterial metabolism.
Transcriptional regulation of the gaiA gene.
Previous results demonstrated that the promoter driving the expression of the gaiA gene is activated intracellularly (58). To gain further knowledge about the potential environmental signals mediating gene activation, a translational fusion was generated between gaiA and the gene coding for β-galactosidase (lacZ), which was used as a reporter. A DNA fragment spanning nucleotides −375 to +56 (with respect to the gaiA ATG start codon) was fused to the lacZ gene present in pUJ9TT (see Materials and Methods), generating plasmid pHOB700. This fragment includes the promoter and upstream regions, containing potential binding sites for regulatory factors, and maintains intact the translational initiation region in order to avoid potential artifacts deriving from affected translational initiation (50). The pHOB700 plasmid was highly unstable in wild-type Salmonella strains, even when passed through an intermediate rec-negative mod+ serovar Typhimurium strain, suggesting that this may be partly due to recombination events. Thus, transcriptional activation studies were performed by using Salmonella strain MT189 (recA1), in which the construct was stable, as a recipient.
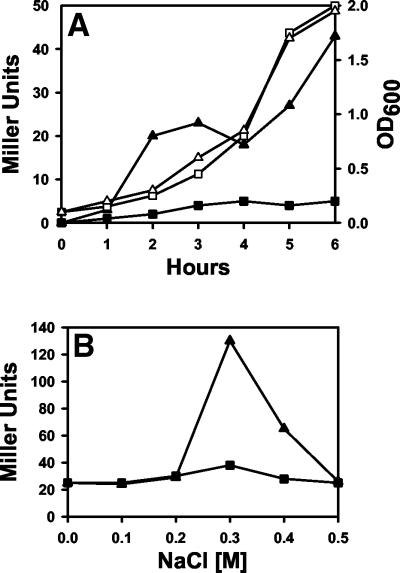
In the initial studies the influence of the growth phase on the expression of the reporter gene was analyzed (Fig. 2A), since this factor seems to play a key role in Salmonella invasion (11, 12, 31, 59). A minor increase in the expression of the reporter was observed at the early-exponential phase, followed by a second increment at the late-exponential and early-stationary phases (Fig. 2A). In contrast, when the effect of changes in osmolarity was analyzed, a clear increment of about sevenfold in the expression of lacZ was observed at 0.3 M NaCl (Fig. 2B). Interestingly, bacterial growth at lower and higher osmolarities resulted in reduced expression. This is in agreement with what was previously observed for the adhesive and invasive phenotypes (optimal at 0.3 M NaCl) and the expression of different virulence factors and type III secretion systems (15, 16, 59). Since the availability of Mg2+ has been also reported to affect Salmonella virulence, via the response regulator PhoP/PhoQ (17), we investigated whether bacterial growth in the presence of different concentrations of Mg2+ affects the expression of the gaiA-lacZ fusion. However, no significant differences were observed (data not shown).
FIG. 2.
Activation of the gaiA promoter in response to different growth conditions. Serovar Typhimurium MT189 bacteria carrying either plasmid pUJ9TT (▪) or plasmid pHOB700 (▴) were grown either in LB medium (A) or in MM63 minimal medium supplemented with different concentrations of NaCl (B), and β-galactosidase production was monitored. Growth rates are indicated by open symbols (optical density at 600 nm [OD600]). Results are expressed as Miller units; standard deviations were lower than 5%.
GaiA is involved in the interactions between serovar Typhi and macrophages.
To assess whether the product encoded by gaiA is involved in the pathogenesis process, a serovar Typhi Ty2 mutant containing a 735-bp in-frame deletion in this gene was generated as described in Materials and Methods. The serovar Typhi ΔgaiA strain did not differ from the parental strain in terms of growth pattern in LB medium, morphology (by light microscopy), or expression of the Vi antigen (data not shown).
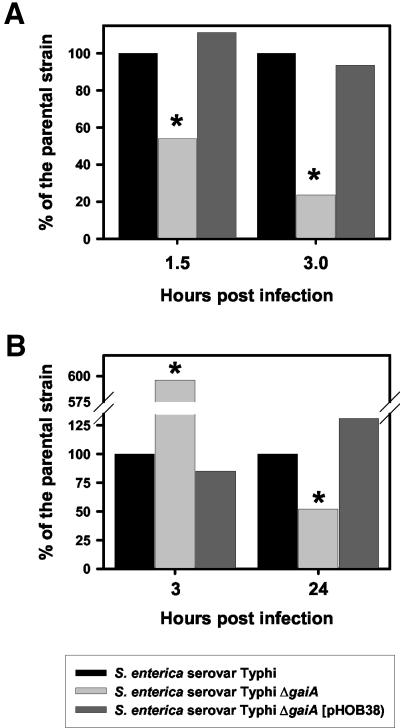
The capacity of the null mutant to infect and survive within epithelial cells was then evaluated. No statistically significant differences were observed in the number of viable intracellular bacteria recovered following 4, 6, 8, and 24 h of infection of Henle 407 epithelial cells (data not shown). Then we analyzed whether the ΔgaiA mutant was able to survive within professional phagocytes. Upon infection of mouse peritoneal macrophages, the serovar Typhi ΔgaiA strain displayed a 46 to 77% reduction (P ≤ 0.05) in the number of viable intracellular bacteria from that for the wild type at 1.5 and 3 h postinfection, respectively (Fig. 3A). To confirm the role played by the product encoded by the ΔgaiA gene in the phenotype observed, a plasmid containing a PCR fragment encompassing the full-length gaiA gene and 360 bp of the regions located upstream of the ATG start codon (pHOB38) was introduced into the serovar Typhi ΔgaiA mutant. The provision of gaiA in trans resulted in full complementation of the mutant phenotype, restoring intracellular survival levels to those of the wild-type parental strain (Fig. 3A). It is unlikely that the differences observed were due to impaired infectivity, since the number of viable bacteria recovered per well after 30 min of infection was in the range of 1 × 105 to 2 × 105 for all strains tested.
FIG. 3.
Interaction of the serovar Typhi ΔgaiA mutant with macrophages. The capacities of the serovar Typhi strain Ty2, its ΔgaiA derivative, and the ΔgaiA mutant complemented with pHOB38 to survive within mouse peritoneal macrophages (A) or monocyte-derived human macrophages (B) were evaluated. The CFU recovered per well was compared with the number of viable bacteria harvested from cells infected with the wild-type strain Ty2. Results are expressed as CFU relative to the values obtained for the parental strain. Asterisks indicate that the differences from the control values were considered significant (P ≤ 0.05).
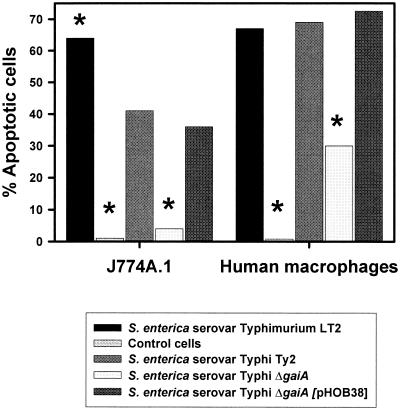
It has been demonstrated that S. enterica serovar Typhimurium is able to trigger apoptosis of infected macrophages (6, 8, 26, 35, 49, 69). This process seems to play a key role also in elicitation of immune responses to Salmonella (67, 69). Since the intracellular survival of the mutant strain was reduced only in macrophages, not within epithelial cells, we decided to assess whether the capacity of the serovar Typhi ΔgaiA mutant to stimulate programmed cell death was also affected. The murine macrophage-like cell line J774A.1 was infected with serovar Typhi Ty2 or its ΔgaiA derivative, and the number of apoptotic cells was determined after 8 h by a terminal deoxynucleotidyltransferase-mediated dUTP-fluorescein nick end labeling (TUNEL) assay (Fig. 4). Approximately 41% of cells infected with serotype Typhi Ty2 underwent apoptosis after 8 h of infection. In contrast, only 4% of the cells infected with the ΔgaiA mutant exhibited signs of apoptosis. The capacity to promote apoptosis was restored in the ΔgaiA mutant complemented with pHOB38.
FIG. 4.
Salmonella-mediated apoptosis of infected cells. The capacities of the serovar Typhi strain Ty2, its ΔgaiA derivative, and the ΔgaiA mutant complemented with pHOB38 to trigger apoptosis in J774A.1 cells and monocyte-derived human macrophages were evaluated and compared with that of the positive-control serovar Typhimurium strain LT2. Results are expressed as percent apoptotic cells. Asterisks indicate that the differences from results for the parental strain S. enterica serovar Typhi Ty2 were considered significant (P ≤ 0.05).
Infected macrophages play a key role in Salmonella infections (21, 41). However, intracellular survival is host dependent. While serovar Typhimurium can survive within both mouse and human macrophages, serovar Typhi survives at reasonable rates only within human macrophages (51, 62). Serovar Typhi also exhibits a particular survival kinetics in human macrophages, which is characterized by an initial phase of bacterial death followed by a second phase of persistent survival. Furthermore, serovar Typhimurium usually causes more apoptosis than serovar Typhi (51). Therefore, it has been proposed that serovar Typhi can survive chronically within human macrophages by causing little damage, thereby favoring systemic dissemination.
Humans are the natural hosts for serovar Typhi. Therefore, to further characterize the role played by the product encoded by the gaiA gene, monocyte-derived human macrophages were infected. Surprisingly, significantly higher numbers of viable bacteria were recovered from cells infected with the Ty2 ΔgaiA mutant than from those infected with the parental strain (P ≤ 0.05) after 3 h of infection (Fig. 3B). This consistent increment in survival was abolished by providing the gaiA gene in trans (Fig. 3B). However, the initial survival pattern was reverted 24 h after infection, with the ΔgaiA mutant exhibiting a slightly lower viability than the wild-type strain. Provision of the gaiA gene in trans resulted in full complementation of the mutant phenotype (Fig. 3B). To better characterize the dynamics of the intracellular survival process for each strain tested, survival indexes (the number of viable bacteria after 24 h divided by the number of viable bacteria after 3 h) were calculated. The survival index of the wild-type strain was 4.2, showing that there was a significant increment in the number of viable bacteria after 24 h. In contrast, the survival index of the ΔgaiA mutant was 0.48, demonstrating that there was a marked reduction in the number of viable bacteria during the course of infection. When the gaiA gene was provided in trans, the phenotype of the deletion mutant reverted, and the survival index was even higher than that of the wild-type strain (6.6).
Thus, it seems that during short-term infection of human macrophages the gaiA gene is not required for intracellular survival, and indeed, its loss causes an apparent abolishment of the initial phase of bacterial death. To evaluate whether this phenotype could be related to differences in the capacity to trigger macrophage apoptosis, we monitored the percentages of apoptotic cells in human macrophages infected with the wild-type and mutant strains. The results obtained (Fig. 4) demonstrated that the ability of the ΔgaiA mutant to trigger apoptosis was significantly reduced from that of the wild-type strain (P ≤ 0.05). When the gaiA gene was provided in trans, full complementation of the mutant phenotype was observed (Fig. 4). Thus, it seems that the difference in survival kinetics displayed by the ΔgaiA mutant in murine and human macrophages is not related to an impaired capacity to trigger apoptosis. Interestingly, it has been demonstrated that spvA, another Salmonella gene which is induced in intracellular bacteria, follows a differential kinetics of induction in both macrophages and epithelial cells, displaying maximal induction (≥100-fold) 6 to 8 h postinfection (65). Considering our results, it is tempting to postulate a potential role for gaiA in bacterial survival within human macrophages during late infection.
S. enterica serovar Typhi is avirulent for mice. This seems to reflect its inability to grow in murine macrophages. However, the use of iron-enriched mucin results in iron overloading, which increases bacterial growth within phagocytic cells (52). In an attempt to further characterize the role of gaiA, the in vivo virulence of the serovar Typhi ΔgaiA mutant was evaluated by using the mouse mucin model. Under these conditions, the LD50 calculated for the ΔgaiA mutant (2.1 × 107 CFU) was approximately 3 orders of magnitude higher than that for the parental strain Ty2 (4.78 × 104 CFU). This result suggests that the presence of a functional gaiA gene is required in vivo for expression of a fully virulent phenotype in the murine mucin model, which agrees with the observed defects of the ΔgaiA mutant in triggering apoptosis and in surviving within murine macrophages. However, we should be cautious when evaluating these data, since mice are not the natural hosts for serovar Typhi.
Salmonella genes which are selectively activated upon infection of eukaryotic cells are likely to be either housekeeping genes or genes encoding novel virulence factors. Preliminary studies suggest that GaiA may be involved in the transport or utilization of carbon sources (data not shown). The cytoplasm of infected cells might be a nonpermissive environment for intracellular pathogens, unless they are able to express special metabolic genes. Although serovar Typhi is a pathogen that resides in membrane-bound compartments of the host cell, its proliferation within these compartments must rely on the acquisition of nutrients that are probably present in the cytoplasm. The potential involvement of GaiA in the transport or utilization of carbohydrates, as well as in bacterial interactions with phagocytes, suggests that it might play an essential role in bacterial nutrition or as a factor coupled to the expression of genes involved in the pathogenesis process. A connection between a nutrition-related function and the induction of virulence genes has been shown for Listeria monocytogenes (18, 46), a pathogen that proliferates in the host cell cytoplasm.
However, with regard to the real role played by the gaiA product, our studies do not reveal what the exact biological function of the GaiA protein during the infection process could be. In fact, direct proof of the involvement of GaiA in pathogenesis in its natural host is still lacking. Serovar Typhi is extremely sensitive to murine macrophages, and the mutant analyzed in this study showed only a slight survival defect at late infection times in monocyte-derived human macrophages. Additional work focused on analysis of the interaction of the serovar Typhi gaiA mutant with other human immune-system cells, such as dendritic cells, or, alternatively, analysis of the capacity of the corresponding serovar Typhimurium mutant to trigger disease in mice might provide further insights into the overall function of this gene during natural infections.
Acknowledgments
We are particularly grateful to K. N. Timmis for his generous support and encouragement and to Bianka Karge, Astrid Müller, Karina Watzke, and Urte Winckler for outstanding technical help.
Part of this work was supported by a grant from the European Community (EU project “Multicomponent Salmonella live vaccines: optimising molecular, cellular and immunological parameters to enhance vaccine safety and immunogenicity,” QLK2-CT-1999-00310).
Editor: D. L. Burns
REFERENCES
- 1.Altekruse, S. F., M. L. Cohen, and D. L. Swerdlow. 1997. Emerging foodborne diseases. Emerg. Infect. Dis. 3:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bhatnagar, R. S., K. Futterer, G. Waksman, and J. I. Gordon. 1999. The structure of myristoyl-CoA:protein N-myristoyltransferase. Biochim. Biophys. Acta 1441:162-172. [DOI] [PubMed] [Google Scholar]
- 4.Blanc-Potard, A. B., and E. A. Groisman. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Brennan, M. A., and B. T. Cookson. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 38:31-40. [DOI] [PubMed] [Google Scholar]
- 7.Chatfield, S., M. Roberts, J. Li, A. Starns, and G. Dougan. 1994. The use of live attenuated Salmonella for oral vaccination. Dev. Biol. Stand. 82:35-42. [PubMed] [Google Scholar]
- 8.Chen, L. M., K. Kaniga, and J. E. Galan. 1996. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 21:1101-1115. [DOI] [PubMed] [Google Scholar]
- 9.Claros, M. G., and G. von Heijne. 1994. TopPredII: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 11.Ernst, R. K., D. M. Dombroski, and J. M. Merrick. 1990. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect. Immun. 58:2014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst, R. K., T. Guina, and S. I. Miller. 1999. How intracellular bacteria survive: surface modifications that promote resistance to host innate immune responses. J. Infect. Dis. 179:S326-S330. [DOI] [PubMed] [Google Scholar]
- 13.Fierer, J., L. Eckmann, F. Fang, C. Pfeifer, B. B. Finlay, and D. Guiney. 1993. Expression of the Salmonella virulence plasmid gene spvB in cultured macrophages and nonphagocytic cells. Infect. Immun. 61:5231-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich, M. J., and R. J. Kadner. 1987. Nucleotide sequence of the uhp region of Escherichia coli. J. Bacteriol. 169:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galan, J. E. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263-271. [DOI] [PubMed] [Google Scholar]
- 16.Galan, J. E., and R. Curtiss III. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 58:1879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García Véscovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 18.Goebel, W., and M. Kuhn. 2000. Bacterial replication in the host cell cytosol. Curr. Opin. Microbiol. 3:49-53. [DOI] [PubMed] [Google Scholar]
- 19.Groisman, E. A., and H. Ochman. 2000. The path to Salmonella. ASM News 66:21-27. [Google Scholar]
- 20.Groisman, E. A., and H. Ochman. 2000. How Salmonella became a pathogen. Trends Microbiol. 5:343-349. [DOI] [PubMed] [Google Scholar]
- 21.Groisman, E. A., and M. J. Saier. 1990. Salmonella virulence: new clues to intramacrophage survival. Trends Biochem. Sci. 15:30-33. [DOI] [PubMed] [Google Scholar]
- 22.Guzman, C. A., M. Rohde, M. Bock, and K. N. Timmis. 1994. Invasion and intracellular survival of Bordetella bronchiseptica in mouse dendritic cells. Infect. Immun. 62:5528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedlung, M., C. Wachtler, E. Johansson, L. Hang, J. E. Somerville, R. P. Darveau, and C. Svanborg. 1999. P fimbriae-dependent, lipopolysaccharide-independent activation of epithelial cytokine responses. Mol. Microbiol. 33:693-703. [DOI] [PubMed] [Google Scholar]
- 24.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284:967-970. [DOI] [PubMed] [Google Scholar]
- 25.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 2000. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 28.Hobohm, U., and C. Sander. 1995. A sequence property approach to searching protein databases. J. Mol. Biol. 251:390-399. [DOI] [PubMed] [Google Scholar]
- 29.Jungnitz, H., N. P. West, M. J. Walker, G. S. Chhatwal, and C. A. Guzman. 1998. A second two-component regulatory system of Bordetella bronchiseptica required for bacterial resistance to oxidative stress, production of acid phosphatase, and in vivo persistence. Infect. Immun. 66:4640-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 31.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. USA 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine, M. M., J. Galen, E. Barry, F. Noriega, S. Chatfield, M. Sztein, G. Dougan, and C. Tacket. 1996. Attenuated Salmonella as live oral vaccines against typhoid fever and as live vectors. J. Biotechnol. 44:193-196. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 93:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakai, K., and M. Kanehisa. 2000. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen, H., J. Engelbrecht, G. von Heijne, and S. Brunak. 1996. Defining a similarity threshold for a functional protein sequence pattern: the signal peptide cleavage site. Proteins 24:165-177. [DOI] [PubMed] [Google Scholar]
- 38.Nimchuk, Z., E. Marois, S. Kjemtrup, R. T. Leister, F. Katagiri, and J. L. Dangl. 2000. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101:353-363. [DOI] [PubMed] [Google Scholar]
- 39.O'Callaghan, D., and A. Charbit. 1990. High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol. Gen. Genet. 223:156-158. [DOI] [PubMed] [Google Scholar]
- 40.Ochman, H., A. S. Geber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohl, M. E., and S. I. Miller. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259-274. [DOI] [PubMed] [Google Scholar]
- 42.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Post, F. A., C. Manca, O. Neyrolles, B. Ryffel, D. B. Young, and G. Kaplan. 2001. Mycobacterium tuberculosis 19-kilodalton lipoprotein inhibits Mycobacterium smegmatis-induced cytokine production by human macrophages in vitro. Infect. Immun. 69:1433-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell, C. J., Jr., C. R. DeSett, J. P. Lowenthal, and S. Berman. 1980. The effect of adding iron to mucin on the enhancement of virulence for mice of Salmonella typhi strain Ty2. J. Biol. Stand. 8:79-85. [DOI] [PubMed] [Google Scholar]
- 45.Reed, L. J., and H. Muench. 1938. A simple method for estimating fifty percent end points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 46.Ripio, M. T., K. Brehm, M. Lara, M. Suarez, and J. A. Vazquez-Boland. 1997. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J. Bacteriol. 179:7174-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowe, B., L. R. Ward, and E. J. Threlfal. 1997. Multidrug-resistant Salmonella typhi: a worldwide epidemic. Clin. Infect. Dis. 24:S106-S109. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed.. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Santos, R. L., R. M. Tsolis, A. J. Baumler, R. Smith III, and L. G. Adams. 2001. Salmonella enterica serovar Typhimurium induces cell death in bovine monocyte-derived macrophages by early sipB-dependent and delayed sipB-independent mechanisms. Infect. Immun. 69:2293-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schauder, B., and J. E. G. McCarthy. 1989. The role of bases upstream of the Shine-Dalgarno region and in the coding sequence in the control of gene expression in Escherichia coli: translation and stability of mRNAs in vivo. Gene 78:59-72. [DOI] [PubMed] [Google Scholar]
- 51.Schwan, W. R., X. Z. Huang, L. Hu, and D. J. Kopecko. 2000. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect. Immun. 68:1005-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sein, J., V. Cachicas, M. I. Becker, and A. E. Ioannes. 1993. Mucin allows survival of Salmonella typhi within mouse peritoneal macrophages. Biol. Res. 26:371-379. [PubMed] [Google Scholar]
- 53.Sessa, W. C., C. M. Barber, and K. R. Lynch. 1993. Mutation of N-myristoylation site converts endothelial cell nitric oxide synthase from a membrane to a cytosolic protein. Circ. Res. 72:921-924. [DOI] [PubMed] [Google Scholar]
- 54.Shan, L., V. K. Thara, G. B. Martin, J. M. Zhou, and X. Tang. 2000. The Pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell 12:2323-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siebers, A., and S. Falkow. 1996. M cells and the pathogenesis of mucosal and systemic infections. Trends Microbiol. 4:22-29. [DOI] [PubMed] [Google Scholar]
- 56.Somerville, J. E., Jr., L. Cassiano, B. Bainbridge, M. D. Cunningham, and R. P. Darveau. 1996. A novel Escherichia coli lipid A mutant that produces an anti-inflammatory lipopolysaccharide. J. Clin. Investig. 97:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Somerville, J. E., Jr., L. Cassiano, and R. P. Darveau. 1999. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 67:6583-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staendner, L. H., M. Rohde, K. N. Timmis, and C. A. Guzman. 1995. Identification of Salmonella typhi promoters activated by invasion of eukaryotic cells. Mol. Microbiol. 18:891-902. [DOI] [PubMed] [Google Scholar]
- 59.Tartera, C., and E. S. Metcalf. 1993. Osmolarity and growth phase overlap in regulation of Salmonella typhi adherence to and invasion of human intestinal cells. Infect. Immun. 61:3084-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tzschaschel, B. D., C. A. Guzman, K. N. Timmis, and V. de Lorenzo. 1996. An Escherichia coli hemolysin transport system-based vector for the export of polypeptides: export of Shiga-like toxin IIeB subunit by Salmonella typhimurium aroA. Nat. Biotechnol. 14:765-769. [DOI] [PubMed] [Google Scholar]
- 61.Valdivia, R. H., D. M. Cirillo, A. K. Lee, D. M. Bouley, and S. Falkow. 2000. mig-14 is a horizontally acquired, host-induced gene required for Salmonella enterica lethal infection in the murine model of typhoid fever. Infect. Immun. 68:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vladonianu, I. R., H. R. Chang, and J. C. Pechere. 1990. Expression of host resistance to Salmonella typhi and Salmonella typhimurium: bacterial survival within macrophages of murine and human origin. Microb. Pathog. 8:83-90. [DOI] [PubMed] [Google Scholar]
- 63.Weston, L. A., and R. J. Kadner. 1987. Identification of uhp polypeptides and evidence for their role in exogenous induction of the sugar phosphate transport system of Escherichia coli K-12. J. Bacteriol. 169:3546-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weston, L. A., and R. J. Kadner. 1988. Role of uhp genes in expression of the Escherichia coli sugar-phosphate transport system. J. Bacteriol. 170:3375-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson, J. A., T. J. Doyle, and P. A. Gulig. 1997. Exponential-phase expression of spvA of the Salmonella typhimurium virulence plasmid: induction in intracellular salts medium and intracellularly in mice and cultured mammalian cells. Microbiology 143:3827-3839. [DOI] [PubMed] [Google Scholar]
- 66.Worley, K. C., B. A. Wiese, and R. F. Smith. 1995. BEAUTY: an enhanced BLAST-based search tool that integrates multiple biological information resources into sequence similarity search results. Genet. Res. 5:173-184. [DOI] [PubMed] [Google Scholar]
- 67.Yrlid, U., and M. J. Wick. 2000. Salmonella-induced apoptosis of infected macrophages results in presentation of a bacteria-encoded antigen after uptake by bystander dendritic cells. J. Exp. Med. 191:613-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, F., S. Inouye, and M. Inouye. 1986. Lipoprotein-28, a cytoplasmic membrane lipoprotein from Escherichia coli. Cloning, DNA sequence, and expression of its gene. J. Biol. Chem. 261:2284-2288. [PubMed] [Google Scholar]
- 69.Zhou, X., N. Mantis, X. R. Zhang, D. A. Potoka, S. C. Watkins, and H. R. Ford. 2000. Salmonella typhimurium induces apoptosis in human monocyte-derived macrophages. Microbiol. Immunol. 44:987-995. [DOI] [PubMed] [Google Scholar]