Abstract
R2 is a site-specific non-long terminal repeat (non-LTR) retrotransposon encoding a single polypeptide with reverse transcriptase, DNA endonuclease and nucleic acid-binding domains. The current model of R2 retrotransposition involves an ordered series of cleavage and polymerization steps carried out by at least two R2 protein subunits, one bound upstream and one bound downstream of the integration site. The role in the retrotransposition reaction of two conserved DNA-binding motifs, a C2H2 zinc finger (ZF) and a Myb motif, located within the N-terminal domain of the protein are explored in this report. These motifs do not appear to play a role in RT or the ability of the protein to bind the R2 RNA transcript. Methylation and missing nucleoside interference-based DNA footprints using polypeptides to the N-terminal domain suggest the ZF and Myb motifs bind to regions −3 to −1 and +10 to +15 with reference to the insertion site. Mutations in these DNA sites or of the N-terminal protein domain blocked binding and the activity of the downstream subunit. Mutations of the protein domain also affected binding of the upstream subunit but not its function, suggesting the primary path to DNA target recognition by R2 involves both upstream and downstream subunits.
INTRODUCTION
Transposable elements have played a significant role in determining the current structure and expression of eukaryotic genomes. One of the most abundant classes of these elements, the non-long terminal repeat (non-LTR) retrotransposons, utilize a simple integration mechanism of reverse transcribing RNA templates directly onto a nick in chromosomal DNA. R2 is a site-specific non-LTR retrotransposon found in 28S rRNA genes of a diverse set of eukaryotes (1,2). R2 retrotransposition is highly specific for both the 28S DNA target site and the R2 RNA template, which has enabled detailed studies of its mechanism of integration (3,4).
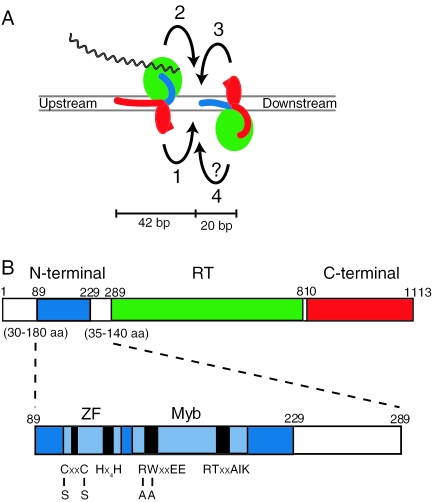
Recent studies of R2 retrotransposition have led to the model in which an R2 homodimer (or possibly larger multimer) asymmetrically bound to target DNA affects integration of the element through a series of ordered catalytic steps (Figure 1A) (5). One subunit binds the DNA region centered 25 bp upstream of the R2 integration site (6), as well as the 3′ end of the R2 RNA transcript to be used for RT (3,5–7). A second R2 subunit binds the DNA region from the integration site to 15 bp downstream of this site (5).
Figure 1.
R2 protein structure and model of retrotransposition. (A) R2 retrotransposition model. The upstream subunit binds to DNA by means of the C-terminal domain of the R2 protein (red arm), and the downstream subunit binds using the N-terminal domain (blue arm). Integration occurs in four ordered steps. The endonuclease of the upstream subunit (red oval) cleaves the bottom DNA strand (1). The reverse transcriptase (green) of the upstream subunit uses the cleaved bottom DNA strand to prime TPRT of the R2 RNA transcript (black wavy line) (2). The endonuclease of the downstream subunit cleaves the top DNA strand (3). The reverse transcriptase of the downstream subunit uses the cleaved top DNA strand to prime synthesis of the second strand displacing the element RNA from the RNA/DNA heteroduplex (4). This fourth step is questioned because it has not been observed in vitro. (B) The R2 ORF. The three domains of the R2 ORF have been colored as in (A). The amino acid positions are those of the R2 protein from B.mori. The variability in the lengths of the protein flanking the conserved N-terminal domain of different R2 proteins is shown. Virtually no length variation exists elsewhere in comparisons of different R2 proteins (14). Shown below is a more detailed diagram of the B.mori R2 N-terminal domain studied in this report. The N-terminal region (blue) contains two nucleic acid-binding domains, a ZF and a Myb. The boundaries of the ZF and Myb domains are shown in light blue with the conserved residue regions shown in black. The residues mutated to make the ZF− and Myb− proteins are shown.
R2 retrotransposition is proposed to proceed via the following steps: (i) the endonuclease of the upstream monomer cleaves the first (bottom) DNA strand, (ii) the reverse transcriptase of the upstream monomer uses the free 3′ OH from the newly created nick to initiate target-primed reverse transcription (TPRT) using the R2 RNA as the template, (iii) the downstream monomer cleaves the second (top) DNA strand, and (iv) the second DNA strand is synthesized. It is not known if R2 or cellular DNA polymerases are responsible for the fourth step, however, the R2 reverse transcriptase is capable of displacing RNA from nucleic acid templates and the second subunit is likely to be in the correct orientation to perform second strand synthesis (5,8) (A. Bibillo and T.H. Eickbush, unpublished data). The basic steps of this TPRT reaction appear to be part of the integration reaction of other non-LTR retrotransposons (9,10) as well as in the integration of SINEs (Alu) and processed pseudogenes (11,12). TPRT is also a critical step in the retrohoming of group II introns (13).
The single open reading frame (ORF) of R2 elements from diverse animals (Figure 1B) contains a central reverse transcriptase domain, a C-terminal domain with a restriction-like endonuclease, and a N-terminal domain with both C2H2 zinc finger (ZF) and Myb-like nucleic acid-binding motifs (14,15). We have shown previously that a 140 amino acid N-terminal polypeptide containing the ZF and Myb motifs accounts for most of the DNase I footprint of the second protein subunit bound to the target site (5). This study further examines the role of the N-terminal Myb and ZF motifs in the retrotransposition of R2. Protein mutagenesis, DNA mutagenesis, more detailed DNA footprint approaches, and assays of the polymerization activities of the R2 were used to examine the involvement of the R2 N-terminal ZF and Myb motifs in DNA-binding, RNA-binding and enzymatic functions of the R2 protein.
MATERIALS AND METHODS
Mutagenesis and purification of full-length R2 protein and peptides
R2 protein was purified to ∼40% homogeneity as described previously (4,6). The ZF mutation (114C/S + 117C/S) and the Myb mutation (151R/A + 152W/A) (see Figure 1B) were generated by QuickChange Site-Directed Mutagenesis (Stratagene) of the R2 expression construct pR260 using primers with point mutations in the appropriate codons (4). The new constructs were named (ZF−)pR260 and (Myb−)pR260. The 3′-untranslated region (3′-UTR) RNA substrate was generated as described previously (6). Complementary DNA oligonucleotides containing the base transversions listed in Figure 4 were used to generate the mutant DNA-binding sites. The oligos spanned from 70 bp upstream of the R2 insertion site to 30 bp downstream.
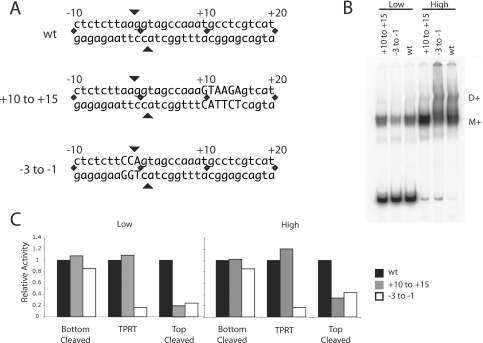
Figure 4.
Mutation of the target DNA at the ZF and Myb-binding sites. (A) DNA substrates used in the binding, cleavage and TPRT reactions. The DNA substrate extended from 70 bp upstream to 30 bp downstream but only the sequences near the R2 insertion site are shown. Mutations of the +10 to +15 region (Myb-binding site) and −3 to −1 (ZF-binding region) are indicated by capital letters. All mutations involved G–T and A–C transversions. (B) Gel shifts of the full-length R2 protein bound to mutant DNA substrates at low (36 fmol) and high (360 fmol) protein concentrations in the absence of dNTPs. Eighty fmol of the DNA substrate and 430 fmol of R2 RNA were present in each binding assay. M+, protein monomer with RNA; D+, protein dimer plus RNA. (C) DNA cleavage and TPRT activity of the R2 protein on the wild-type (WT), +10 to +15 mutant and −3 to −1 mutant DNA substrates. The incubation conditions were identical to that in (B). After the incubation the fraction of DNA cleaved was determined on denaturing polyacrylamide gels, while EMSA gels were used to the calculate the fraction of DNA bound. Cleavage reactions were conducted in both the presence and absence of dNTPs and the results averaged. Activity is reported as per bound unit of DNA with the activity observed on the WT DNA substrate set at 100% (black bar). Gray bars, data for the +10 to +15 mutant DNA substrate; white bars, data for the −3 to −1 DNA substrate.
DNA corresponding to the 89–229 and 89–289 polypeptides were generated by PCR using the pR260 plasmid as the template DNA (4). The PCR fragment was cloned into the pET28a vector (Novagen) with the His6 tag in-frame at the amino terminus. The pET28a construct was placed into BL21(DE3) RIL codon + bacteria (Stratagene) for inducible expression with Isopropyl-β-d-thiogalactopyranoside (IPTG). The construct for the ZF− polypeptide was generated in an identical fashion except that the DNA template used to generate the PCR fragment was the ZF− pR260 plasmid. The expressed 89–229 and 89–289 polypeptides were purified over talon resin columns to near 90% homogeneity.
Enzymatic assays of reverse transcriptase activities
Wild-type, ZF− and Myb− R2 proteins were assayed for processivity, their ability to polymerize through duplex nucleic acids, and end-to-end template jumping as described previously (8,16) (see Supplementary Data).
DNA-binding and footprinting assays
Substrate DNAs were 100 bp and spanned from 50 bp upstream to 50 bp downstream or from 70 bp upstream to 30 bp downstream of the R2 insertion site. The former substrate was used to footprint the bottom-strand (Figures 2 and 3) and in Figures 5 and 6. The latter substrate was used to footprint the top-strand (Figures 2 and 3) and in Figure 4. R2 protein-binding, cleavage, and TPRT assays were performed in 50 mM Tris–HCl (pH 8.0), 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, 11% glycerol, 0.1 mg/ml BSA, 0.01% Triton X-100, and with or without 25 µM dNTP as described previously (5). Reactions involving full-length R2 protein were carried out at 37°C for 30 min while reactions involving R2 polypeptides were carried out at 37°C for 15 min. Methylation interference footprints and missing nucleoside footprints were performed following established protocols (17–19) with a modification level of one modification per DNA molecule. The DNA-binding reactions (400 fmol DNA) used for footprint analysis contained 35–65% of the DNA substrate bound by protein with bound DNA separated from free DNA on a native polyacrylamide gels as described previously (5,6).
Figure 2.
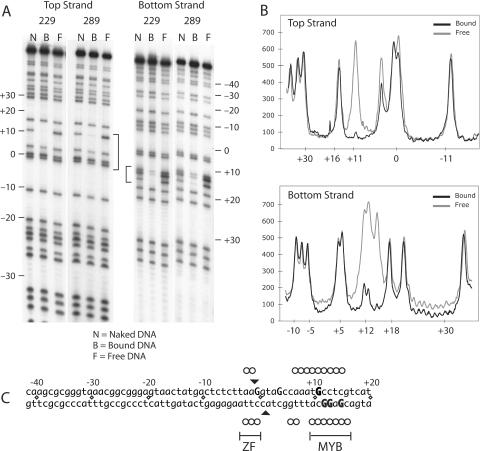
Methylation interference footprints of N-terminal R2 polypeptides. (A) Methylation interference footprints. The lanes are marked N, for naked DNA (no polypeptide in reaction); B, for the polypeptide bound fraction of DNA; and F, the fraction of DNA that did not bind the polypeptide (free). Footprint analysis was done either with the top-strand or the bottom-strand labeled on the 5′ end. The numbers along the gel represent the relative position of phosphate/nucleotide position on the target DNA with respect to the central R2 cleavage dyad. Negative numbers are given to phosphate positions upstream of the insertion site, as conventionally written, and positive numbers are positions downstream of the insertion site see (C). Bands in the N lanes correspond to all G residues in the sequence. Brackets between the two panels identify the footprint region. (B) Densitometry scans of the footprinted region for the 229 polypeptide footprint in (B). Top and bottom-strands are reported in the 3′-5′ direction. Black lines, scans of bound B lanes, gray lines, scans of unbound F lanes. Backbone phosphates are numbered on the x-axis. (C) DNA sequence of the 28S rRNA gene region bound by R2 proteins. Black triangles are the positions of the R2 cleavage sites on the two strands. Guanosine residues interfering with the ability of the polypeptide to bind to target DNA when methylated are indicated in uppercase (bold text, strongly interfering sites; plain text, weakly interfering sites). Missing nucleoside interference footprints identified in Figure 3 are indicated on the target DNA sequence by open circles.
Figure 3.
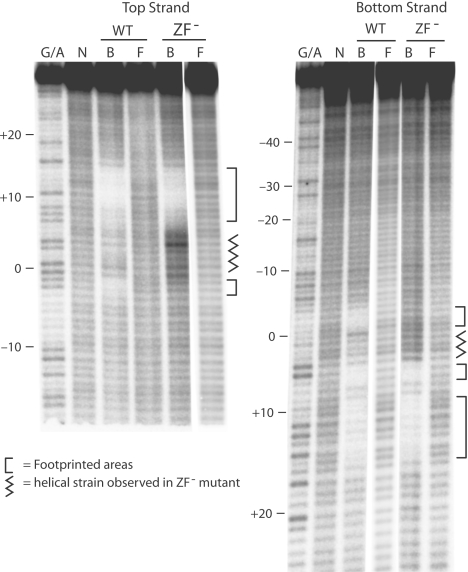
Missing nucleoside footprints of N-terminal R2 polypeptides. Missing nucleoside footprint of WT and ZF− 229 polypeptides. The footprint analysis was conducted separately for the top or bottom-strands labeled at their 5′ end. G/A lanes, guanosine plus adenine DNA ladder; B, polypeptide bound fraction of DNA; and F, fraction of DNA that did not bind the polypeptide (free).
Figure 5.
EMSA of WT, Myb− and ZF− R2 proteins. Each reaction contained ∼13 fmol of top-strand labeled substrate DNA, 430 fmol of RNA and 13 to 120 fmol of WT, Myb− or ZF− R2 protein (triangles). The protein–DNA–RNA complexes formed with WT protein have been described previously (5) and correspond to M+, protein monomer with RNA; D-, protein dimer without RNA; D+, protein dimer with RNA; and ΔM+, protein monomer with RNA after both DNA strands are cleaved.
Figure 6.
DNA cleavage and TPRT activities of the Myb− mutant R2 protein. In each panel the WT and Myb− R2 proteins are compared at three protein concentrations. The proteins were equilibrated by RT activity in standard primer extension reactions (Materials and Methods). The diagrams compare the ability of the WT (black squares) and Myb− (gray diamonds) proteins to bind target DNA in EMSA assays (A), cleave the DNA on the bottom-strand (B), perform TPRT (C) and cleave the DNA on the top-strand (D). DNA-binding is reported as the fraction of DNA bound by protein as measured by EMSA gels. DNA cleavage is reported as the fraction of DNA cleaved divided by the fraction of bound DNA. TPRT is reported as the fraction of DNA that had undergone TPRT divided by the fraction of DNA that had been cleaved on the bottom-strand.
RESULTS
Binding of the N-terminal R2 polypeptides to target DNA
Comparison of the R2 ORF from divergent arthropods (14) revealed sequences with similarity to ZF and Myb-binding motifs near the N-terminus of the protein encoded by all R2 elements (residues 89 to 229 in Figure 1B). In the most abundant lineage of insect R2 elements the length of the ORF N-terminal of the conserved segment varied from 30 to 180 residues, while the length between the conserved N-terminal segment and the first conserved region of the RT domain varied from 35 to 140 amino acid. Comparison of these sequences from different R2 elements suggested that there had been little selective constraint on either of these regions of the encoded proteins (14,20,21).
Polypeptides corresponding to the 140 amino acid conserved region of the Bombyx mori R2 element (4) and a 200 amino acid peptide that also included the 60 amino acid connection to the RT domain were synthesized. These polypeptides were designated by the last amino acid residue included in the polypeptide: 229 and 289, respectively. The 229 and 289 polypeptides could be shown to bind to target DNA in electrophoretic mobility shift assays (EMSA) (Supplementary Figure 1A).
To determine the location of the contact points between the N-terminal peptides and the target DNA N7-methylated guanosine interference footprints were conducted. Figure 2A compares three fractions of DNA: methylated DNA that had not been exposed to the polypeptide (naked DNA and lanes labeled N), methylated DNA that had been incubated with the polypeptide and then fractionated into bound (labeled B) and free (labeled F) fractions on EMSA gels. In this assay, N7-methylated guanosines that interfere with the ability of the polypeptide to bind to the major groove of the target DNA should be under-represented in the bound fraction and over-represented in the free fraction. Scans of the B and F lanes of the 229 polypeptide footprint are presented in Figure 2B. In the various panels of this figure, the position of the DNA backbone phosphates have been numbered relative to the R2 cleavage dyad with negative numbers corresponding to sequences upstream of the DNA cleavage site (with respect to the transcription of the 28S gene), and positive numbers representing downstream sequences (see Figure 2C).
On the bottom-strand there were interfering guanosine residues at nucleotide positions +15, +13 and +12. On the top-strand there was one strongly interfering guanosine residue at position +11 and two weaker at positions −1 and +4. The methylation interference footprints of the two polypeptides suggested that the R2 peptides bound to the DNA in the major groove both near the cleavage sites and around position +13. The 229 and 289 polypeptides yielded similar footprints suggesting the protein sequences downstream of the ZF and Myb motifs had little affect on DNA-binding. Each of the methylated guanosine residues which interfered with the ability of the polypeptides to bind to target DNA has been marked as an uppercase ‘G’ on the DNA sequence presented in Figure 2D.
As a second approach to monitor protein/DNA contacts between target DNA and the N-terminal domain of the R2 protein, missing nucleoside footprints were carried out using the 229 polypeptide. Like the methylation footprint discussed above, the missing nucleoside footprint is an interference-based assay except that instead of blocking binding via a methyl group in the major groove, random nucleosides are removed from the target DNA leading to strand cleavage (17). Again bound (B) and free (F) fractions of the modified DNA were compared to DNA not exposed to the protein (N). As shown in Figure 3 there were two regions of interference located on the top-strand from −3 to −2 and from +10 to +15, as well as two regions on the bottom-strand from −3 to −1 and from +10 to +16. There was moderate interference at positions +7, +8 and +9 on the top-strand and +6 and +7 on the bottom-strand. The positions of these interfering nucleosides are marked with open circles on the DNA target sequence in Figure 2C.
Because ZF and Myb-binding domains bind short regions of a DNA helix (22,23), the ZF and Myb motifs of the R2 protein were expected to each bind one of the two separate zones of footprinting revealed by the methylated-G or missing nucleoside experiments This supposition was directly tested by generating a polypeptide in which the C2H2 motif of the ZF was changed to S2H2 (see Figure 1B). The mutant (ZF−) polypeptide readily bound target DNA (see Supplementary Figure 1) and footprinted by missing nucleoside interference (Figure 3). Of the two major footprints only the region from +6 to +16 was detected with the ZF− polypeptide. This finding suggested that in the normal (wild-type) peptide the region from −3 to −1 makes contact with the ZF motif, while the region between +10 and +16 makes contact with the Myb domain. It is interesting to note that DNA targets with missing nucleosides in the region between 0 and +6 on the top or bottom-strand were preferentially bound by the ZF− polypeptide. This finding suggests that binding of the Myb domain introduces stress into the DNA helix that is relieved by binding of the ZF motif or by removal of an upstream nucleoside.
Mutating the target DNA at the ZF or Myb-binding sites
As an alternative approach to study the interaction of the R2 protein with its target DNA the two binding sites identified in Figures 2 and 3 were mutated. As shown in Figure 4A, DNA targets were generated containing transversions in the region from −3 to −1 or from +10 to +15. The binding of full-length R2 protein to these mutated target sites was then compared relative to that of the original DNA target (Figure 4B). The binding reactions were conducted at a low protein concentration (20% of the wild-type target bound) where most of the complexes corresponded to protein monomers, and at a high protein concentration (100% of wild-type target bound) where the complexes corresponded to both protein monomers and dimers. At the low protein concentration binding of the R2 protein to the +10 to +15 mutant DNA was similar to that of the wild-type DNA, while binding to the −3 to −1 mutant DNA was slightly reduced. At the high protein concentration, the +10 to +15 DNA mutant supported only low levels of dimer formation compared to wild-type, while the −3 to −1 DNA mutant generated higher levels of a continuous smear extending from the position of the monomer complex to the well of the gel.
In Figure 4C the DNA cleavage and TPRT activities associated with R2 integration are compared on the three target DNAs. Enzymatic activities were assayed after the reaction by separation of the purified DNA products on denaturing gels (5). All activities are presented relative to the amount of DNA bound by protein and with the activity supported by the wild-type DNA target set at 100%. In the case of the +10 to +15 mutant DNA, bottom-strand cleavage and TPRT reactions were conducted at levels similar to the wild-type DNA target, while top-strand cleavage was significantly reduced. These results support the model that one protein subunit binds upstream of the integration site and conducts both the bottom-strand cleavage and TPRT, while a second subunit binds downstream of the integration site and cleaves the top DNA strand.
In the case of the −3 to −1 mutant DNA target, interpretation of the results are more complex because these mutations are near the integration site. The −3 to −1 mutant DNA target also supported high levels of bottom-strand cleavage, while top-strand cleavage was reduced to levels similar to that supported by the +10 to +15 mutant DNA. Unlike the +10 to +15 mutant DNA, the TPRT reaction supported by the −3 to −1 mutant DNA was only 20% of the level of the other DNA targets. It is not possible to resolve whether this decrease was due to an involvement of the ZF motif in the TPRT reaction or whether the DNA mutations interfered with the ability of the RT domain to utilize the cleaved target DNA as primer.
Mutating the ZF and Myb protein motifs: reverse transcriptase functions
To evaluate the role of the R2 N-terminal domain in a integration reaction point mutations were generated in either the ZF domain (114C/S + 117C/S) or the Myb domain (151R/A + 152W/A) of the full-length R2 protein (see Figure 1B). The ZF mutations should eliminate the binding of a Zn++ cation and thus disrupt the structure of the motif (22). The Myb mutations involved two highly conserved residues (14) one corresponding to a large hydrophobic residue found in all characterized Myb motifs. Mutation of this hydrophobic residue has been shown to disrupt DNA-binding by Myb proteins (23,24). both ZF and Myb mutations were shown to have significant effects on DNA-binding (next section).
In order to determine if these mutations affected RNA-binding the catalytic properties of the reverse transcriptase of the mutant proteins (ZF− and Myb−) were compared to WT protein (WT). The R2 reverse transcriptase exhibits several catalytic properties that distinguish it from retroviral reverse transcriptases: (i) high processivity, (ii) the ability to polymerize through duplexed nucleic acid regions (strand displacement) and (iii) homology independent template switching (end-to-end jumping) (8,16,25). These unique properties of the R2 reverse transcriptase were hypothesized to be the result of an extended protein/RNA template interface, which would increase the stability of the polymerization complex and destabilize duplex regions of the RNA ahead of the active site. The ZF and Myb N-terminal nucleic acid-binding motifs are possible candidates for these extended protein–RNA template interactions. Direct comparison of the mutant proteins to wild-type protein suggested that the N-terminal domain does not interact with the RNA template during RT (see Supplementary Figure 2).
Mutating the ZF and Myb protein motifs: DNA-binding and cleavage functions
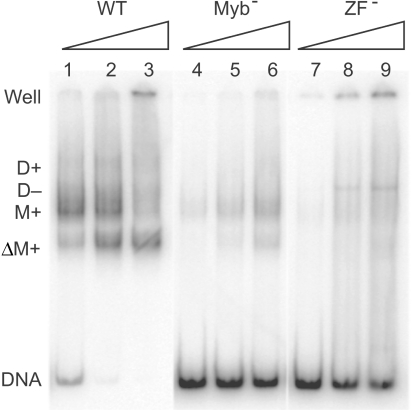
We next tested whether the mutations in the ZF and Myb motifs affected the ability of the full-length R2 protein to bind target DNA. These studies were conducted in the presence of the 250 nt 3′-UTR RNA to mimic a TPRT reaction. As described previously (5) the protein–DNA complexes observed with the WT protein (lanes 1–3) included protein monomers with the RNA (M+), protein dimers with (D+) or without (D−) the RNA, and protein monomers with RNA after double-stranded cleavage of the target DNA (ΔM+). In the latter complex, the downstream DNA and protein had disassociated from the protein–upstream DNA complex. Protein complexes remaining within the ‘well’ of the gel could also be observed at the highest protein concentration, which was a result of the excess R2 protein-binding to two target DNA molecules thus forming large protein:DNA networks.
The DNA–protein complexes formed by the Myb− protein (lanes 4–6) was significantly less than that of the wild-type protein with only about 50% of the DNA bound in complexes even at the highest protein concentration. A defined monomer complex but no dimer complex was observed. In the case of the ZF− protein (lanes 7–9), the level of complex formation was again significantly reduced relative to WT protein, and most of the bound DNA migrated as a diffuse smear from the ‘well’ down to the unbound DNA. The only distinct complex appeared about the position of a D-complex. The diffuse smear generated by the ZF− protein indicated an inability of this protein to form stable complexes with the target DNA. It is likely that the misfolded ZF motif destabilized the R2 protein conformations required for specific monomer and dimer formation and thus a large set of unstable protein–DNA complexes were formed.
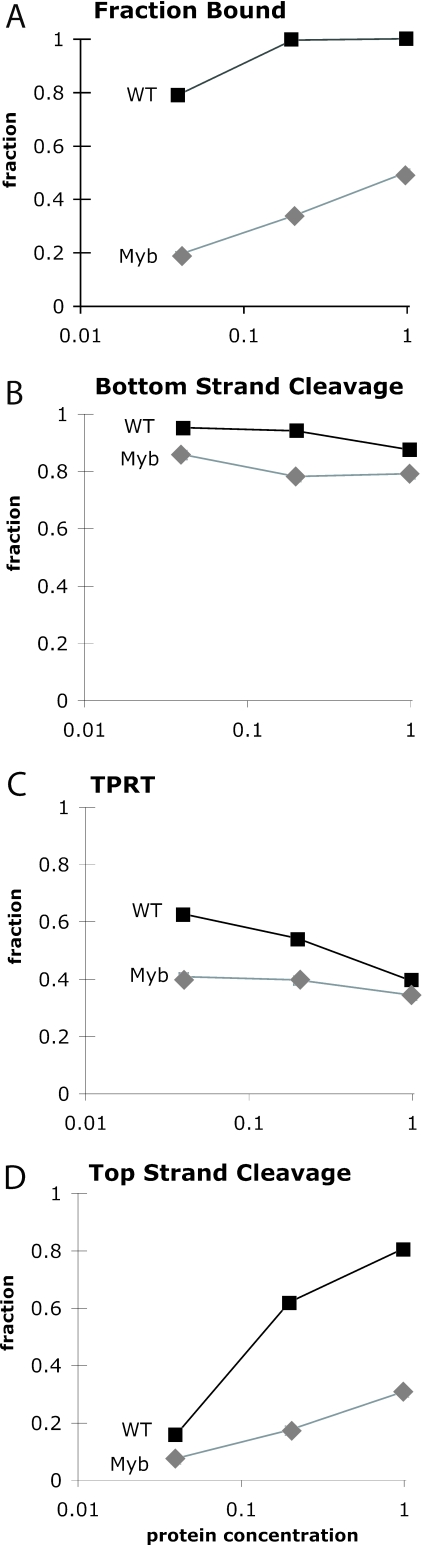
The Myb− protein was next tested for its ability to conduct the various enzymatic steps involved in the R2 integration reaction (Figure 6). The levels of WT and Myb− protein were equilibrated by their RT activity in primer extension assays (see previous section). The ZF− protein was also tested, but this mutant protein gave only low levels of cleavage and TPRT activity (data not shown) consistent with its inability to form stable DNA–protein complexes (Figure 5). All enzymatic activities for the WT and Myb− proteins in Figure 6 are presented relative to the amount of protein that was bound to the target DNA on EMSA gels at that concentration (Figure 6A). The amount of bottom-strand cleavage per unit of bound DNA for the Myb− protein was nearly as high as with the WT protein (average 81% cleaved over the three concentrations compared to an average of 92% for WT) (Figure 6B). The Myb− protein was also able to efficiently conduct the TPRT reaction: an average 40% of the DNA bound by the Myb− protein underwent TPRT compared with 54% for WT protein (Figure 6C). The decrease in TPRT level at higher protein concentrations of WT was due to the larger fraction of DNA being cleaved on both strands before TPRT could initiate (Figure 6D). Finally, the WT and Myb− protein differed most dramatically in the level of top-strand cleavage. Even at the highest protein concentration only 30% of the top DNA strand was cleaved in the Myb− reaction compared to 80% for the WT protein (Figure 6D).
These findings suggested that the Myb− protein was less able to initially bind the target DNA, even as a monomer, but once this subunit was bound it was able to conduct both bottom-strand cleavage and TPRT. The ability of the Myb− protein to conduct TPRT suggested the Myb motif was not involved in the binding of the R2 protein with the R2 RNA template to allow its specific utilization in a TPRT reaction. The dramatic inhibition of top-strand cleavage was consistent with the inability of the Myb− protein to form the dimer complex (see also Figure 5).
DISCUSSION
R2 elements have been inserting into the 28S rRNA gene site since the origin of arthropods (1) and possibly since the protostome/deuterostome divergence (2). The target 28S DNA sequence has not changed throughout this long history, all R2 lineages insert precisely between the same 2 bp, and non-R2 sequences have never been found inserted into this target site. Thus the R2 protein subunits undergo highly specific protein–nucleic acid interactions involving both the R2 RNA transcript and target DNA.
The highly conserved ZF motif located near the N-terminal end of all R2 proteins has the same order and spacing of cysteine, histidine and large hydrophobic residues found in the C2H2 ZF domain of many extensively studied DNA-binding proteins [reviewed in (22)]. Major DNA contact of these ZF motifs is by means of a α-helix starting between the second cysteine and the first histidine residues of the motif. The amino acids at −1, 3 and 6 of the helix are positioned to make contact with three consecutive bases in the DNA, thereby providing the sequence-specificity of the binding. These three residue positions in the R2 protein are highly conserved, consistent with the model that they are critical to the base contacts: Thr/Ser (position −1), Gly (position 3) and Val/Leu (position 6) (14). Based on the missing nucleoside footprints (Figure 3) and the methyl-G interference of position −1 but not +1 (Figure 2) the R2 ZF appears to bind the sequence AAG/TTC, the 3 bp immediately upstream of the insertion site.
Myb-binding motifs are ∼50 amino acid in length and contain as their core structure three α-helical domains with the third helix inserted in the major groove of DNA (23,26,27). The N-terminal domain of R2 proteins reveals sequence similarity in the region corresponding to the beginning of both the first and third α-helical domains of Myb motifs (14). Large hydrophobic residues associated with the first and third helixes of the Myb motifs are also conserved in R2 proteins, and mutation of the first hydrophobic residue of the R2 Myb motif significantly reduced DNA-binding similar to that of mutations in the Myb proteins (24). Finally, the R2 motif appears to be resting in the major groove of the DNA as methyl-G residues at position +10, +11, +12 and +15 interfere with the binding of the R2 peptide.
For most DNA-binding proteins at least two ZF or two Myb motifs are required for specific protein recognition. The multiple ZF or Myb motifs each bind short consecutive regions along the DNA, essentially wrapping around the DNA helix in the major groove (22,26). While two DNA-binding motifs are present in R2, the protein differs in that it utilizes single ZF and Myb motifs to bind regions of the DNA over 10 bp apart (centered at −2 and +12, see Figure 2C). Based on molecular models of the footprints, instead of wrapping around the DNA helix, the R2 polypeptide appears to cross the minor groove to gain access to major grooves on adjacent helical turns. This crossing of the minor groove may explain the weaker missing nucleoside contacts at positions +7, +8, +9 on the top-strand and +6, +7 on the bottom-strand (Figure 3) (28). Before reaching the ZF-binding site the R2 polypeptide may travel for a few base pairs along the major groove in loose association with the top-strand, as suggested by the methyl-G interference at position +4.
We also tested the role of the N-terminal domain in the interactions of the R2 protein with RNA. The ZF and Myb motifs play no direct role in the high processivity of the R2 RT, the ability of the RT to displace an RNA helix from an RNA template, and the ability of the RT to jump from the 5′ end of one template to the 3′ end of a second RNA template (Supplementary Figure 2). The RT domain of the R2 protein is considerably larger than the RT of retroviruses containing a number of extra ‘fingers’ or extensions in the right-hand structure of RT domains (29). It seems likely that these additional regions of RT domain, and not the N-terminal domain, provide the extra binding of the R2 protein to the RNA template during polymerization.
A second, more specific, interaction of the R2 protein with RNA involves the ability of the protein to utilize in the TPRT reaction only RNA transcripts that contain the 3′-UTR of the R2 element (3). The N-terminal domain of R2 protein also does not appear to be involved in these specific interactions with the R2 RNA transcript. The isolated N-terminal domain could not be shown to bind this RNA in EMSA experiments (data not shown). More significantly, once the Myb− protein was bound to the target site it was able to support high levels of TPRT (Figure 6C). Unfortunately the ZF− protein did not bind the target DNA well enough to enable a direct test of its involvement in the TPRT reaction. However, we believe it unlikely that the R2 ZF specifically binds R2 RNA, because while proteins containing multiple C2H2 ZFs (e.g. TFIIIa) have been shown to bind both DNA and RNA (30,31), the same ZFs are not involved in the binding of both [reviewed in (32)].
Finally, the experiments in this report revealed that mutations in the ZF and Myb protein motifs diminished the ability of the upstream R2 subunit to bind to the target DNA (Figure 5), even though this upstream interaction does not involve the N-terminal domain. This finding suggests that the primary path to target recognition by the upstream R2 subunit is through a higher order complex involving the downstream subunit. Such a model is also supported by our previous study which demonstrated that binding by the R2 protein was reduced to DNA target substrates only containing sequences upstream of the insertion site (6). Support for this model can also be found in EMSA assays conducted as a function of time in the presence of excess DNA and RNA substrates (5). At the earliest time points of these assays the R2 protein was distributed in a series of complexes with DNA and RNA that included monomer, dimer and larger multimers. Only over time did this series of complexes become a uniform band containing the upstream monomer, suggesting that at least in vitro the downstream subunit or subunits tend to disassociate from the original complexes. This participation of both upstream and downstream subunits in the initial binding of the R2 protein to the target site does not appear to require specific protein recognition of the downstream sequences. As shown in Figure 4 both the −3 to −1 and +10 to +15 mutant DNA targets supported normal levels of binding by the upstream monomer. Thus the experiments in this report add support to our protein dimer model for R2 integration. The N-terminal domain is responsible for binding of the downstream subunit, which in turn aids binding of the upstream subunit. The N-terminal domain of the upstream subunit is not involved in the binding of this subunit to DNA or in the TPRT reaction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Karin Tetzlaff Averbeck and Danna Eickbush for critical reading of the manuscript, Bill Burke for generating the original ZF mutation, and Junqiang Ye for generating the Myb mutation. This work was supported by the National Institutes of Health grant GM42790 to T.H.E. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Malik H.S., Burke W.D., Eickbush T.H. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 2.Kojima K.K., Fujiwara H. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol. Biol. Evol. 2004;21:207–217. doi: 10.1093/molbev/msg235. [DOI] [PubMed] [Google Scholar]
- 3.Luan D.D., Eickbush T.H. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 5.Christensen S.M., Eickbush T.H. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol. Cell Biol. 2005;25:6617–6628. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen S., Eickbush T.H. Footprint of the retrotransposon R2Bm protein on its target site before and after cleavage. J. Mol. Biol. 2004;336:1035–1045. doi: 10.1016/j.jmb.2003.12.077. [DOI] [PubMed] [Google Scholar]
- 7.Ruschak A.M., Mathews D.H., Bibillo A., Spinelli S.L., Childs J.L., Eickbush T.H., Turner D.H. Secondary structure models of the 3′ untranslated regions of diverse R2 RNAs. RNA. 2004;10:978–987. doi: 10.1261/rna.5216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibillo A., Eickbush T.H. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J. Biol. Chem. 2002;277:34836–34845. doi: 10.1074/jbc.M204345200. [DOI] [PubMed] [Google Scholar]
- 9.Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 10.Anzai T., Takahashi H., Fujiwara H. Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)(n) by endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol. Cell Biol. 2001;21:100–108. doi: 10.1128/MCB.21.1.100-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nature Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 12.Esnault C., Maestre J., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nature Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 13.Belfort M., Derbyshire V., Parker M.M., Cousineau B., Lambowitz A.M. Mobile Introns:pathways and proteins. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 761–783. [Google Scholar]
- 14.Burke W.D., Malik H.S., Jones J.P., Eickbush T.H. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 1999;16:502–511. doi: 10.1093/oxfordjournals.molbev.a026132. [DOI] [PubMed] [Google Scholar]
- 15.Yang J., Malik H.S., Eickbush T.H. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl Acad. Sci. USA. 1999;96:7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bibillo A., Eickbush T.H. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J. Biol. Chem. 2004;279:14945–14953. doi: 10.1074/jbc.M310450200. [DOI] [PubMed] [Google Scholar]
- 17.Hayes J.J., Tullius T.D. The missing nucleoside experiment: a new technique to study recognition of DNA by protein. Biochemistry. 1989;28:9521–9527. doi: 10.1021/bi00450a041. [DOI] [PubMed] [Google Scholar]
- 18.Guille M.J., Kneale G.G. Methods for the analysis of DNA–protein interactions. Mol. Biotechnol. 1997;8:35–52. doi: 10.1007/BF02762338. [DOI] [PubMed] [Google Scholar]
- 19.Kingston R.E. DNA–protein interactions. In: Ausubel F.M., editor. Current Protocols in Molecular Biology. Vol. 2. Hoboken, NJ: John Wiley and Sons, Inc.; 1993. pp. 12.2.1–12.4.16. [Google Scholar]
- 20.George J.A., Eickbush T.H. Conserved features at the 5′ end of Drosophila R2 retrotransposable elements: implications for transcription and translation. Insect Mol. Biol. 1999;8:3–10. doi: 10.1046/j.1365-2583.1999.810003.x. [DOI] [PubMed] [Google Scholar]
- 21.Kojima K.K., Fujiwara H. Long-term inheritance of the 28S rDNA-specific retrotransposon R2. Mol. Biol. Evol. 2005;22:2157–2165. doi: 10.1093/molbev/msi210. [DOI] [PubMed] [Google Scholar]
- 22.Wolfe S.A., Nekludova L., Pabo C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 23.Ogata K., Morikawa S., Nakamura H., Hojo H., Yoshimura S., Zhang R., Aimoto S., Ametani Y., Hirata Z., Sarai A., et al. Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb. Nature Struct. Biol. 1995;2:309–320. doi: 10.1038/nsb0495-309. [DOI] [PubMed] [Google Scholar]
- 24.Saikumar P., Murali R., Reddy E.P. Role of tryptophan repeats and flanking amino acids in Myb–DNA interactions. Proc. Natl Acad. Sci. USA. 1990;87:8452–8456. doi: 10.1073/pnas.87.21.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bibillo A., Eickbush T.H. The reverse transcriptase of the R2 non-LTR retrotransposon: continuous synthesis of cDNA on non-continuous RNA templates. J. Mol. Biol. 2002;316:459–473. doi: 10.1006/jmbi.2001.5369. [DOI] [PubMed] [Google Scholar]
- 26.Ogata K., Morikawa S., Nakamura H., Sekikawa A., Inoue T., Kanai H., Sarai A., Ishii S., Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 27.Ogata K., Kanai H., Inoue T., Sekikawa A., Sasaki M., Nagadoi A., Sarai A., Ishii S., Nishimura Y. Solution structures of Myb DNA-binding domain and its complex with DNA. Nucleic Acids Symp. Ser. 1993;29:201–202. [PubMed] [Google Scholar]
- 28.Dixon W.J., Hayes J.J., Levin J.R., Weidner M.F., Dombroski B.A., Tullius T.D. Hydroxyl radical footprinting. Methods Enzymol. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- 29.Eickbush T.H. Origin and evolutionary relationships of retroelements. In: Morse S.S., editor. Evolutionary Biology of Viruses. NY: Raven Press, Ltd; 1994. pp. 121–157. [Google Scholar]
- 30.Christensen J.H., Hansen P.K., Lillelund O., Thogersen H.C. Sequence-specific binding of the N-terminal three-finger fragment of Xenopus transcription factor IIIA to the internal control region of a 5S RNA gene. FEBS Lett. 1991;281:181–184. doi: 10.1016/0014-5793(91)80388-j. [DOI] [PubMed] [Google Scholar]
- 31.Searles M.A., Lu D., Klug A. The role of the central zinc fingers of transcription factor IIIA in binding to 5S RNA. J. Mol. Biol. 2000;301:47–60. doi: 10.1006/jmbi.2000.3946. [DOI] [PubMed] [Google Scholar]
- 32.Matthews J.M., Sunde M. Zinc fingers—folds for many occasions. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.