Abstract
Botulinum neurotoxins (BoNTXs) produced by Clostridium botulinum are among the most poisonous substances known. Of the seven types of BoNTXs, genes for type C1 and D toxins (BoNTX/C1 and D) are carried by bacteriophages. The gene for exoenzyme C3 also resides on these phages. Here, we present the complete genome sequence of c-st, a representative of BoNTX/C1-converting phages. The genome is a linear double-stranded DNA of 185,682 bp with 404-bp terminal direct repeats, the largest known temperate phage genome. We identified 198 potential protein-coding regions, including the genes for production of BoNTX/C1 and exoenzyme C3. Very exceptionally, as a viable bacteriophage, a number of insertion sequences were found on the c-st genome. By analyzing the molecular structure of the c-st genome in lysogens, we also found that it exists as a circular plasmid prophage. These features account for the unstable lysogeny of BoNTX phages, which has historically been called “pseudolysogeny.” The PCR scanning analysis of other BoNTX/C1 and D phages based on the c-st sequence further revealed that BoNTX phages comprise a divergent phage family, probably generated by exchanging genomic segments among BoNTX phages and their relatives.
Keywords: bacteriophage, botulinum neurotoxin, insertion sequence, plasmid prophage
Botulinum neurotoxins (BoNTXs) produced by Clostridium botulinum are among the most poisonous substances known (1). BoNTX-producing strains cause food poisoning with high mortality and fetal infant intestinal infection, known as infant botulism. BoNTXs are classified into seven groups (A, B, C1, and D-G), based on antigenicity. Each type of BoNTX is produced as a large polypeptide and converted to a dichain molecule composed of L and H chains by bacterial or host proteases. The H chain is responsible for the binding of the toxin to the presynaptic membrane and for the translocation of the L chain into the cytosol. The L chain is a metalloprotease that specifically cleaves SNARE proteins, components of the neuroexocytosis apparatus, leading to the irreversible blockade of neurotransmitter release at neuromuscular junctions, resulting in flaccid paralysis (2, 3).
BoNTXs in culture medium exist as a high-molecular-weight protein complex called a progenitor toxin, in which BoNTXs are associated with two nontoxic components designated as hemagglutinin (HA) and nontoxic nonhemagglutinin component. HA consists of four subcomponents: HA1, HA2, HA3a, and HA3b (4). Although the composition of nontoxic components exhibits a certain variation according to the type of toxin, they protect the BoNTX molecules from gastric juice and intestinal digestive enzymes and facilitate the binding and adsorption of the toxins through the upper intestinal tract (4, 5). Several studies have demonstrated that BoNTX genes are clustered with genes for nontoxic components and BotR, an alternative σ factor for the BoNTX gene cluster (4, 6).
Of the seven types of BoNTXs, genes for type C1 (BoNTX/C1) and type D (BoNTX/D) are carried by bacteriophages, which were discovered in the early 1970s (7-11). These phages are categorized into three groups according to their conversion spectra; phages from strains C-Stockholm (C-ST) and C-468, those from strains D-1873 and C-203, and those from strains D-South African and D-4947 (12). These groups differ also in antigenicity, although they share a similar morphology. BoNTX phages were later found to encode exoenzyme C3, an ADPribosyltransferase of ρ GTPases (13). It is also known that the lysogeny of BoNTX phages is unstable. Lysogens can be cured easily by culturing them in media containing acridine orange or anti-phage sera. Nontoxigenic derivatives emerge at a high frequency, even without such treatments. This phenomenon has historically been called pseudolysogeny (9, 10, 14).
Prophages are common in many bacteria and often carry virulence genes and other lysogenic conversion genes into host organisms. A number of such virulence gene-converting phages have already been sequenced (15-18). However, despite the long research history of BoNTX phages, we have very poor understanding of their genetic features. We know only that their genomes are large dsDNAs (19). Here, we report the complete genome sequence of c-st, a representative of BoNTX/C1 phages, isolated from strain C-ST.
Methods
Phages, Bacterial Strains, and Preparation of DNAs. All of the C. botulinum strains used were from our laboratory stock (11, 12). Type C strains included C-ST, C-203, C-468, and C-6813. Type D included D-1873, D-4947, and D-South African. Strain (C)-AO2 is a phage-cured derivative of C-ST (7). Bacterial DNAs were prepared by using a genomic DNA purification kit (Takara Bio, Otsu, Japan) from the cells cultivated anaerobically overnight at 37°C in cooked meat medium (Becton Dickinson). The c-st phage was reisolated from C-ST, and propagated on (C)-AO2 in LYG medium (1% lactalbumin/2% yeast extract/0.5% glucose/0.15% cysteine hydrochloride, pH 7.2). The phage genomic DNA was isolated from phage particles by pulsed-field gel electrophoresis (PFGE) as follows. The phage lysate was first treated with RNase and DNase I (Takara Bio) at a final concentration of 1 μg/ml for 30 min at 37°C and incubated with 1 M NaCl for 16 h at 4°C. After removing cell debris by centrifugation at 11,000 × g for 20 min at 4°C, phage particles were recovered by polyethylene glycol precipitation (6,000, Nacalai Tesque, Kyoto), resuspended in 10 mM Tris·HCl (pH 8.0), and embedded in 1% agarose plugs (CleanCut Agarose, Bio-Rad). Plugs were treated with 0.1% proteinase K (Roche Applied Science) and 0.5% N-lauroylsarcosine (Nacalai Tesque) for 24 h at 50°C and subjected to PFGE. PFGE was performed on a CHEF Mapper apparatus (Bio-Rad) by using 1% agarose gels (pulsed-field certified agarose, Bio-Rad) in 0.5× TBE (Tris/borate/EDTA) at 6 V/cm at 14°C. Pulse times were ramped from 6.75 to 15.54 s for 34.28 h, with a linear increase in pulse intervals. Finally, the phage DNA migrating at the 186-kb position was recovered from the gel by using the Prep-A-Gene DNA purification kit (Bio-Rad).
Sequencing, Annotation, and Computer Analyses. The genome sequence of c-st was determined by the random shotgun sequencing method as described in refs. 20 and 21. The genome was sequenced to an average coverage of 13.7× (5,100 reads in total) and at a 2× minimum coverage (at least once in each strand). The sequences of each end of the c-st genome were determined by direct sequencing of the 1.1-kb AatII fragment derived from the left end and the 1.9-kb SalI fragment from the right end.
Potential protein-coding regions (ORFs) ≥150 bp were identified by using the program genome gambler 1.51 (22), and each ORF was reviewed manually for the presence of a ribosomal binding sequence. Functional annotation was based on homology searches against the GenBank nonredundant protein sequence database by using the program blastp (23). The sequencher DNA sequencing software (Gene Codes, Ann Arbor, MI) was used for sequence assembly and multiple sequence alignment. The cumulative GC skew was analyzed as described by Grigoriev in ref. 24. The sequences reported in this article have been deposited in the GenBank database under the accession nos. AP008983 (c-st), AB217840, and AB217841 (the upstream and downstream regions of the BoNTX/D gene cluster in d-1873, respectively).
PCR Amplification. PCR amplification was carried out by using the LA long PCR kit or the Ex TaqPCR kit (Takara Bio) in the following setting: preheating (96°C, 1 min), 30 cycles of denaturation (96°C, 1 min)/annealing (50°C, 1 min)/extension (66°C), and an additional extension (66°C, 5 min). Extension time was varied according to expected product sizes (1 min/kb). PCR products were analyzed on 1% or 2% agarose gels. DNA sequences of PCR products were determined by the primer-walking method. All of the primers used in this study are listed in Table 1, which is published as supporting information on the PNAS web site.
Southern Hybridization Analyses. After cultivating C. botulinum strains overnight at 37°C in cooked meat medium, cells were collected by centrifugation at 8,000 × g for 10 min at 4°C, washed twice with 10 mM Tris·HCl (pH 8.0), and embedded in 1% agarose plugs. The plugs were treated first with 1.2 mg/ml lyzozyme (Sigma-Aldrich) and 0.5% N-lauroylsarcosine at 37°C for 16 h and then with proteinase K and N-lauroylsarcosine as described above. The cellular DNAs prepared by this procedure and the phage-particle DNA, which was also embedded in agarose plugs, were digested with appropriate restriction enzymes, and subjected to PFGE. DNA fragments were transferred onto nylon membranes (Hybond-N plus, Amersham Pharmacia), and hybridization and signal detection were done by using the ECL direct nucleic acid labeling and detection system (Amersham Pharmacia) according to the manufacturer's instruction. All hybridization probes were prepared by PCR.
Results
Reisolation of the c-st Phage. To prepare the phage DNA for genome sequencing, we first propagated the c-st phage, which was originally isolated from strain C-ST and had been kept in our laboratory by repeated propagations on strain (C)-AO2. However, the phage DNA preparation was found to be highly heterogeneous DNA molecules ranging from 150 to 180 kb in size (data not shown). Because the phage was propagated from a single plaque, it is likely that, after multiple propagations, the phage genome had become structurally unstable by some genetic alteration so as to provoke highly frequent genome rearrangements. Therefore, we reisolated the c-st phage from strain C-ST for the genome sequencing. The DNA preparation obtained from the reisolated c-st phage contained a single major DNA fragment of 186 kb, although a small amount of a 175-kb band was still observed in the PFGE analysis (data not shown).
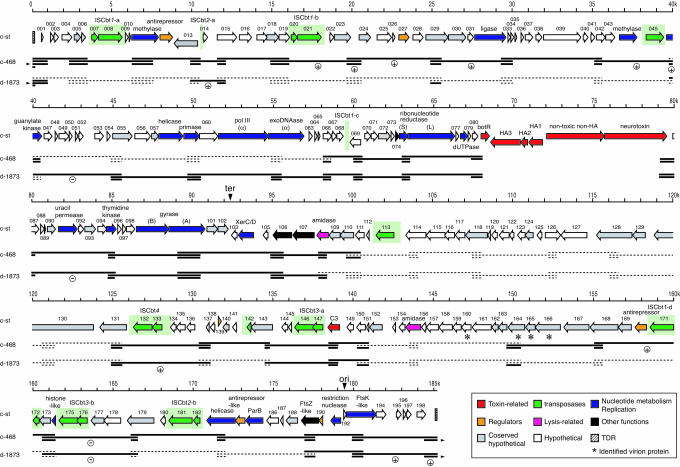
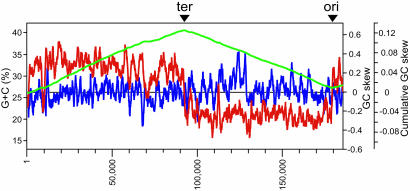
General Features of the c-st Genome. The genome of c-st is a linear 185,682-bp dsDNA with terminal direct repeats (TDRs) of 404 bp. The average G + C content is 26.2%, slightly lower than those of sequenced Clostridium species (25-27), Clostridium acetobutylicum (30.9%), Clostridium perfringens (28.6%), and Clostridium tetani (28.6%). We identified 198 ORFs on the phage genome (CST001-198) but no tRNA gene. The c-st genome was divided into three parts according to the orientation of ORFs (Fig. 1). Most ORFs on the left half of the genome (Region L, CST001-102) and the right most end (Region RR, CST193-198) were transcribed in the left-to-right direction but those on the right half (Region RL, CST103-191) in the right-to-left direction. The GC skew analysis suggested that the RL/RR and L/RL junctions correspond to the replication origin and terminus of c-st, respectively (Fig. 2).
Fig. 1.
Genome organization of the c-st phage and the summary of PCR scanning analysis of other BoNTX phages. Horizontal arrows represent ORFs identified on the c-st genome, and ORF numbers or gene products are indicated above each arrow. ORFs were categorized into eight groups according to their predicted functions. TDRs are indicated by striped boxes, IS elements by green boxes, and the putative replication origin (ori) and terminus (ter) by vertical arrows. Four virion component proteins identified by micosequencing analysis of the c-st virion proteins are indicated by asterisks. Results of PCR scanning analysis of phages c-468 and d-1873 are shown below the ORF map. Horizontal thick lines represent segments where PCR products were obtained, and broken lines represent segments where no products were obtained. The segments that yielded PCR products larger than those of c-st are indicated by (+) and smaller by (-). Because circular plasmid prophages were examined in this PCR scanning analysis, segments encompassing the TDRs were portioned to both ends (the connection indicated by horizontal arrowheads).
Fig. 2.
Base composition and GC skew analyses of the c-st genome. The average G + C percent, the GC-skew (G - C/G + C) in the forward strand, and the cumulative GC-skew are represented by blue, red, and green lines, respectively. All were calculated by using a window size of 1,000 bp and a step size of 100 bp. ter, terminus; ori, origin.
Of the identified 198 ORFs, only 57 were functionally assigned, based on the sequence homology to known proteins, whereas more than half (102 ORFs) shared no significant sequence homology with any proteins in databases. The remaining 39 ORFs exhibited sequence similarity to the proteins of unknown function (see Table 2, which is published as supporting information on the PNAS web site). Genes for the production of BoNTX/C1 complex (CST081-086) were clustered at the 68-80-kb position, whereas the C3 gene (CST148) was located separately at the 139-kb position.
Among the 95 ORFs with sequence similarity to known proteins, 26 exhibited some similarity to the gene products of SPβ, a large (134 kb) temperate phage lysogenized in Bacillus subtilis strain 168 (28). Although the level of their sequence similarities to the SPβ counterparts was low (20-30%), the relative locations on the genomes were well conserved (see Fig. 5, which is published as supporting information on the PNAS web site). The c-st phage is, thus, phylogenetically, but distantly, related to SPβ. In fact, although both are siphoviruses, their tails have distinct structures (see Fig. 6, which is published as supporting information on the PNAS web site).
The c-st phage encodes a relatively larger number of proteins involved in DNA processing and nucleotide metabolism (Fig. 2). Among the 57 functionally assigned ORFs, 15 were for various kinds of DNA-processing events and 6 were for nucleotide metabolism. Most of them, probably comprising early operons, resided on Region L (Fig. 2). On the other hand, most ORFs on Region RL are of unknown function. However, four virion component proteins, identified by our preliminary micosequencing analysis of virion proteins, were found on this region (CST160 and 164-166). Our preliminary ImmunoGold staining analysis using specific antisera further indicated that CST160 is a sheath protein, and CST165 is a head protein. Two amidase genes were also encoded on this region, suggesting that genes on Region RL comprise late operons.
Insertion Sequence (IS) Elements. A remarkable feature of the c-st genome was the abundance of IS elements (Fig. 2). We identified a total of 12 copies including 5 truncated or degenerated ones. They were classified into seven types. All were previously unidentified IS elements belonging to the IS200/605 family that lack terminal inverted repeats (29). Among these, the structures of four types (ISCbt1-4) were defined by comparing the sequences of IS-inserted regions in c-st with those of analogous unoccupied regions in other BoNTX phages (see Fig. 7, which is published as supporting information on the PNAS web site). ISCbt1 and ISCbt2 were inserted site-specifically into the TTAC and ATACAT sequences, respectively, without target-site duplication and ISCbt3 also site-specifically to the AAGGAG sequence.
Lysogenization as a Circular Plasmid. The unstable lysogenic state of BoTNX phages has long been known, but the mode of lysogenization has not yet been elucidated. The genomic features of c-st, however, raised a possibility that the phage is present as a circular replicon in lysogens. (i) Most ORFs were bidirectionally oriented, as often observed in the circular chromosomes of low GC Gram-positive bacteria, and clear transitions of ORF direction occurred at the putative replication origin and terminus. (ii) A XerC/D family protein that mediates the resolution of daughter chromosomes/plasmids was located in the very vicinity of the putative replication terminus. (iii) ParB and FtsK/SpoIIIE-like proteins, which each may be involved in chromosome/plasmid partition and DNA segregation or translocation, were also encoded in the vicinity of the putative replication origin.
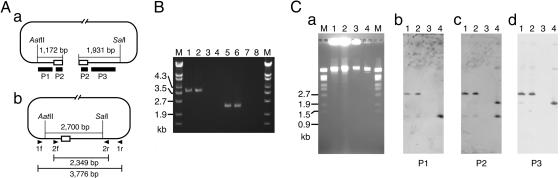
To detect a circular form of the c-st genome, we first analyzed the phage DNA in the lysogen by PCR using two sets of primers; 1f/1r and 2f/2r (Fig. 3). In each set, one primer targeted to the left end of the linear phage genome and the other to the right end. Both primer sets yielded PCR products from the cellular DNAs of strain C-ST and a c-st-lysogen of strain (C)-AO2 (referred to as (C)-AO2:c-st) but not from that of noninfected (C)-AO2 (Fig. 3B), indicating that both ends of the linear c-st genome are connected to each other in the lysogen. Direct sequencing of the PCR products revealed that this end-connected DNA molecule contains only one copy of TDR, suggesting that it was created by recombination between the two TDRs located at each end of the c-st genome.
Fig. 3.
Structures of the c-st genome in lysogens and phage particles. (A) Locations of hybridization probes and PCR primers on the linear and circular forms of the c-st genome are shown. Among the AatII and SalI sites on the c-st genome, only AatII and SalI sites located closest to the right and left ends, respectively, are shown. Open boxes represent TDRs. (B) Cellular DNAs from strains C-ST, (C)-AO2:c-st, and (C)-AO2 and the phage-particle DNA were examined by PCR using primer pairs 1f/1r (lanes 1-4) and 2f/2r (lanes 5-8). (C)-AO2:c-st is a c-st-lysogen derived from (C)-AO2. Lane M, size makers; lanes 1 and 5, C-ST; lanes 2 and 6, (C)-AO2:c-st; lanes 3 and 7, (C)-AO2; lanes 4 and 8, phage particle. (C) The same set of DNA preparations as in B was digested by AatII and SalI, separated on 1% agarose gels (a), and subjected to the Southern hybridization analyses using probes P1 (b), P2 (c), and P3 (d). Lanes M and 1-4 are as in B.
We next examined the phage particle DNA and the cellular DNAs of the lysogens by Southern hybridization analysis (Fig. 3C). In this analysis, each DNA preparation was double-digested by AatII and SalI and analyzed by three hybridization probes (Fig. 3A), P1 (specific to the region between the left TDR and the far left AatII site), P2 (TDR), and P3 (the region between the far right SalI site and the right TDR). In the phage-particle DNA, the P1 and P2 probes hybridized to 1.2- and 1.9-kb fragments derived from the left and right ends, respectively, of the linear phage DNA and P3 to both. In contrast, all probes hybridized to a single 2.7-kb fragment in the DNA preparations from lysogens, confirming that lysogens contain only an end-connected form of the c-st genome.
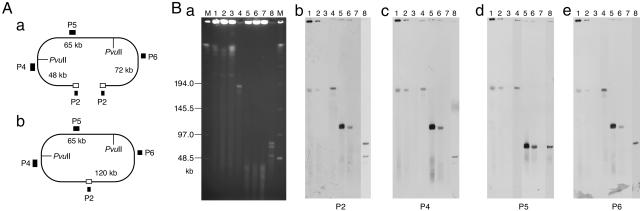
Finally, to determine whether the c-st genome is integrated into the chromosome or not in the lysogens, another series of Southern hybridization analyses were performed by using four probes (P2, P4, P5, and P6) and two restriction enzymes (ApaI and PvuII). ApaI does not digest the phage DNA but the host chromosome, whereas PvuII digestion of the linear phage DNA yields three fragments of 48, 65, and 72 kb, which can be detected specifically by probes P4, P5, and P6, respectively (Fig. 4). When digested by ApaI, a 186-kb signal was detected by all probes, not only in the phage particle but also in the lysogens. In the lysogens, however, much stronger signals were detected at the well position. These strong signals mostly disappeared when the DNAs were digested by PvuII, suggesting that they were derived from a circular form of DNA that does not migrate into the PFGE gel. Weak 186-kb signals detected in the ApaI-digested DNAs of lysogens were most likely from linear phage DNA molecules generated by mechanical or nonspecific digestion. In the PvuII-digested DNAs from lysogens, probes P2, P4, and P6, which hybridize to the left and/or the right PvuII fragments of the linear phage genome, detected a 120-kb fragment derived from a circularized phage genome. Importantly, probe P5 yielded a 65-kb signal, which was derived from the 65-kb internal PvuII fragment, not only in the phage particle but also in the lysogens, indicating that c-st is not integrated into the host chromosome but present as a circular plasmid in the lysogens.
Fig. 4.
Southern hybridization analysis of the c-st DNAs from lysogens and phage particles. (A) Locations of PvuII sites and hybridization probes and the linear and circular forms of the c-st genome are shown. (B) Cellular DNAs from strains C-ST, (C)-AO2:c-st, and C-AO2 and the c-st phage-particle DNA were digested with ApaI (lanes 1-4) or PvuII (lanes 4-8), separated by PFGE (a), and subjected to Southern hybridization analysis using probes P2 (b), P4 (c), P5 (d), and P6 (e). Lane M, size makers; lanes 1 and 5, C-ST; lanes 2 and 6, (C)-AO2:c-st; lanes 3 and 7, C-AO2; lanes 4 and 8, phage particle.
Genomic Diversity of BoNTX Phages. To know whether the genomic features observed for c-st are conserved in other BoNTX phages, we examined the genomes of phages c-468 (another C1 phage from strain C-468) and d-1873 (a BoNTX/D phage from strain D-1873) by PCR scanning (20, 30) using a set of PCR primers covering the whole c-st genome (Fig. 1; and see Table 3, which is published as supporting information on the PNAS web site). Unexpectedly, many of these primers, prepared based on the c-st sequence, did not yield PCR products not only in d-1873 but also in c-468, indicating that these phages are significantly diverged from c-st in genome structure and/or sequence. However, a 40-kb region encompassing the upstream of the BoNTX gene cluster to the downstream of the putative replication terminus (map position 58-98 kb) were conserved in both phages, although some small insertions and deletions were detected. A region containing the replication origin and TDRs and the C3 gene-containing region were also conserved. We have sequenced the upstream and downstream regions of the BoNTX/D gene cluster in d-1873 (5 kb each), and compared them with the c-st sequences (see Fig. 8, which is published as supporting information on the PNAS web site). The data indicated that, although small deletions and replacements have taken place, both regions were highly conserved between the two phages, exhibiting 97-98% nucleotide sequence identity.
Other parts of the two BoNTX phage genomes showed a high level of variation. When we compared the two phages, more segments were amplified from c-468 than from d-1873, in accordance with the fact that c-st and c-468 possess a similar converting spectrum and antigenicity (12). Of interest was that conserved and divergent regions exhibited a mosaic distribution on these phage genomes. For example, the first 40-kb region was well conserved in c-468, but only a few small segments were amplified from this region in d-1873. In contrast, the next 18-kb region (map position 40-58 kb) was well conserved in d-1873 but not in c-468. The right half of the genome was much more diverged compared with the left half, but the mosaic distribution of conserved and divergent regions was also apparent, particularly in c-468.
Discussion
In this study, we determined the complete genome sequence of c-st, a representative of BoNTX/C1-converting phages. The c-st genome is a linear 186-kb dsDNA with 404-bp TDRs, the largest known temperate-phage genome. The phage encodes 198 proteins. They include a relatively large number of proteins for DNA processing and nucleotide metabolism that are assumed to be required for the replication of the large c-st phage genome. Subsequent molecular analyses of the phage genome revealed that this phage exists as a circular plasmid prophage in the lysogen and does not integrate into the host chromosome. This mode of lysogenization appears to be related to the unstable lysogeny of BoNTX phages.
Temperate phages are generally integrated into host chromosomes and stably maintained as prophages in lysogens. However, a few exceptional phages are known to exist extrachromosomally as circular or linear plasmid prophages. Such plasmid prophages require some special mechanisms to ensure the stable inheritance in their lysogens. The circularized 93-kb genome of phage P1, the most extensively characterized among such phages (31), is stably maintained as a low-copy plasmid by multiple mechanisms: partitioning of the plasmid into daughter cells by the ParAB system, multimer resolution by the Cre-lox system, and postsegregational killing by the Phd-Doc toxin-antitoxin system (32-34). The c-st phage also encodes a XerC/D family protein that probably resolves the plasmid multimer (35) and ParB- and FtsK/SpoIIIE-like proteins that may be involved in plasmid partitioning or segregation (36, 37). However, the unstable lysogeny of c-st suggests that these proteins do not maintain the c-st plasmid as efficiently as do the P1 systems.
The c-st phage differs from P1 in the mechanism of genome circularization as well. The P1 genome is “circularly permutated” and “terminally redundant,” generated by rolling-circle replication and genome packaging by a processive “headful packaging” mechanism. After being injected into the host cell, the P1 phage genome is circularized by Cre/lox-mediated site-specific recombination or homologous recombination between terminal redundant sequences (38). In contrast, the c-st genome is circularized by the recombination between the 404-bp TDRs. The presence of such TDR sequences and a lack of circular permutation in c-st indicate that the packaging mechanism also differs from that of P1. In terms of the possession of TDRs, c-st rather resembles the T7 phage family, where TDRs are involved in concatemer formation, although T7 is a virulent phage and no circularization step in the replication process has been described (39).
Another notable genomic feature of c-st is the abundance of IS elements. The c-st phage genome contains 12 copies that can be classified into seven types that occupy as much as 10% of the c-st genome. In general, genes on phage genomes are well organized, so that they are efficiently expressed according to the genetic program for propagation. Thus, multiple IS transpositions should be disadvantageous for bacteriophages, and, therefore, phage genomes would not favor IS insertions. In fact, IS elements are very rarely found on the genomes of viable bacteriophages; among the 284 phage genomes so far sequenced (www.ncbi.nlm.nih.gov/genomes/static/phg.html), only eight contain IS elements (P1, HK022, Sf6, phiE125, Bcep22, phiAT3, LP6, and RM 378, one copy in each). Of the 12 IS elements identified in c-st, 7 were structurally intact. Furthermore, their insertion sites were often unoccupied on other BoNTX phage genomes (Fig. 7), suggesting that they recently entered c-st and so are likely to be functional. Although it is not clear why BoNTX phages permit such multiple IS insertions, their transpositions to some c-st genome regions, which are essential for the plasmid functions, can impair the plasmid stability. The prophage instability may be partly attributable to these IS elements.
PCR scanning analyses of the genomes of phages c-468 and d-1873 revealed that a high level of genomic diversity exists not only between the BoNTX/C1 and BoNTX/D phages but also among the BoNTX/C1 phages (Fig. 2). Distribution of conserved and divergent regions on the genomes displayed a mosaic pattern, suggesting that these genomes are genetic mosaics generated by exchanging genomic segments among the BoNTX phages and their relatives. This type of genomic mosaicism is common in nearly every group of tailed-phages that has been examined, and this is a topic of much current discussion in the field of virus evolution (40). In the case of BoNTX phages, the genome mosaicism appears to be reflected on their heterogeneity in converting spectrum and antigenicity (12). IS elements may have played some role also in the process of diversification of the BoNTX phages.
On the other hand, a region containing the putative replication origin, TDRs, and a region containing the replication terminus are conserved in the three BoNTX phages, suggesting that the mechanisms for circularization, replication, and lysogenization predicted for c-st are probably shared by other BoNTX phages. Two regions, each containing the BoNTX gene cluster and the C3 gene, are also well conserved. Of particular importance is that the genomic sequences of BoNTX gene-flanking regions are highly conserved, even between different type of toxins (C1 and D) (Fig. 8) and may provide a good opportunity to generate a hybrid phage in which only a part of the BoNTX gene is exchanged. Genes encoding such mosaic toxins have actually been identified (41).
We have very poor understanding of the mechanisms of dissemination or evolution of other types of BoNTX genes. Strains producing BoNTXs are quite heterogeneous: C. botulinum strains, which are further grouped into four biotypes, and some strains of Clostridium butyricum and Clostridium bratii. Tetanus toxin produced by C. tetani is also closely related to BoNTXs. Genomic locations of the genes are also variable; BoNTX/A, B, and E are on chromosomes, whereas BoNTX/G, C. butyricum toxin, and tetanus toxin are on plasmids (42-44). The genomic location of the BoNTX/F gene is still ambiguous. How were the BoNTX family members disseminated and how did they diverge among these bacteria? Considering their extremely high toxicity and potential application to bioterrorism, these are important issues to be elucidated. As a consequence, the presence of a large number of IS elements on the BoNTX phage genomes is again intriguing in that IS elements can mediate the inter-replicon transfer of toxin genes. Lysogenization of BoNTX phages as circular plasmids may imply that they have some genetic relationship to the plasmids encoding other types of toxins. Although no recognizable sequence homology was observed between c-st and the tetanus-toxin-encoding plasmid (27), it would be worthwhile to analyze the genome sequences of BoNTX-encoding plasmids.
Supplementary Material
Acknowledgments
We thank Dr. T. Murata for valuable suggestions, Akemi Yoshida for technical assistance, and Yumiko Hayashi for language assistance. This work was supported by the Japan Society for the Promotion of Science Research for the Future Programs (97L00101 and JSPS-RFTF00L01411); Grants-in-Aid for Scientific Research and the 21st Century Center of Excellence (COE) Program from the Ministry of Education, Culture, Sports, Science and Technology, Japan; and grants from the Japan Health Sciences Foundation and the National Project on Protein Structural and Functional Analyses.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BoNTX, botulinum neurotoxin; IS, insertion sequence; PFGE, pulsed-field gel electrophoresis; TDR, terminal direct repeat.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AP008983, AB217840, and AB217841).
References
- 1.Gill, M. G. (1982) Microbiol. Rev. 46, 86-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niemann, H., Blasi, J. & Jahn, R. (1994) Trends Cell Biol. 4, 179-185. [DOI] [PubMed] [Google Scholar]
- 3.Schiavo, G., Matteoli, M. & Montecucco, C. (2000) Physiol. Rev. 80, 717-766. [DOI] [PubMed] [Google Scholar]
- 4.Oguma, K., Inoue, K., Fujinaga, Y., Yokota, K., Watanabe, T., Ohyama, T., Takeshi, K. & Inoue, K. (1999) J. Toxicol. 18, 17-34. [Google Scholar]
- 5.Fujinaga, Y., Inoue, K., Nomura, T., Sasaki, J., Marvaud, J. C., Popoff, M. R., Kozaki, S. & Oguma, K. (2000) FEBS Lett. 467, 179-183. [DOI] [PubMed] [Google Scholar]
- 6.Raffestin, S., Dupuy, B., Marvaud, J. C. & Popoff, M. R. (2005) Mol. Microbiol. 55, 235-249. [DOI] [PubMed] [Google Scholar]
- 7.Inoue, K. & Iida, H. (1970) Jpn. J. Microbiol. 14, 87-89. [DOI] [PubMed] [Google Scholar]
- 8.Inoue, K. & Iida, H. (1971) Jpn. J. Med. Sci. Biol. 24, 53-56. [PubMed] [Google Scholar]
- 9.Eklund, M. W., Poysky, F. T., Reed, S. M. & Smith, C. A. (1971) Science 172, 480-482. [DOI] [PubMed] [Google Scholar]
- 10.Eklund, M. W., Poysky, F. T. & Reed, S. M. (1972) Nat. New Biol. 235, 16-17. [DOI] [PubMed] [Google Scholar]
- 11.Oguma, K., Iida, H. & Inoue, K. (1973) Jpn. J. Microbiol. 17, 425-426. [DOI] [PubMed] [Google Scholar]
- 12.Oguma, K., Iida, H., Shiozaki, M. & Inoue, K. (1976) Infect. Immun. 13, 855-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauser, D., Gibert, M., Eklund, M. W., Boquet, P. & Popoff, M. R. (1993) J. Bacteriol. 175, 7260-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oguma, K. (1976) J. Gen. Microbiol. 92, 67-75. [DOI] [PubMed] [Google Scholar]
- 15.Casjens, S. (2003) Mol. Microbiol. 49, 277-300. [DOI] [PubMed] [Google Scholar]
- 16.Canchaya, C., Proux, C., Fournous, G., Bruttin, A. & Brussow, H. (2003) Microbiol. Mol. Biol. Rev. 67, 238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd, E. F. & Brussow, H. (2002) Trends Microbiol. 10, 521-529. [DOI] [PubMed] [Google Scholar]
- 18.Brussow, H., Canchaya, C. & Hardt, W.-D. (2004) Microbiol. Mol. Biol. Rev. 68, 560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujii, N., Oguma, K., Yokosawa, N., Kimura, K. & Tsuzuki, K. (1988) Appl. Environ. Microbiol. 54, 69-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama, K., Takashima, K., Ishihara, H., Shinomiya, T., Kageyama, M., Kanaya, S., Ohnishi, M., Murata, T., Mori, H. & Hayashi, T. (2000) Mol. Microbiol. 38, 213-231. [DOI] [PubMed] [Google Scholar]
- 21.Murata, T., Ohnishi, M., Ara, T., Kaneko, J., Han, C. G., Li, Y. F., Takashima, K., Nojima, H., Nakayama, K., Kaji, A., et al. (2002) J. Bacteriol. 184, 3194-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakiyama, T., Takami, H., Ogasawara, N., Kuhara, S., Kozuki, T., Doga, K., Ohyama, A. & Horikoshi, K. (2000) Biosci. Biotechnol. Biochem. 64, 670-673. [DOI] [PubMed] [Google Scholar]
- 23.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grigoriev, A. (1998) Nucleic Acids Res. 26, 2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nolling, J., Breton, G., Omelchenko, M. V., Makarova, K. S., Zeng, Q., Gibson, R., Lee, H. M., Dubois, J., Qiu, D., Hitti, J., et al. (2001) J. Bacteriol. 183, 4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu, T., Ohtani, K., Hirakawa, H., Ohshima, K., Yamashita, A., Shiba, T., Ogasawara, N., Hattori, M., Kuhara, S. & Hayashi, H. (2002) Proc. Natl. Acad. Sci. USA 99, 996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruggemann, H., Baumer, S., Fricke, W. F., Wiezer, A., Liesegang, H., Decker, I., Herzberg, C., Martinez-Arias, R., Merkl, R., Henne, A., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazarevic, V., Dusterhoft, A., Soldo, B., Hilbert, H., Mauel, C. & Karamata, D. (1999) Microbiology 145, 1055-1067. [DOI] [PubMed] [Google Scholar]
- 29.Chandler, M. & Mahillon, J. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowittz, A. M. (Am. Soc. Microbiol., Washington, DC) pp. 305-366.
- 30.Ohnishi, M., Terajima, J., Kurokawa, K., Nakayama, K., Murata, T., Tamura, K., Ogura, Y., Watanabe, H. & Hayashi, T. (2002) Proc. Natl. Acad. Sci. USA 99, 17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobocka, M. B., Rose, D. J., Plunkett, G., III, Rusin, M., Samojedny, A., Lehnherr, H., Yarmolinsky, M. B. & Blattner, F. R. (2004) J. Bacteriol. 186, 7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordstrom, K. & Austin, S. J. (1989) Annu. Rev. Genet. 23, 37-69. [DOI] [PubMed] [Google Scholar]
- 33.Austin, S., Ziese, M. & Sternberg, N. (1981) Cell 25, 729-736. [DOI] [PubMed] [Google Scholar]
- 34.Lehnherr, H., Maguin, E., Jafri, S. & Yarmolinsky, M. (1993) J. Mol. Biol. 233, 414-428. [DOI] [PubMed] [Google Scholar]
- 35.Blakely, G., Colloms, S., May, G., Burke, M. & Sherratt, D. (1991) New Biol. 3, 789-798. [PubMed] [Google Scholar]
- 36.Begg, K. J., Dewar, S. J. & Donachie W. D. (1995) J. Bacteriol. 177, 6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, L. J. & Errington, J. (1994) Science 264, 572-575. [DOI] [PubMed] [Google Scholar]
- 38.Sternberg, N., Hamilton, D., Austin, S., Yarmolinsky, M. & Hoess, R. (1981) Cold Spring Harbor Symp. Quant. Biol. 45, 297-309. [DOI] [PubMed] [Google Scholar]
- 39.Hausmann, R. (1988) in The Bacteriophages, ed. Calender, R. (Plenum, New York) Vol. 1, pp. 259-289. [Google Scholar]
- 40.Hendrix, R. (2003) Curr. Opin. Microbiol. 6, 506-611. [DOI] [PubMed] [Google Scholar]
- 41.Moriishi, K., Koura, M., Abe, N., Fujii, N., Fujinaga, Y., Inoue, K. & Ogumad, K. (1996) Biochim. Biophys. Acta 1307, 123-126. [DOI] [PubMed] [Google Scholar]
- 42.Eklund, M. W., Poysky, F. T., Mseitif, L. M. & Strom, M. S. (1988) Appl. Environ. Microbiol. 54, 1405-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hauser, D., Gibert, M., Boquet, P. & Popoff, M. R. (1992) FEMS Microbiol. Lett. 99, 251-256. [DOI] [PubMed] [Google Scholar]
- 44.Hauser, D., Gibert, M., Marvaud, J. C., Eklund, M. W. & Popoff, M. R. (1995) Toxicon 33, 515-526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.