Abstract
Many organs are composed of branched networks of epithelial tubes that transport vital fluids or gases. The proper size and shape of tubes are crucial for their transport function, but the molecular processes that govern tube size and shape are not well understood. Here we show that three genes required for tracheal tube morphogenesis in Drosophila melanogaster encode proteins involved in the synthesis and accumulation of chitin, a polymer of N-acetyl-β-d-glucosamine that serves as a scaffold in the rigid extracellular matrix of insect cuticle. In all three mutants, developing tracheal tubes bud and extend normally, but the epithelial walls of the tubes do not expand uniformly, and the resultant tubes are grossly misshapen, with constricted and distended regions all along their lengths. The genes are expressed in tracheal cells during the expansion process, and chitin accumulates in the lumen of tubes, forming an expanding cylinder that we propose coordinates the behavior of the surrounding tracheal cells and stabilizes the expanding epithelium. These findings show that chitin regulates epithelial tube morphogenesis, in addition to its classical role protecting mature epithelia.
Keywords: branching morphogenesis, tracheal system, Drosophila, tube shape, mandril
Many organs, including the lungs, kidney, liver, and vascular system, are composed of branched networks of epithelial (or endothelial) tubes that transport vital fluids or gases, and the proper size and shape of the tubes are crucial for their transport function. Although there has been progress recently in elucidating the molecular basis of some of the early steps in the development of branched tubular networks, including branch budding and tube formation (1-4), very little is understood about the molecular processes that govern tube size and shape. The Drosophila tracheal (respiratory) system, with its simple cellular structure, an epithelial monolayer without surrounding support cells and accessible development and genetics, provides a valuable model system to address fundamental questions of branching morphogenesis, including how tube size and shape are controlled (5).
During development of the Drosophila tracheal system, branches bud sequentially from an epithelial sac consisting of ≈80 cells in each hemisegment (6). Two primary branches extend toward and fuse with branches from neighboring segments, forming the dorsal trunk that spans the length of the animal, whereas others extend and ramify on internal tissues. Once the network is established, tubes expand postmitotically to reach their characteristic lengths and diameters (7). A genetic screen identified eight genes required for tube expansion (7), four of which have been cloned (8-11). All four encode claudins or other components of septate junctions, the insect equivalent of vertebrate tight junctions (12, 13). Mutations in seven other septate junction genes also affect tube expansion (9-11, 13, 14), demonstrating that these junctions not only seal the tracheal epithelium but also regulate its size and shape. Here, we describe another molecular class of tube expansion genes that demonstrate an unexpected role in tracheal tube morphogenesis for chitin, an extracellular polymer of N-acetyl-β-d-glucosamine that is well known in its function as a scaffold for rigid insect cuticle (15).
Methods
Fly Stocks. All stocks are described at www.flybase.org and were obtained from the Bloomington Drosophila Stock Center (Indiana University, Bloomington) or Szeged Drosophila Stock Center (Szeged, Hungary), unless otherwise indicated. cystick13717, gnarled l (3)04846ex59, knk1, kkv1, Df(2L)eya10, and tigX have been described (7, 16-18). The GAL4-upstream activating sequence (UAS) system (19) was used to express transgenes in the developing tracheal system, by using breathless-GAL4 (20), UAS-GFP, and other UAS responders described below. Flies were raised on corn meal agar media, and embryos were collected on molasses or apple juice agar plates. Crosses and embryo aging were carried out at 25°C and 60% humidity.
Isolation of cystic Alleles. Male dp cn bw flies fed with 25 mM ethyl methanesulfonate (21) were mated en masse to virgin female Sco/CyO bw flies. Individual F1 males of genotype dp* cn bw/CyO bw were screened for a lethal cystic mutation by crossing to cystick13717/CyO bw females. For noncomplementing lines, balanced lines were established and presence of a mutation confirmed by complementation tests with other cystic alleles.
Genetic Mapping and DNA Sequencing. Recombination of cystick13717 chromosome with an aldpbprcnpxsp chromosome localized cystic to the dp-b interval (25A-35D5). In a subsequent mapping cross between dp cysticLM1 cn bw/CyO and wgSp-1/CyO, we recovered 47 recombinant chromosomes that were either dp+ wg+ or dp wgSp-1. Single-nucleotide polymorphisms (SNPs) that distinguish the parental chromosomes were identified in the region by genomic DNA sequencing and used to map the recombination breakpoints.
Endpoints of chromosomal deficiencies were molecularly mapped by PCR amplification of short genomic fragments located around the deletion endpoint. Endpoints of the cystic03851 deletion were determined by sequencing genomic DNA isolated by plasmid rescue of the neighboring P element insertion and confirmed by PCR mapping.
Sequencing of candidate genes was done by PCR amplification of coding exons from heterozygous genomic DNA. Screening of candidate genes for mutations by temperature gradient gel electrophoresis (Reveal Mutation Discovery platform, SpectruMedix, State College, PA) (22) was done on PCR products amplified from heterozygous genomic DNA derived from cystic alleles backcrossed to the parental dp cn bw chromosome and compared with control PCR products from dp cn bw homozygous DNA.
Misexpression of knk and kkv in Vivo. UAS-knk was generated by PCR amplification of the knk ORF in cDNA RE24065 (Drosophila Genomics Resource Center, Indiana University, Bloomington) using primers Knk-5′-NotI (5′-GCGCGGCCGCAGTTCACTCAGATCGCTTGGA) and Knk-3′-XhoI (GCCTCGAGTCTAAAGTACAACACAGTTCT). The resulting amplicon was cloned into NotI-XhoI sites of pUAST (19). UAS-kkv was made in a similar way using the RE32455 cDNA and primers Kkv-5′-NotI (GCGCGGCCGCTGGGTTTAGCGAAGACAACGA) and Kkv-3′-XhoI (GCCTCGAGATCCATCAAATCATCAATGAT). The knk-GFP genomic construct was generated by PCR of Canton-S genomic DNA using primers Knk-genomic-5′-BamHI (ATGAGAGTAAATTATAAACCATGC) and Knk-genomic-3′-HindIII (AAGCTTGTTCTGGAGCACGAGCCACGG) to amplify the knk genomic locus including ≈3 kb upstream of the transcription start site. This was ligated to a DNA fragment containing the 3′ UTR of knk obtained by PCR amplification of RE32455 cDNA using primers Knk-3′UTR-HindIII (AAGCTTTGACTGTTTTCTCATTCTTAG) and Knk-3′UTR-XhoI (CTCGAGGTGCTAAGTTACAACCTTTGCGTGT) and inserted in pBluescript. The resultant construct was digested with HindIII, and a PCR fragment containing GFP coding region was inserted in-frame at the 3′ end of the knk coding region. The entire construct was inserted into pCaSpeR4 between the BamHI and XhoI sites. Protein coding sequences of all constructs were confirmed by DNA sequencing. Constructs were introduced into y w animals by P element transformation. At least three transformant lines for each construct were tested and gave similar results. UAS-knk #6 and #9 (chromosome II insertions), UAS-kkv #6 and #9 (chromosome II), and knk-GFP #1 and #2 (chromosome III) were used here. knk-GFP #1 at 63A2 was recombined onto a ru1 h1 th1 st1 knk1 cu1 sr1 es ca1 chromosome, and resultant chromosomes containing both P[knk-GFP] and knk1 loci were used to test its rescue activity.
Immunohistochemistry, Chitin Staining, and in Situ Hybridization. Embryos were collected and fixed as described (6). Tracheal lumen was visualized by using rabbit TL-1 antisera (1:3,000), mAb 2A12 (1:5), and rabbit AS55 (1:300) (23), all of which recognize anonymous luminal antigens, or a Bacillus circulans chitinase A1 chitin-binding-domain/maltose-binding-protein fusion protein conjugated to rhodamine or fluorescein (1:500, New England Biolabs). Other primary antibodies used were: rabbit anti-Neurexin IV (1:1,000) (24), rabbit anti-Coracle (1:500) (25), rat anti-E-cadherin (1:20) (26), rabbit anti-GFP (1:500, Molecular Probes), and monoclonals anti-Crumbs (1:20) (27) and anti-Na-K ATPase α-subunit (1:300) (28). Primary antibodies were detected with biotin-conjugated secondary antibodies (Jackson ImmunoResearch) and avidin-biotin-diamino benzidine (DAB) immunohistochemistry (Vector Laboratories). Secondary antibodies conjugated to fluorescein, Cy3, or Cy5 fluorophores (Jackson ImmunoResearch) were used for laser-scanning confocal microscopy. In situ hybridization of whole-mount embryos (29) was done with digoxigenin-labeled cDNAs RE24065 (knk), RE32455 (kkv), and RE31673 (cystic).
Live Imaging of Tracheal Expansion. Embryos were collected at room temperature overnight, genotyped by GFP expression from the balancer chromosome, and staged by gut morphology. Early stage 14 embryos were mounted in a thin layer of halocarbon oil in a CoverWell imaging chamber gasket (Molecular Probes) and placed on a glass slide. A Z series of confocal fluorescence images (0.44-μm optical sections) was collected every 10 min. Laser power was adjusted to avoid saturation of the signal. Z section montages were assembled with photoshop 7.0 (Adobe Systems, San Jose, CA).
Electron Microscopy. One-hour egg lays of control dp cn bw flies, dp cysticLM1 cn bw/CyO arm-GFP and dp cysticLM42 cn bw/CyO arm-GFP flies were collected on apple juice agar plates and allowed to develop at 25°C. Embryos were dissected from the chorion, genotyped by GFP fluorescence, and staged by gut morphology. Embryos were fixed for 30 min in 4% paraformaldehyde/2% glutaraldehyde/100 mM sodium phosphate (pH 6). Vitelline membranes were dissected away and fixative removed by washing with cold phosphate buffer. Postfixation was 1% OsO4 for 2 h at 4°C, then 0.5% uranyl acetate overnight. Embryos were dehydrated in ethanol (50%, 70%, 95%, and 100%), infiltrated with EMbed 812 resin (Electron Microscopy Sciences, Fort Washington, PA), and cast into molds. Sections (70-90 nm) were cut at ≈2/3 egg length using a Leica (Deerfield, IL) Ultracut S microtome and visualized with a JEOL JEM-1230 transmission electron microscope. Images were collected with a digital camera and digital micrograph software (Gatan, Pleasanton, CA).
Results and Discussion
The extant mutation (k13717) in the cystic tube expansion gene is a recessive mutation that dramatically alters tracheal tube diameter and shape in homozygous embryos (7). cystick13717 is a P[lacZ]-induced mutation, but the identified transposon insertion is unlinked to the cystic phenotype. To facilitate genetic analysis and mapping of cystic, we screened 4,394 ethyl methanesulfonate-mutagenized chromosomes and identified 20 recessive alleles that failed to complement the lethality of cystick13717. Fifteen mutations (LM1, LM16, LM23, LM24, LM29, LM33, LM35, LM42, LM44, LM45, LM47, LM49, LM51, LM55, and LM59) severely disrupted tracheal tube morphology, showing constrictions and cyst-like expansions along the tracheal tubes in homozygous embryos, similar to the original cystic allele (Class III alleles, Fig. 1 D and E). Class II homozygotes (LM21 and LM32) displayed mild tracheal defects, with slight constrictions in the tracheal lumen (Fig. 1C). Class I homozygotes (LM7, LM15, and LM19) had normal tracheal morphology (Fig. 1B). Transheterozygous mutant combinations (LM42/LM1, LM 51/k13717, k13717/LM21, LM16/LM21, and LM16/LM15) displayed phenotypes similar to, or intermediate between, those of the corresponding homozygotes. The tracheal phenotypes of LM1, LM24, and LM42 hemizygous embryos [mutation in trans to cystic deficiency Df(2L)BSC6] were not more severe than those of the corresponding homozygotes, indicating that Class III alleles are null (amorphic) or strong loss-of-function mutations. General embryonic morphology and gut morphology appeared normal under differential interference contrast microscopy, except for an impenetrant dorsal closure defect observed in Class III homozygotes. No defects were detected in heterozygous embryos, indicating that the mutations are fully recessive.
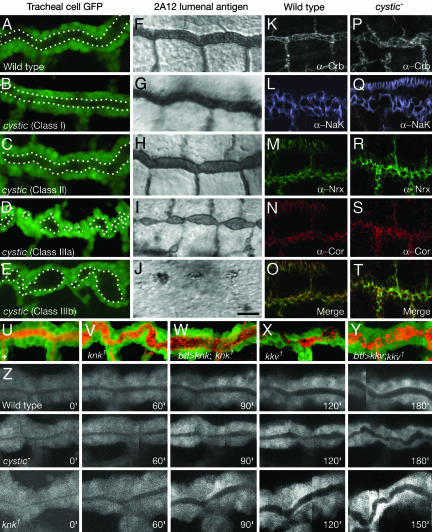
Fig. 1.
cystic, knk, and kkv mutants disrupt tracheal tube expansion. (A-T) Tracheal phenotype and marker expression in cystic mutants. (A-E) Portion of dorsal trunk in stage 16 control cystic+ embryo (A) and representative homozygous cystic mutants [LM15 (B); LM32 (C); LM1 (D); LM42 (E)]. Embryos carry btl-GAL4 and UAS-GFP transgenes to label tracheal cells with GFP. White dots outline lumen. (F-J) Same as A-E, except lumen is directly visualized by mAb 2A12 staining of similar stage embryos. Because 2A12 antigen is reduced in stronger cystic alleles, histochemical staining reactions for animals in H-J were extended to enhance the residual 2A12 staining. Class I alleles have normal tube morphology and 2A12 staining. Class II alleles show slight constrictions in tube diameter and reduced 2A12 staining. Class IIIa and IIIb mutants display severe constrictions and expansions along the length of the dorsal trunk and greatly reduce (IIIa) or almost completely eliminate (IIIb) 2A12 staining. (K-T) Longitudinal confocal optical sections through dorsal trunks of stage 16 control cystic+ (K-O) and Class III cystic- (LM1, P, R, S, T; LM42, Q) embryos immunostained for Crumbs (apicolateral membrane marker), Na+-K+ ATPase α subunit (NaK; basolateral), Neurexin IV (Nrx; septate junction) and Coracle (Cor; septate junction) as indicated. Expression of epithelial polarity markers is not altered in cystic mutants. The band of staining near the top of Q is epidermis. (U-Y) Tracheal phenotype of knk and kkv mutants. Tracheal cells are labeled with GFP (green) as in A-E. Lumen is labeled with mAb 2A12 (red). (U) Control wild-type (knk+ kkv+) dorsal trunk. (V) knk1 homozyote. Note lumen constrictions and expansions similar to Class III cystic alleles (D, E). (W) btl-GAL4, UAS-knk; knk1. Tracheal expression of Knk ameliorates knk tube morphogenesis phenotype. However, 2A12 staining is more punctate than normal. (X) kkv1 homozygote. Note lumen constrictions and expansions similar to Class III cystic alleles. (Y) btl-GAL4, UAS-kkv; kkv1. Tracheal expression of Kkv partially ameliorates kkv tube morphogenesis phenotype. (Z) Dynamic imaging of dorsal trunk expansion in wild-type, cysticLM51/k13717, and homozygous knk1 mutant embryos carrying btl-GAL4 and UAS-GFP transgenes to visualize tracheal cells. Series show an ≈3-h period beginning at stage 14 as dorsal trunk expansion begins. Because mutant tubes are contorted, each image is a montage of optical sections showing z plane of maximal luminal diameter. (Scale bar: 10 μm for A-J and U-Y, 15 μm for K-T, ≈7.5 μm for Z.)
Early events in tracheal development, including tracheal sac formation and primary branch budding and outgrowth, appeared normal in Class III cystic mutants. Defects were detected at late stage 14 (≈11 h after egg lay), when the dorsal trunk begins to dilate. Imaging of tube expansion in live embryos showed that in wild type, the dorsal trunk lumen gradually and uniformly dilates to three times its original diameter over a several-hour period (Fig. 1Z). In Class III cystic mutants, expansion occurred unevenly. Portions of the tube dilated poorly, whereas other regions expanded too much (Fig. 1Z). As expansion proceeded, the dorsal trunk became progressively more misshapen, with constricted and distended regions all along its length.
Expression of the 2A12 and AS55 luminal markers, which normally begin to be expressed just before tube expansion, was also severely compromised in cystic mutants. In wild type, 2A12 antigen first appears in the tracheal epithelium at stage 14 in cytoplasmic punctae or vesicles and then accumulates along the apical (luminal) surface during stage 15 (≈12 h after egg lay) as tubes expand (7). Although the punctate vesicular pattern was observed at stages 14 and early 15 in all cystic mutants examined, in strong alleles, 2A12 antigen accumulation at the luminal surface was markedly reduced (Class IIIa alleles; Fig. 1I) or absent (Class IIIb alleles; Fig. 1J). Likewise, AS55 antigen, which normally begins accumulating at the lumen surface at stage 14 (7), was severely reduced or absent in Class III alleles (data not shown). By contrast, expression levels of epithelial polarity markers including apicolateral Crumbs and E-cadherin, basolateral Na+-K+ ATPase, and septate junction proteins Neurexin IV, Coracle, and Fasciclin III appeared normal in cystic mutants (Fig. 1 K-T and data not shown).
cysticLM1 was mapped to a 300-kb region on chromosome II by meiotic recombination (Fig. 2A). Deficiency mapping further localized cysticLM1 to an ≈8-kb interval that overlaps the location of a 61-kb deletion adjacent to the P[lacZ] insertion on the cystic03851 chomosome, a previously undescribed P element-induced allele of cystic (G. Beitel and M.A.K., unpublished data). The interval contained two candidate genes, CG9531 and CG9535 (Fig. 2 A). Sequencing of their coding regions in cystic alleles identified deletion, nonsense, or missense mutations in CG9535 in all 15 alleles analyzed (Fig. 2B), and the severity of the molecular defect corresponded to the strength of the tracheal phenotype (see below). No changes were found in the coding region of the other candidate gene in the cystic alleles analyzed (LM1, LM16, and LM42), and only two insignificant nucleotide changes were found among ≈105 nucleotides sequenced or surveyed by temperature gradient gel electrophoresis in the five genes neighboring the candidate interval for the 17 cystic alleles analyzed. We conclude that CG9535 is cystic.
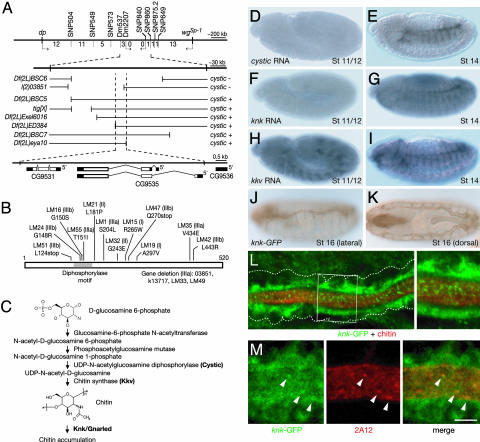
Fig. 2.
cystic, knk, and kkv encode chitin biosynthesis pathway genes expressed in developing tracheal system. (A) Recombination mapping of cystic locus. Visible markers and SNPs used for mapping in cytogenetic interval 25A-28A on chromosome II are indicated. Dotted arrows indicate the direction of cysticLM1 mutation relative to SNP. Values given are the number of chromosomes in which recombination occurred between flanking SNPs. The region between SNPs Dm537 and Dm860 is expanded below the recombination map. The structures of chromosomal deficiencies that remove portions of this region (gaps in lines) and their complementation results with cystic mutations (LM1, LM16, and LM42) are indicated. cystic interval defined by the overlapping deficiencies is expanded below showing structures of the two candidate genes (CG9531 and CG9535) in interval. (B) Changes in CG9535 (UDP-n-acetylglucosamine diphosphorylase) coding sequence in cystic alleles. Allele name, phenotypic class, and coding change are indicated. A isoform is shown; B isoform lacks residues 1-37. (C) Chitin synthesis pathway in insects highlighting steps proposed to be mediated by Cystic, Kkv, and Knk/Gnarled. (D-I) Whole-mount in situ hybridization of embryos at stages indicated showing expression patterns of cystic, knk, and kkv mRNAs. Tracheal expression is first detected at stage 11 for kkv and late-stage 12 or early 13 for cystic and knk. At stage 14 (E, G, and I), all three genes are expressed in tracheae, with highest expression levels in the dorsal trunk. kkv and knk mRNA are also expressed in several other tissues, including foregut, hindgut, and epidermis (data not shown). (J and K) Lateral (J) and dorsal (K) views of stage 16 embryo carrying P[knk-GFP] genomic rescue construct, immunostained for GFP. Knk-GFP is expressed in the dorsal trunk and, at lower levels, in the transverse connectives and lateral trunk, similar to the endogenous gene (I and data not shown). (L and M) Closeup of dorsal trunk of stage 15 embryos carrying P[knk-GFP] genomic rescue construct, double-stained for GFP and chitin (L) or GFP and 2A12 antigen (M). White box, region enlarged in (Inset at right). White dots show position of basal surface of tracheal epithelium. Note that Knk-GFP accumulates toward apical (luminal) side of cells. It colocalizes with 2A12 at apical surface (M) but does not overlap chitin (L). Note punctae of Knk-GFP at plasma membrane or just within lumen (L and Inset) that colocalize with 2A12 punctae (arrowheads in M). Images in L and M are single (0.84-μm) confocal sections. (Scale bar: ≈75 μm for D-K, 7.5 μm for L, 5 μm for M.)
cystic encodes the Drosophila homolog of UDP-N-acetylglucosamine diphosphorylase (EC 2.7.7.23), which catalyzes formation of UDP-N-acetylglucosamine and is the penultimate enzyme in the chitin synthesis pathway (15) (Fig. 2C). The enzyme is highly conserved in eukaryotes: Cystic is 51% identical to human UAP1/AGX1 (30). The structure and enzymology of this family of enzymes have been extensively studied, and residues important for substrate binding and catalysis are conserved in Cystic (30) (Fig. 5A, which is published as supporting information on the PNAS web site). The three missense mutations (LM16, LM24, and LM55) in the diphosphorylase consensus motif located in the substrate-binding pocket (Figs. 2B and 5B) and the deletion and frameshift mutations (LM33, LM47, LM49, LM51, 03851, and k13717) all have a strong tracheal phenotype, consistent with their assignment as null alleles. The other strong alleles (LM1, LM35, and LM42) alter other conserved residues in the substrate-binding site (Fig. 5 A and B). cystic is expressed in the developing tracheal system beginning at late stage 12/early 13 and continuing through stages 14 and 15 when tube expansion occurs (Fig. 2 D and E and data not shown).
gnarled is another tracheal tube expansion gene with a phenotype similar to strong cystic mutants (7). gnarled maps to cytologic interval 85E11-F16, the same region as knickkopf (knk), a gene implicated in epidermal cuticle development (16, 31). knk1 mutants displayed a tracheal tube expansion defect indistinguishable from that of gnarledex59, and knk1 failed to complement gnarledex59 for tracheal phenotype and viability (Fig. 1 V and Z and data not shown). Expression of a UAS-knk transgene in the developing tracheal system under btl-GAL4 control rescued the tracheal defects and restored viability to both gnarledex59 and knk1 mutants (Fig. 1W and data not shown). We conclude that gnarled and knk are the same gene.
knk RNA is expressed in the developing tracheal system in the same pattern as cystic (Fig. 2 F and G). knk encodes a novel protein (31). To investigate its cellular localization, we generated a genomic rescue construct with the coding sequence for GFP inserted at the 3′ end of the knk coding region. The Knk-GFP fusion gene was expressed in a similar pattern to the endogenous gene (Fig. 2 J and K) and rescued knk1 to viability. Knk-GFP localized diffusely toward the apical (luminal) surface of tracheal cells, where chitin is deposited (Fig. 2L). There were also discrete punctae of Knk-GFP at the apical surface or just within the tracheal lumen that colocalized with 2A12 punctae (Fig. 2M). Although knk is required for chitin accumulation in the lumen (see below), Knk-GFP protein localized near but did not overlap the distribution of chitin (Fig. 2L). This suggests a role for Knk in targeting or secretion of chitin or chitin synthesis enzymes.
krotzkopf verkehrt (kkv) encodes a chitin synthase homolog that, like knk, is required for epidermal cuticle development (16, 31, 32). kkv is expressed in the developing tracheal system (Fig. 2 H and I), and kkv mutants have a tracheal tube expansion defect similar to knk and cystic (Fig. 1X and data not shown). Expression of a UAS-kkv transgene in the developing tracheal system ameliorated the kkv tube expansion defect (Fig. 1Y).
The expression of chitin synthesis genes in developing tracheal cells suggested that chitin might be produced during tube expansion. Indeed, probing wild-type embryos with a rhodamine-conjugated chitin-binding protein revealed chitin in the tracheal lumen at stage 13, just before tube expansion begins (Fig. 3A). Chitin continued to accumulate throughout the period of tube expansion (Fig. 3 B-D). Confocal optical sectioning showed that chitin was not restricted to the apical epithelial surface as it is in mature tracheal tubes and other epithelia (refs. 15 and 33, and see below). Rather, it formed an expanding cylinder that filled almost the entire luminal space (Fig. 3 A-D Insets). No chitin was detected in the developing tracheal system of cystic and kkv mutants (Fig. 3 E-H). Some chitin was detected in knk mutants at the onset of tube expansion but not later in the process (Fig. 3 I and J). Thus, chitin is synthesized and forms an expanding cylinder in the lumen of developing tracheal tubes, and this process is disrupted in the expansion mutants.
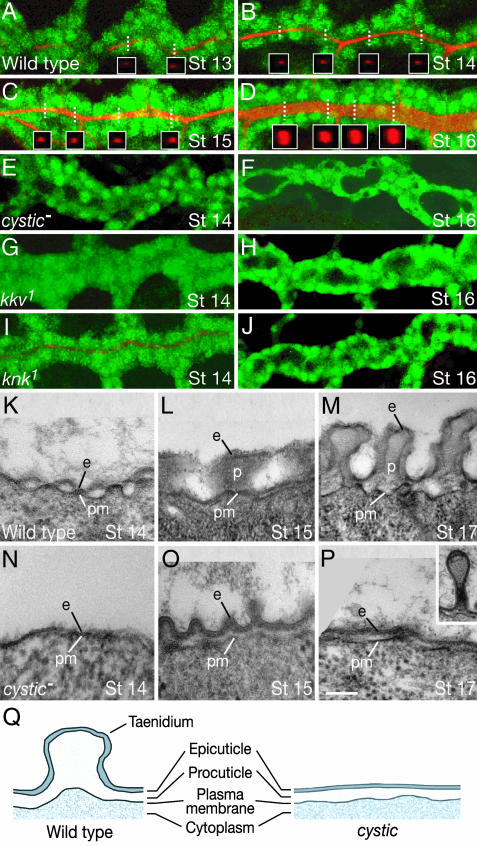
Fig. 3.
Chitin accumulation in tracheal lumen during tube expansion and effect of cystic, knk, and kkv mutations. (A-J) btl-GAL4, UAS-GFP embryos of indicated genotype and age double-stained for GFP (green) and chitin (red) and visualized by confocal microscopy. Longitudinal optical sections through dorsal trunk are shown. In wild type (A-D), chitin is first detected at late-stage 13 in discontinuous patches in dorsal trunk lumen. From stage 14 on, chitin staining is continuous and expands as the dorsal trunk dilates. By stage 15, chitin is present in all tracheal branches. Insets in A-D show reconstructed cross sections of chitin staining in the optical z axis at the x-y positions indicated by dotted white lines; note that chitin forms a solid cylinder. In cystic and kkv homozygous mutants, no chitin is detected (E-H). In knk homozygotes, some chitin is detected initially (I) but is undetectable after stage 14 (J). (K-P) Ultrastructure of trachea in wild-type and cystic mutants. (K-M) TEM cross sections through dorsal trunks of wild-type (dp cn bw) embryos at stages indicated. Epicuticle (e) begins to be deposited on the apical plasma membrane (pm) at stage 14, when the lumen is narrow. By stage 15, as the lumen expands, ridges form in the procuticle (p), and by stage 17, the ridges are recognizable as taenidia. (N-P) TEM sections through homozygous cysticLM1 dorsal trunks as above. No procuticle, large ridges, or taenidia are detected. Inset in P shows cuticular polyp that extends into lumen. These are seen occasionally at stage 17 and may represent abortive attempts at taenidia formation. (Scale bar: 10 μm for A-J, 0.2 μm for K-P.) (Q) Diagram of tracheal cuticle in wild-type and cystic mutants. Procuticle and protruding taenidia, where chitin is concentrated, are severely attenuated or absent in cystic mutants.
Analysis of tracheal tubes during the expansion period by transmission electron microscopy (TEM) did not detect the luminal chitin cylinder, presumably because this dynamic form of chitin is not preserved under the TEM fixation and staining conditions. However, the TEM analysis did reveal the presence of a thin extracellular matrix lining the apical (luminal) surface of the tracheal epithelium before expansion begins (Fig. 3K). In newly expanded tubes, small ridges formed in the matrix (Fig. 3L). These later become taenidia, the circumferential ridges in tracheal cuticle that contain a procuticle chitin core (33) (Fig. 3 M and Q). In Class III cystic mutants (LM1 and LM42), the thin matrix formed but the ridges did not, and the mature tracheal epithelium had a thin epicuticle but virtually no procuticle or taenidia (Fig. 3 N-Q). These results imply that there is a second, more conventional, form of chitin, stable to TEM fixation and staining, which begins to accumulate during or just after tube expansion and forms the taenidial rings of tracheal cuticle. We did not detect ultrastructural defects in epidermal cuticle in the cystic mutants, and only subtle defects were detected in epidermal cuticle in whole-mount cuticle preparations (data not shown); this suggests that other gene(s) provide cystic function in the epidermis, or there is a small maternal contribution of cystic that is sufficient for epidermal but not tracheal function of the gene.
Chitin is one of the most abundant biopolymers in nature. It has long been known to serve as a scaffold for the stiff cuticle that lines and protects exposed epithelia, including epidermal and tracheal epithelia (15, 33). Our results provide strong evidence that chitin also functions in epithelial morphogenesis. Chitin synthesis genes are expressed during tracheal development, and chitin accumulates throughout the lumen of tracheal tubes, forming an expanding cylinder whose growth parallels that of the surrounding epithelium. In mutants lacking chitin, expansion is uncoordinated, with some regions of the epithelium dilating poorly while others overexpand, forming grossly misshapen tubes with constrictions and dilatations all along their length. Although these results strongly implicate chitin in tracheal morphogenesis, the data do not exclude the possibility that the chitin synthesis genes have functions other than their roles in chitin synthesis that could cause or contribute to the tracheal phenotype.
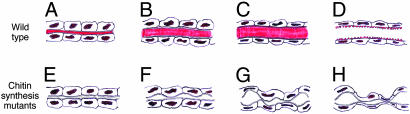
We propose that the expanding chitin cylinder in the luminal matrix coordinates the behavior of the surrounding tracheal cells and stabilizes the expanding epithelium. This ensures that tracheal tubes expand progressively and uniformly (Fig. 4). As tubes reach the appropriate size, a developmental transition must occur that arrests expansion and alters chitin structure or synthesis to form taenidial rings and the rigid cuticle that protects the apical surface of the mature tracheal epithelium. This transition might involve a change in the chitin secretion mechanism or in its assembly into fibrils or its association with matrix proteins. Ultimately, the central chitin cylinder must be degraded or remodeled and cleared along with other luminal contents as the tubes fill with gas and become functional for respiration several hours later. To test the proposed roles of chitin in the model, it will be necessary to find ways to selectively alter or abolish the different forms of chitin.
Fig. 4.
Model of chitin function in tracheal tube expansion. Schematic longitudinal sections through an expanding tracheal tube in wild type (A-D) and mutants lacking chitin (E-H). Chitin is highlighted in red. An expanding cylinder of chitin in the lumen coordinates the behavior of the surrounding tracheal cells and stabilizes the expanding epithelium (A-C). As tubes reach their appropriate size (C), a transition occurs that arrests expansion and alters chitin structure or synthesis, forming taenidial rings (ridges in apical extracellular matrix) in the rigid tracheal cuticle that lines and protects the epithelium (D). The central chitin cylinder and other luminal contents are degraded and removed by the time the tubes fill with gas several hours later. In mutants lacking chitin, early steps in tube morphogenesis are normal (E), but expansion occurs unevenly, and the tubes become progressively more misshapen (F-H). Cuticle forms but lacks taenidia.
Although chitin is rare in vertebrates (34, 35), many vertebrate tubes contain related carbohydrates or uncharacterized fibrillar material at their apical surface or throughout the lumen (36-39). Although it is not known whether these substances function in tube morphogenesis, there is evidence that at least some of these molecules, including chondroitin sulfate and hyaluronic acid, can influence related morphogenesis processes such as epithelial invagination (40-42). There is also a long history in the field of tissue engineering of using cylinders (“mandrils”) made of synthetic and/or biological materials as templates for constructing and shaping blood vessels for clinical use (43). It will be important to investigate the structure and function of the luminal contents of developing vertebrate tubes to determine whether they contain cylinders like chitin and engineered mandrils that coordinate the morphogenesis of the surrounding epithelium.
Supplementary Material
Acknowledgments
We thank Greg Beitel for advice, discussions, and strains; SpectruMedix for temperature gradient electrophoresis analysis; John Perrino for EM help; and Tom Bunch (University of Arizona, Tucson), Nancy Bonini (University of Pennsylvania, Philadelphia), and Tadashi Uemura (Kyoto University) for strains and reagents. This work was supported by a grant from the National Institutes of Health (to M.A.K.) and fellowships from the American Heart Association (to W.P.D.), the National Science Foundation (to B.L.), and the Human Frontier Science Program (to S.L.). M.A.K. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: W.P.D., B.L., K.S., S.L., L.M., and M.A.K. designed research; W.P.D., B.L., K.S., S.L., and L.M. performed research; W.P.D., B.L., K.S., S.L., L.M., and M.A.K. analyzed data; and W.P.D., B.L., K.S., S.L., and M.A.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SNP, single-nucleotide polymorphism; TEM, transmission electron microscopy.
References
- 1.Hogan, B. L. & Kolodziej, P. A. (2002) Nat. Rev. Genet. 3, 513-523. [DOI] [PubMed] [Google Scholar]
- 2.O'Brien, L. E., Zegers, M. M. & Mostov, K. E. (2002) Nat. Rev. Mol. Cell. Biol. 3, 531-537. [DOI] [PubMed] [Google Scholar]
- 3.Affolter, M., Bellusci, S., Itoh, N., Shilo, B., Thiery, J. P. & Werb, Z. (2003) Dev. Cell 4, 11-18. [DOI] [PubMed] [Google Scholar]
- 4.Lubarsky, B. & Krasnow, M. A. (2003) Cell 112, 19-28. [DOI] [PubMed] [Google Scholar]
- 5.Ghabrial, A., Luschnig, S., Metzstein, M. M. & Krasnow, M. A. (2003) Annu. Rev. Cell. Dev. Biol. 19, 623-647. [DOI] [PubMed] [Google Scholar]
- 6.Samakovlis, C., Hacohen, N., Manning, G., Sutherland, D. C., Guillemin, K. & Krasnow, M. A. (1996) Development (Cambridge, U.K.) 122, 1395-1407. [DOI] [PubMed] [Google Scholar]
- 7.Beitel, G. J. & Krasnow, M. A. (2000) Development (Cambridge, U.K.) 127, 3271-3282. [DOI] [PubMed] [Google Scholar]
- 8.Behr, M., Riedel, D. & Schuh, R. (2003) Dev. Cell 5, 611-620. [DOI] [PubMed] [Google Scholar]
- 9.Paul, S. M., Ternet, M., Salvaterra, P. M. & Beitel, G. J. (2003) Development (Cambridge, U.K.) 130, 4963-4974. [DOI] [PubMed] [Google Scholar]
- 10.Llimargas, M., Strigini, M., Katidou, M., Karagogeos, D. & Casanova, J. (2004) Development (Cambridge, U.K.) 131, 181-190. [DOI] [PubMed] [Google Scholar]
- 11.Wu, V. M., Schulte, J., Hirschi, A., Tepass, U. & Beitel, G. J. (2004) J. Cell Biol. 164, 313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knust, E. & Bossinger, O. (2002) Science 298, 1955-1959. [DOI] [PubMed] [Google Scholar]
- 13.Wu, V. M. & Beitel, G. J. (2004) Curr. Opin. Cell. Biol. 16, 493-499. [DOI] [PubMed] [Google Scholar]
- 14.Hemphala, J., Uv, A., Cantera, R., Bray, S. & Samakovlis, C. (2003) Development (Cambridge, U.K.) 130, 249-258. [DOI] [PubMed] [Google Scholar]
- 15.Merzendorfer, H. & Zimoch, L. (2003) J. Exp. Biol. 206, 4393-4412. [DOI] [PubMed] [Google Scholar]
- 16.Jurgens, G., Wieschaus, E., Nusslein-Volhard, C. & Kluding, H. (1984) Wilhelm Roux's Arch. Dev. Biol. 193, 283-295. [DOI] [PubMed] [Google Scholar]
- 17.Bonini, N. M., Leiserson, W. M. & Benzer, S. (1998) Dev. Biol. 196, 42-57. [DOI] [PubMed] [Google Scholar]
- 18.Bunch, T. A., Graner, M. W., Fessler, L. I., Fessler, J. H., Schneider, K. D., Kerschen, A., Choy, L. P., Burgess, B. W. & Brower, D. L. (1998) Development (Cambridge, U.K.) 125, 1679-1689. [DOI] [PubMed] [Google Scholar]
- 19.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401-415. [DOI] [PubMed] [Google Scholar]
- 20.Shiga, Y., Tanaka-Matakatsu, M. & Hayashi, S. (1996) Dev. Growth Differ. 38, 99-106. [Google Scholar]
- 21.Ashburner, M. (1989) Drosophila (Cold Spring Harbor Laboratory, Plainview, NY).
- 22.Li, Q., Liu, Z., Monroe, H. & Culiat, C. T. (2002) Electrophoresis 23, 1499-1511. [DOI] [PubMed] [Google Scholar]
- 23.Reichman-Fried, M., Dickson, B., Hafen, E. & Shilo, B. Z. (1994) Genes Dev. 8, 428-439. [DOI] [PubMed] [Google Scholar]
- 24.Baumgartner, S., Littleton, J. T., Broadie, K., Bhat, M. A., Harbecke, R., Lengyel, J. A., Chiquet-Ehrismann, R., Prokop, A. & Bellen, H. J. (1996) Cell 87, 1059-1068. [DOI] [PubMed] [Google Scholar]
- 25.Fehon, R. G., Dawson, I. A. & Artavanis-Tsakonas, S. (1994) Development (Cambridge, U.K.) 120, 545-557. [DOI] [PubMed] [Google Scholar]
- 26.Oda, H., Uemura, T., Harada, Y., Iwai, Y. & Takeichi, M. (1994) Dev. Biol. 165, 716-726. [DOI] [PubMed] [Google Scholar]
- 27.Tepass, U., Theres, C. & Knust, E. (1990) Cell 61, 787-799. [DOI] [PubMed] [Google Scholar]
- 28.Lebovitz, R. M., Takeyasu, K. & Fambrough, D. M. (1989) EMBO J. 8, 193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tautz, D. & Pfeifle, C. (1989) Chromosoma 98, 81-85. [DOI] [PubMed] [Google Scholar]
- 30.Peneff, C., Ferrari, P., Charrier, V., Taburet, Y., Monnier, C., Zamboni, V., Winter, J., Harnois, M., Fassy, F. & Bourne, Y. (2001) EMBO J. 20, 6191-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ostrowski, S., Dierick, H. A. & Bejsovec, A. (2002) Genetics 161, 171-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moussian, B., Schwarz, H., Bartoszewski, S. & Nusslein-Volhard, C. (2005) J. Morphol. 264, 117-130. [DOI] [PubMed] [Google Scholar]
- 33.Noirot, C. & Noirot-Timothee, C. (1982) in Insect Ultrastructure, eds. King, R. C. & Akai, H. (Plenum, New York), Vol. 1, pp. 351-381. [Google Scholar]
- 34.Wagner, G. P., Lo, J., Laine, R. & Almeder, M. (1993) Experientia 49, 317-319. [Google Scholar]
- 35.Semino, C. E., Specht, C. A., Raimondi, A. & Robbins, P. W. (1996) Proc. Natl. Acad. Sci. USA 93, 4548-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pries, A. R., Secomb, T. W. & Gaehtgens, P. (2000) Pflügers Arch. 440, 653-666. [DOI] [PubMed] [Google Scholar]
- 37.Martins Mde, F. & Bairos, V. A. (2002) Int. Rev. Cytol. 216, 131-173. [DOI] [PubMed] [Google Scholar]
- 38.Solursh, M. & Morriss, G. M. (1977) Dev. Biol. 57, 75-86. [DOI] [PubMed] [Google Scholar]
- 39.Folkman, J. & Haudenschild, C. (1980) Nature 288, 551-556. [DOI] [PubMed] [Google Scholar]
- 40.Lane, M. C., Koehl, M. A., Wilt, F. & Keller, R. (1993) Development (Cambridge, U.K.) 117, 1049-1060. [DOI] [PubMed] [Google Scholar]
- 41.Haddon, C. M. & Lewis, J. H. (1991) Development (Cambridge, U.K.) 112, 541-550. [DOI] [PubMed] [Google Scholar]
- 42.Hwang, H. Y., Olson, S. K., Esko, J. D. & Horvitz, H. R. (2003) Nature 423, 439-443. [DOI] [PubMed] [Google Scholar]
- 43.Daly, C. D., Campbell, G. R., Walker, P. J. & Campbell, J. H. (2004) Front. Biosci. 9, 1915-1924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.