Abstract
Functional genomics has become a major focus in the study of microbial pathogenesis. This study used a functional genomic tool, differential display reverse transcription-PCR, to identify a transcriptional profile of Cryptococcus neoformans cells as they produced meningitis in an immunosuppressed host. This serial global gene expression during infection allowed for the identification of up- and down-regulated genes during infection. During this profiling, a single gene for the enzyme isocitrate lyase (ICL1) was found to be up regulated at 1 week of infection in a rabbit meningitis model and during a time of maximum host cellular response. The finding suggested that this enzyme and the glyoxylate shunt pathway are important to this yeast's energy production during infection. However, site-directed icl1 mutants had no apparent virulence defect in two animal models and no growth defect within macrophages. These observations suggest that although the yeast responded to a certain environmental cue(s) by an increase in ICL1 expression during infection, this gene was not necessary for progression of a C. neoformans infection. Compounds that specifically target only ICL1 are unlikely to cripple C. neoformans growth in vivo.
The basidiomycete Cryptococcus neoformans has become an attractive model yeast for molecular pathogenesis studies and identification of drug targets. It possesses a series of important features which make it a particularly useful molecular model pathogen. First, it is an important human pathogen whose incidence of life-threatening infections dramatically increased during the human immunodeficiency virus pandemic, thus making its appearance as a worldwide opportunistic infection (38). This infection can occur in both immunocompetent and immunocompromised individuals. Current therapeutic regimens are helpful in the management of infections, but the need for prolonged therapy in some patient populations and the frequent occurrence of failures demonstrate that further fungicidal drug regimens remain an important clinical need (7). Second, it is a sexual yeast with the ability to perform genetic crosses that allow for the use of genetic recombination and offers the ease of working with a haploid genome. Third, there is a firm understanding of the pathophysiology and immunology of this infection which adds insight into the relevance of the host (6, 39). Fourth, there are several excellent, robust animal models in which the proof of concept for virulence studies can be examined and used for the study of different treatment strategies. Fifth, there are already a series of genes in C. neoformans associated with infection and their quantitative and qualitative impact on virulence has been described through gene disruption experiments. These genes include those for the following: capsule formation (8-11, 27), melanin production (20, 31, 49), high-temperature growth (36, 48), purine metabolism (47), myristylation (32), topoisomerase (54), signal transduction (2, 3, 12, 18, 41, 55), alpha-mating type locus (30), phospholipase (13), urease (14), and mannosylation (57). In addition to a single gene disruption strategy, a more global approach has used signature-tagged mutagenesis to identify virulence genes (40). Furthermore, it has also been shown that with the use of a double-gene knockout, genes can act synergistically to complete the virulence phenotype (55). It is clear that a further understanding of the molecular virulence composite of C. neoformans can be achieved by using one or more genes at a time.
In conjunction with the direct strategy of selecting a specific gene(s) or phenotype(s), another approach to identify potential genes involved in the virulence composite is to use the array of functional genomic techniques for transcriptional profiling. For instance, these techniques can focus on isolation and identification of infection site-specific expressed genes in C. neoformans. The hypothesis to be tested is that the genes turned on or off at the site of infection and their regulators are essential for the survival of the yeast in the host or that these genes identify certain genetic pathways or networks which are necessary to produce infection and disease. In other words, the yeast can help direct the focus of the investigations.
Since the goal in our studies is to identify potential gene targets for antifungal drug development and to reduce the complexity of gene expression profiling, we chose to limit our examination of C. neoformans gene expression to acute meningitis with active yeast proliferation within the central nervous system. In this acute stage of infection, the yeast is surviving and growing within the subarachnoid space and must respond to a limited but dynamic immune response. It is this stage in which most disseminated cryptococcal infections are diagnosed and treated. However, it should be emphasized that the C. neoformans gene expression profile might be significantly different during chronic infection or with other stages of infection, such as during dormancy or reactivation, or at other body sites of infection.
C. neoformans infection can be ideally studied in a rabbit model of cryptococcal meningitis which has correlated both pathologically and clinically with human disease (45). This site-specific model system allows for global C. neoformans gene expression profiling in the subarachnoid space, by direct removal of aliquots of yeast cells and testing of yeast gene expression over days of infection. We used reverse transcription-PCR (RT-PCR) with differential display analysis (ddRT-PCR) to identify a global profile of genes being expressed at the site of infection. Then, a specifically regulated gene was chosen and a null mutant was created to determine its importance for the virulence composite. This gene (ICL1), which encodes the enzyme isocitrate lyase, was induced at 1 week of infection in the subarachnoid space. Isocitrate lyase is the main controlling protein in the glyoxylate shunt pathway in yeasts (4, 5, 21). In other yeasts, the pathway is known to be regulated by environmental concentrations of glucose (52) and the main function of the protein is to produce energy when glucose is limited and two carbon molecules are more readily available in the environment (28, 35). ICL1 was isolated, sequenced, and characterized, and icl1 null mutants were created. ICL1's inducible expression in vivo did not correlate with its impact on the virulence composite.
MATERIALS AND METHODS
Strains and media.
C. neoformans strain H99, a serotype A, MATα clinical isolate, and a fluoroorotic acid (FOA)-resistant H99 (ura5) strain (H99-1) were used in this study. The icl1 mutant strains (H99-11, H99-23) were created from this FOA-resistant strain. Strains were routinely grown on yeast extract-peptone-dextrose (YEPD) medium. However, yeast nitrogen base (YNB) supplemented with 2% glucose or 4% (wt/vol) acetate (YNB-acetate) was used for detection of ICL1 expression or acetate utilization, respectively.
Preparation of genomic DNA.
High-molecular-weight genomic DNA from C. neoformans was prepared as previously described (15). Genomic DNA for PCR was isolated from the C. neoformans strains as follows: yeast cells were grown to mid- to late-log phase in 10 ml of YEPD broth at 30°C, pelleted and washed three times in sterile distilled water, and resuspended in 0.5 ml of TENTS (10 mM Tris [pH 7.5], 1 mM EDTA [pH 8.0], 100 mM NaCl, 2% Triton X-100, 1% sodium dodecyl sulfate). Then, 0.2 ml of 0.5-mm-diameter glass beads and 0.5 ml of phenol-chloroform (1:1) were added and samples were vortexed for 2 min. After centrifugation for 10 min in a Microfuge apparatus at 13,000 rpm, the aqueous phase was transferred to a fresh tube, 2 volumes of 100% ethanol was added, and samples were placed at −20°C for 10 min. Precipitated DNA was collected, resuspended in 0.5 ml of Tris-EDTA (pH 8.0) containing 10 μg of RNase A/ml, and incubated at 37°C for 20 min. The DNA was then extracted once with phenol-chloroform, reprecipitated, washed with 70% ethanol, resuspended in 100 μl of Tris-EDTA, and stored at −20°C.
RNA extraction and ddRT-PCR.
Total RNA was prepared as described in the FastRNA Kit-Red protocol (BIO 101, La Jolla, Calif.), and first-strand synthesis of cDNA was performed using the protocol prescribed for the GenHunter RNA Image Kit (GenHunter, Nashville, Tenn.). First-strand synthesis was performed using anchored primers oligo(dT-dA), oligo(dT-dC), and oligo(dT-dG). ddRT-PCR was performed with the 13-mer arbitrary primer H-AP-1 (5′-AAGCTTGATTGCC-3′), yielding multiple bands (Fig. 1). Visualization of the mRNA transcripts was performed using 33P (New England Nuclear, Boston, Mass.) in the PCR mix. PCR conditions were 95°C for 2 min (1 cycle); 94°C for 30 s, 40°C for 50 s, and 72°C for 2 min (35 cycles); and 72°C for 5 min (1 cycle). The cloning and sequencing of display fragments were performed with the pCR-Trap cloning system (GenHunter) according to manufacturer's instructions.
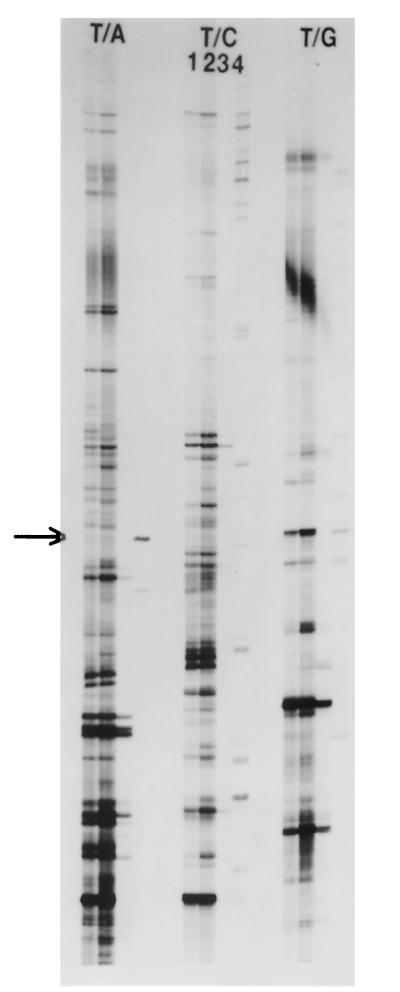
FIG. 1.
RT-PCR differential display analysis of mRNA from C. neoformans. Lanes T/A, T/C, and T/G show the results for each individual anchored oligo(T) primer with second primer H-AP-1. Lane T/C 1, in vitro yeast cells grown at 30°C; lane T/C 2, in vitro yeast cells grown at 37°C; lane T/C 3, in vivo yeast cells taken from CSF of rabbits at day 2 of infection; and lane T/C 4, in vivo yeast cells taken from CSF of rabbits at day 7 of infection. The arrow indicates a band representing detection of ICL1 expression from yeast cells at day 7 of infection.
For isolation and sequencing of the entire ICL1 gene, the following strategy was used. The EMBL3 genomic DNA library of C. neoformans has been previously described (15). The genomic library of H99 in EMBL3 was screened by using the ICL1-amplified transcript isolated from the differential display of C. neoformans. Positive plaques were purified through three rounds of repeated screening using the plate lysate method, as described by Sambrook et al. (50). The genomic clones were further analyzed by restriction enzyme digestion with BamHI, EcoRI, SalI, and XhoI. Two restriction enzymes were chosen, a SalI-restricted fragment of 7 kb and XhoI, producing two fragments of 5 and 2.3 kb. These fragments were then subcloned separately into pBluescript SK(−) plasmid. Double-stranded DNA from the SalI clone was sequenced in both directions and shown to contain the entire ICL1 gene. Sequences from the Stanford C. neoformans database (http://www-sequence.stanford.edu/group/C.neoformans/index.html) were aligned to recreate the JEC 21 ICL1 (serotype D) gene.
Molecular biological techniques.
DNA probes were labeled with [32P]dCTP (New England Nuclear), using the random primers labeling kit (Gibco-BRL, Gaithersburg, Md.) with 0.5 kb of ICL1 gene or the entire ICL1 gene as template.
Southern and Northern blotting, plaque lifts, and hybridization of nylon filters were performed according to Sambrook et al. (50). DNA was sequenced by the dideoxy chain termination method (51) using Sequenase version 2.0 (USB, Life Science, Cleveland, Ohio), or sequencing performed by the Duke University Sequencing Facility.
Karyotype analysis of H99 was performed according to the method of Perfect et al. (44).
Construction of the ICL1 gene disruption cassette by PCR.
By using a PCR overlap strategy for making disruption constructs (15, 25), three PCR fragments were generated (see Fig. 5). Each fragment was generated in a separate PCR with the PFU Taq polymerase (Stratagene, La Jolla, Calif.), using H99 genomic DNA or a plasmid isolate of the H99 genomic URA5 gene. Conditions for the PCR were as follows: 95°C for 2 min (one cycle); 95°C for 15 s, 55°C for 15 s, and 72°C for 2 min (35 cycles); and a final extension at 72°C for 2 min (one cycle). Note that the extension time at 72°C allows for 1 min per 1,000 bp.
FIG. 5.
Gene disruption of ICL1 and analysis. (A) PCR overlap knockout construct. Separate PCRs for 5′ P/ICL1, URA5, and 3′ ICL1 and then PCR overlap with equal concentrations of product producing ICL/URA5 were performed. (B) PCR detection of a ICL/URA5 insertion at the wild-type locus. Lane 3 shows the wild type (H99) with a 1.9-kb fragment. icl1 knockout mutants with an insertion of URA5 at the ICL1 locus (lanes 2, 4, 5, and 6) were expected to have a 400-bp-larger fragment, and a transformant with both endogenous and ectopic constructs is present in lane 1. Lanes 2 and 4 represent H99-11 and H99-23 strains. (C) Southern analysis of H99 and two icl mutants (H99-11 and H99-23). Restriction enzymes were ClaI (lanes 1 to 3), BamHI (lanes 4 to 6), and EcoRI (lanes 7 to 9). The Southern analysis of H99 (lanes 1, 4, and 7), H99-11 (lanes 2, 5, and 8), and H99-23 (lanes 3, 6, and 9) shows a single copy, and the expected size difference is seen as ClaI fragments between the wild type and the icl mutants.
Generation of construct fragments was performed as follows. (i) The 5′ P/ICL1 region was generated by using a primer site designated T2 (5′CTCTATCTTCCTTCTCCGCCC3′), which was 600 bp upstream of the ATG start site, and a primer site designated ICL12C (5′GGTCGAGCAACTTCGCTCCCCGCCCTCACCACGAGC3′), which was 400 bp downstream of the ATG start site. The ICL12C primer is composed of 18 bp of the 5′ P/ICL1 fragment at the 3′ end and 18 bp of the 5′ region of the URA5 gene. (ii) The URA5 insert (1,760 bp) was generated by using the primers URA1C (5′GCTCGTGGTGAGGGCGGGGAGCGAAGTTGCTCGACC3′) and URA4C (5′GCGGTTGATAGCGCATTGGGCTTGCCTCCAGGAGGTGG3′). URA1C is the complementary sequence of primer ICL12C (see above). URA4C is the complementary sequence of primer ICL13C (see below). This fragment allowed for a deletion within ICL1 of approximately 1,360 bp. (iii) The 3′ICL1 fragment (700 bp) was generated by primers ICL13C (5′CCACCTCCTGGAGGGAAGCCCAATGCGCTATCAACCGC3′), which was the complementary sequence of the URA4C primer, and IC5 (5′CCAGCGGTGGATGAGACACC 3′) (20 bp), which was the end of the ICL1 gene just upstream of the stop codon.
The three PCR fragments were gel purified, and the concentration was adjusted to give 25 ng/μl. Equal amounts of each fragment (2 μl) were added to a single PCR tube, and the outermost primers (T2 and IC5) were used with ExTaq (Fisher Scientific, Atlanta, Ga.) to generate the ICL1 knockout construct. PCR conditions for the knockout construct were as follows: 95°C for 2 min (1 cycle); 95°C for 15 s, 55°C for 15 s, and 72°C for 4 min (35 cycles); and a final extension at 72°C for 5 min (1 cycle).
Transformation of C. neoformans.
The PCR construct ICL1/URA5 was used to transform the FOA-resistant C. neoformans H99 (ura5 mutant) strain by using biolistic delivery of DNA, following the protocol described previously (46). URA+ prototrophic transformants were selected on URA dropout medium containing 1 M sorbitol at a temperature of 30°C. Transformants were patched on URA dropout plates and then streaked onto YNB-acetate plates to determine acetate utilization at 30°C. Genomic DNA preparations of the selected URA+ acetate-negative mutant transformants were used for both PCR and Southern blot analysis to determine whether insertion of the URA5 gene had occurred at the wild-type ICL1 gene locus.
In vitro growth kinetics of C. neoformans icl1 null mutants versus wild-type H99.
The in vitro growth rate assays were performed at 30°C and 37°C, in YNB high-glucose (2%), YNB low-glucose (0.2%), and YNB-acetate (2%) media. At 2, 8, 18, 24, and 48 h, quantitative counts were performed. Strains were also grown for phenotype analysis on dopamine, low-iron minimal medium agar, and minimal medium broth with 0.2% oleic acid as the sole carbon source.
Animal models. (i) Rabbit cryptococcal meningitis model.
New Zealand White rabbits weighing 2 to 3 kg were housed in separate cages and provided with water and Purina rabbit chow ad libitum. H99 and icl mutants (H99-11 and H99-23) were prepared by growth for 48 h at 30°C in YNB broth supplemented with 2% glucose. The cells were pelleted, washed once, and suspended in phosphate-buffered saline (PBS) at a cell density of 3.3 × 108 cells/ml. After sedation with ketamine (Fort Dodge Laboratories, Fort Dodge, Iowa) and xylazine (Vedco, St. Joseph, Mo.), approximately 108 viable cells of each strain in a volume of 0.3 ml were inoculated intracisternally into three rabbits per strain that had received an intramuscular injection of 1.2 mg of betamethasone acetate-phosphate (Schering-Plough, Kenilworth, N.J.) 1 day earlier and then daily during infection. Rabbits were sedated with ketamine and xylazine on days 4, 7, 11, and 14 after inoculation, and cerebrospinal fluid (CSF) was withdrawn. Quantitative yeast cultures were performed by diluting the CSF in PBS, plating on YEPD agar, and incubating at 30°C for 72 h and by counting colonies. For isolation of yeast cells to obtain RNA from in vivo cells for ddRT-PCR and confirmatory RT-PCR of ICL1, 2 × 109 H99 yeast cells from YEPD agar plates were grown for 72 h at 30°C, suspended in PBS, and inoculated intracisternally into 10 rabbits. Rabbits were immunosuppressed with daily doses of 5 mg of hydrocortisone administered intramuscularly. CSF yeast cells were collected at days 2, 4, and 7 of infection. After being withdrawn from subarachnoid space, yeast cells were placed directly on ice, washed twice in sterile water, and stored at −70°C until RNA was isolated.
(ii) Inhalation murine model.
Cryptococcal strains were used to infect 4- to 6-week-old female A/JCr mice (NCI/Charles River Laboratories) by nasal inhalation as previously described (13). Ten mice per strain were inoculated with 5 × 104 yeast cells of H99 and icl1 null mutants H99-11 and H99-23 in a volume of 50 μl (introduced into the nares after sedation with pentobarbital) and hung from a string by their front incisors. The mice were monitored twice daily and fed ad libitum. Any mice that were moribund or appeared to be in pain were sacrificed by CO2 inhalation.
All animal protocols were approved by the Duke University Laboratory Animal Committee.
Macrophage growth assay.
The MH-S murine alveolar macrophage cell line (American Type Culture Collection, Manassas, Va.) was maintained in RPMI 1640 medium containing 10% fetal calf serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 4.5 g of glucose/liter, 1.5 g of bicarbonate/liter, and 0.05 mM 2-mercaptoethanol and penicillin-streptomycin at 37°C with 5% CO2. Macrophages were harvested from monolayers by using 0.25% trypsin-0.03% EDTA, and cell viability was determined by trypan blue exclusion and by counting in a hemacytometer. The macrophage concentration was adjusted to 105 viable cells/ml. And in experiments using activated macrophages, the cells were primed with 50 units of murine gamma interferon/ml and stimulated with 0.3 μg of lipopolysaccharide/ml just prior to mixing yeasts. One hundred microliters of the macrophage suspensions was placed into 96-well plates. Cryptococci (H99, H99-11, and H99-23) were washed three times in PBS, counted, and placed in tissue culture medium at 105/ml. Yeast suspension (100 μl) was added to the MH-S cells at a multiplicity of infection (effector to target) ratio of 1:1. Control wells containing only macrophages and yeasts were included in all experiments. All wells contained 10 μg of 18B7 (immunoglobulin G1 anti-glucuronoxylomannan monoclonal antibody)/ml as an opsonin. The macrophage-yeast mixtures were allowed to incubate for 30 min to 1 h and were then washed with three exchanges of culture medium to remove extracellular yeasts. At 4 and 24 h, quantitative cultures were performed by aspirating the medium from each well and lysing macrophages with two exchanges of 100 μl of 1% sodium dodecyl sulfate, and the contents were combined and plated on YPD agar with chloramphenicol. All experiments were done in triplicate and repeated.
Statistics.
Student's t test was used in evaluating the yeast colony counts from rabbits and macrophage monolayers. Survival data from the mouse experiments were analyzed by using a Kruskal-Wallis test. A P value of ≤0.05 was considered significant.
Nucleotide sequence accession number.
The sequence data of the C. neoformans ICL1 gene (H99, serotype A) are available from GenBank/EMBL/DDBJ under accession no. AF 455253.
RESULTS
Differential gene expression.
With the use of differential gene expression studies, we obtained a transcriptional profile of yeast cells under specific environmental and in vivo conditions. In Fig. 1, the use of ddRT-PCR with the anchored primers and the H-AP-1 primer shows the partial transcripts of H99 cells under in vitro conditions (lane 1 at 30°C and lane 2 at 37°C). The lanes were loaded with the same amount of RNA from each temperature, and the transcriptional profiles were similar. We observed fewer than 30 potential transcript differences between the two temperatures. One transcript difference between the two conditions was identified as an up-regulated cytochrome oxidase C subunit 1 gene (COX1), which was found at 37°C but not at 30°C. This differential gene regulation was confirmed by Northern analysis. Lanes 3 and 4 represent transcripts from yeast cells directly removed from the CSF on days 2 and 7 of infection. Following CSF aspiration from each animal, yeast cells from 10 rabbits were directly placed on ice and then pooled together. To minimize stress on yeast cells, they were washed two times with cold water (4°C), which lysed host cells, and then yeasts were frozen until RNA isolation. This processing of the yeast cells should have had minimal influence on their RNA transcription. However, amounts of total RNA from these yeast cells were difficult to measure because RNA yields from in vivo cells were low. We suspect this finding was caused by a combination of factors in vivo, including low RNA synthesis levels, large polysaccharide capsules, and the limited number of yeast cells (between 106 and 108 CFU) compared to those found under in vitro conditions (108 to 109 CFU). For days 2, 4, and 7 of infection, total viable yeast counts from the CSF from each animal were similar when pooled, with concentrations ranging from 6.5 × 107 CFU at day 2 to 2 × 107 CFU at day 7. It can be seen by the display gel of the in vivo cells (Fig. 1) that there was a dynamic change in the transcriptional profile of C. neoformans as it met the stresses of prolonged infection and of CSF cellular responses, whose evolution has been described previously (42, 43, 45). The gene expression profile of C. neoformans appears dramatically different from that of in vitro-grown cells, and the transcripts changed from day 2 to day 7 of infection. Multiple partial transcripts were cloned and sequenced from the RT-PCR display, and our interest was particularly focused on one highly up-regulated transcript observed at day 7 (see arrow in Fig. 1). It was not present at day 2 of infection and was not observed under in vitro growth conditions. We therefore repeated the RT-PCR at days 4 and 7 of infection, at which times the total numbers of yeast cells from CSF were similar. As previously shown in the screening differential display gel, the transcript by RT-PCR was present at day 7 of infection but was not detected from yeast cells at day 4 of infection (Fig. 2).
FIG. 2.
RT-PCR for ICL1 expression in yeast cells taken from CSF on days 4 and 7 of infection. Specific RT-PCR results for the ICL1 transcript on RNA isolated from yeast cells at day 4 (left) are compared to those from day 7 of infection (right).
Characterization of ICL1.
The partial gene transcript was isolated from the differential display gel, cloned, and sequenced. When a search was conducted in GenBank, the nucleotide sequence matched isocitrate lyase genes from other fungal species. This partial transcript was used as a probe to isolate and sequence the full-length gene from a genomic library of H99. A cDNA clone was identified and sequenced to help identify intronic borders. The nucleotide sequence of ICL1 from serotype A H99 is similar (>98% identical) to a copy of the JEC21 ICL1 in the database at Stanford (http://www-sequence.stanford.edu/group/C.neoformans/index.html) of the serotype D (JEC21) C. neoformans genome sequence. The ICL1 gene is approximately 2,200 bp in size. It contains seven introns with average length of 80 bp. Intron delineation followed a typical configuration (GTNNGT-CTRAY-YAG, where hyphens indicate start of intron, loop, and end of intron, respectively). It is interesting that in the majority of introns, a sequence of (C/G)AAA preceded either the GTNNGT or the CAG sequence. We also found two introns which did not conform to the above arrangement. One intron had the sequence GTNNCG, while another had the termination sequence TAG. After restriction enzyme digestion, a Southern blot analysis of H99 genomic DNA demonstrated that ICL1 was a single-copy gene, unlike Saccharomyces cerevisiae, which has a second isocitrate lyase gene (24). A search of genomic databases at Stanford, The Institute for Genomic Research, and Duke did not reveal a second copy or a similar homolog. A chromosome blot was probed with ICL1, which confirmed a single location of the gene on the second smallest chromosome of H99 (46).
The deduced amino acid sequence of H99 ICL1 was compared to those of other fungal species (Fig. 3). The C. neoformans ICL1 was found to be most homologous with that of another basidiomycete, Coprinus cinereus, and was followed in rank of similarity by those of Neurospora crassa, Aspergillus nidulans, S. cerevisiae, Candida albicans, and Yarrowia lipolytica. The 5′ region of the isocitrate lyase protein in C. neoformans had a wide range of variation in amino acid sequence compared to those of other fungi, which is typical of this protein. The homologous regions are the amino acid stretches of 86 to 142, 161 to 269, 366 to 388, 404 to 430, 455 to 464, and 502 to 537. The conserved region SPS (amino acids 425 to 427) is highly maintained between fungal species and has been identified as the active site of the enzyme for binding the substrates of isocitrate and glyoxylate (29).
FIG. 3.
The deduced amino acid sequence of C. neoformans ICL1 and its comparison with other fungal ICL1s. Conserved areas within the coding region between these fungi are noted. Cn, C. neoformans; Co, Coprinus cinereus, As; Aspergillus nidulans; Ca, Candida albicans.
ICL regulation.
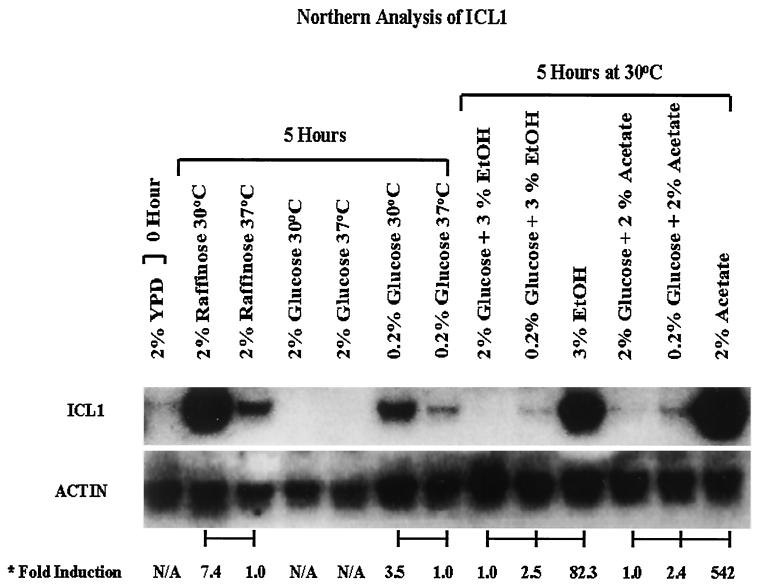
ICL1 expression was further analyzed by setting up the following in vitro conditions to elucidate its regulation. The Northern blot analysis in Fig. 4 shows the following expression effects of certain environmental stimuli. The concentration of glucose has profound effects on ICL1 expression. At low glucose concentrations (i.e., 0.2%), which might approach levels that are found in biological fluids, ICL1 is minimally expressed. With 2% glucose in the medium, transcripts of ICL1 are not detected. This suppression appears to be specific for glucose, since another sugar, raffinose, appears to actually induce ICL1 production. There seems to be a three- to fourfold induction of ICL1 in yeast cells exposed to 30°C compared to that at 37°C. ICL1 is highly induced in the presence of two carbon compounds such as ethanol and acetate, but these highly stimulatory molecules can be suppressed in their inducing abilities in the presence of elevated glucose concentrations, since 2% glucose can completely suppress the ability of these compounds to induce ICL1 (Fig. 4).
FIG. 4.
Northern analysis of ICL1 expression under specific in vitro conditions. Gene expression was measured after exposure to different temperatures (30°C and 37°C), sugars, or two carbon molecules for 5 h. The severalfold induction of expression under specific conditions was compared to ICL expression of cells prior to inducing conditions and corrected for actin expression by phosphorimager intensity measurements.
Creation of icl1 null mutants.
With this understanding of ICL1 structure and regulation, both in vitro and in vivo, our next focus was to correlate its expression with a possible phenotype; thus, a null mutant was constructed. A PCR overlap strategy was performed in which the 1,760 bp URA5 was placed within the coding region of ICL1, which removed approximately 1,300 bp of the internal ICL1 gene (Fig. 5A). The construct ICL1/URA5 was biolistically delivered into an FOA-resistant H99 strain, H99-1. Thirty-four URA5-positive transformants were taken from the auxotrophic selection plates and screened for growth on acetate plates. Seventeen of the transformants selected were URA5 positive but could not grow on acetate. A PCR was performed with primers from the ICL1 gene which flanked the area where the URA5 gene was inserted. This strategy was designed to detect the insertion of the URA5 gene at the ICL1 locus; a replacement of the locus should produce a PCR product which is approximately 400 bp larger than that of the wild-type locus. In Fig. 5B, the results of a PCR with primers to identify the ICL1 locus show transformants and the wild-type H99 strain. Four transformants displayed the anticipated ∼400-bp increase in size for the homologous insertion of the URA5 at the wild-type locus. One transformant possessed both the intact endogenous gene and the disruption construct. For two of the transformants, which we have designated icl1::URA5 mutants H99-11 and H99-23, we had Southern blot analysis performed to ensure that these mutants represented single homologous events (Fig. 5C). Mutants H99-11 and H99-23 were further characterized and did not grow on plates with acetate as the only carbon source (Fig. 6). However, when grown in YEPD broth and compared to the parent strain (H99) over 72 h at 30°C or 37°C, their growth rates were identical. Furthermore, H99-11 and H99-23 were not viable after growth for 7 days in medium with 0.2% oleic acid as the only carbon source, whereas wild-type (H99) cells proliferated approximately 100-fold from the original inoculum. When examined for other possible phenotypes related to virulence, the mutant strains grew at high temperatures (39°C) at the same rate as the parent strain, melanized similarly on dopamine plates, and produced similar size capsules when grown on low-iron minimal medium.
FIG. 6.
Growth on YEPD plate (A) and acetate plate (B). All strains grow on nutrient medium (A), but when medium contains acetate as the only carbon source (B), the icl mutants will not grow (lower two quadrants of panel B).
Outcome of infection with icl1 mutants.
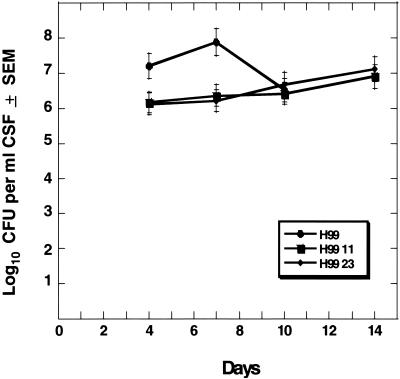
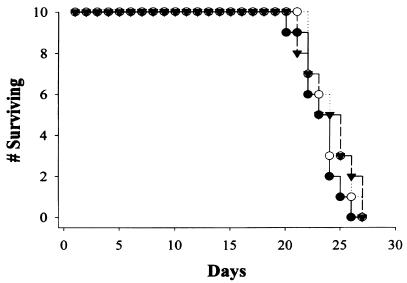
H99-11 and H99-23 were inoculated into animals in two animal models to determine whether isocitrate lyase function was required for or impacted the virulence composite of C. neoformans. The wild-type strain (H99) and the two icl1 mutants were inoculated into the subarachnoid space of immunosuppressed rabbits. As shown in Fig. 7, quantitative counts were measured from days 4 to 14 of infection. At days 4 and 7, there was a slight increase in numbers of wild-type yeast cells compared to those of both icl mutants. However, only at day 4 of infection did one mutant show a significant difference in numbers of yeast cells compared to those of wild-type cells (P = 0.04), but by day 10 of infection there was no difference in yeast numbers between any two of the strains. In fact, when CSF yeast counts for the two icl1 mutant strains were measured at 2 weeks of infection compared to 1 week of infection, the numbers of yeasts continued to increase in the subarachnoid space. In order to further analyze the impact of icl1 mutants on infection, we then used an inhalation model of murine cryptococcosis. This model represents a separate animal species, another method of infection which places different demands on the infecting fungus, and a different endpoint measurement of virulence. As shown in Fig. 8, there were no significant differences in survival between wild-type H99, H99-11, and H99-23.
FIG. 7.
Quantitative yeast counts of H99 and null mutants H99-11 and H99-23 in CSF of rabbits. There were three animals per group. Infection with wild-type H99 was measured to day 10 of infection, and infection with H99-11 and H99-23 was measured to day 14 of infection. Differences in yeast counts between groups at days 4, 7, and 10 of infection were not significant (P > 0.05) except for that in the comparison of H99 versus H99-11 at day 4 (P = 0.04).
FIG. 8.
Murine survival of inhalation cryptococcosis. There were 10 mice in each group, and animals were evaluated daily. There was no difference in survival between groups (P = 0.8). ○, H99-11; •, H99-23; and ▾, H99.
Impact of icl1 mutations on intracellular growth.
As shown in Table 1, H99, H99-11, and H99-23 grew almost 100-fold inside macrophages over 24 h without an impact of macrophage activation on yeast growth. No growth defect was detected for H99-11 and H99-23 compared to wild-type H99.
TABLE 1.
C. neoformans growth inside macrophages over 24 h
| Strain | Mean log10 CFU/well (± SD) at:c
|
|||
|---|---|---|---|---|
| 4 H
|
24 H
|
|||
| Restinga | Activatedb | Restinga | Activatedb | |
| H99 | 3.24 ± 0.10 | 3.20 ± 0.05 | 5.09 ± 0.03 | 5.02 ± 0.13 |
| H99-11 | 3.25 ± 0.05 | 3.12 ± 0.12 | 5.03 ± 0.09 | 5.02 ± 0.08 |
| H99-23 | 3.23 ± 0.53 | 3.15 ± 0.03 | 5.04 ± 0.11 | 5.08 ± 0.12 |
Unstimulated macrophage effectors.
Gamma interferon-lipopolysaccharide-activated macrophage effectors.
No differences in counts between each pair of strains (P > 0.05). SD, standard deviation.
DISCUSSION
Investigators of functional genomics seeking global genetic screens for virulence (40) have a series of tools, such as cDNA library subtraction, serial analysis of gene expression, in vivo expression technology, ddRT-PCR, and microarray technology, for use in comprehensively examining global gene expression in fungal pathogens under a variety of conditions. This study demonstrates that one of these systems, ddRT-PCR, can be used to perform transcriptional profiling of C. neoformans as infection progresses within the host. It is clear that C. neoformans transcriptional profiles change in the central nervous system during the progression of infection, and ddRT-PCR can be used to identify these regulated genes.
These qualitative and quantitative genomic techniques are extremely powerful for use in attempts to understand the yeast response to a changing environment from several aspects. First, the in vivo transcriptional profiling circumvents concerns about trying to dynamically replicate specific host environments in vitro. Second, results from transcript identification allow new insights into potential genetic factors which might not have been predicted a priori. Third, with a genetically tractable yeast, a gene expression profile can be examined in relationship to the actual gene function and phenotype under similar environmental conditions. It is often assumed that if a gene is expressed or specifically regulated under a particular set of conditions, then that gene or its linked genes are important for the growth and/or survival of the organism under those conditions. However, this assumption is not always correct. For example, a disconnection between expression and phenotype for S. cerevisiae has been vividly illustrated in a study in which a functional genomic screen for gene expression had a parallel analysis of gene deletion strains for phenotype (59). Results in this study showed that little correlation existed between biological growth and gene expression data. This fact makes it essential that the importance of a fungal gene's expression profile for the organism's growth and/or survival be matched to its null mutant phenotype. In fact, in this study no correlation was found between ICL1 gene-induced expression at the infection site and its importance to the total virulence composite.
ICL1 is a gene which encodes a relatively conserved protein that critically controls the glyoxylate pathway used for utilization of two carbon molecules (C2) for energy. Isocitrate lyase cleaves isocitrate to succinate and glyoxylate. In other systems, isocitrate lyase and the entire glyoxylate pathway have been shown to be enhanced under conditions of low glucose and low oxygen tension (57), in the presence of acetate and high temperature (26), and during intracellular growth (26, 37). On the other hand, the enzyme can be inhibited by metabolites such as succinate, 3-phosphoglycerate, or phosphoenolpyruvate. In C. neoformans, icl1 mutants could not grow on plates with acetate as a sole carbon source, and while the wild-type strain could grow in a fatty acid (oleic acid) medium, icl1 mutants died in this medium. These phenotype results are consistent with isocitrate lyase's importance in providing energy from C2 compounds and fatty acids in its environment, such as may occur in the intracellular environment of a macrophage. It is also clear from our studies that the C. neoformans ICL1 gene is highly up regulated by ethanol and acetate but suppressed by high concentrations of glucose. Unlike the dimorphic fungal pathogen Penicillium marneffei, in which ICL1 expression is increased at mammalian temperatures (N. G. Haycocks, C. R. Cooper, and M. R. McGinnis, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. F34, 2001), ICL1 expression is not induced at 37°C. In fact, we found a slight decrease in ICL1 expression at 37°C compared to 30°C. This finding demonstrates that there has been a divergence in ICL1 response to certain environmental stimuli within fungi.
There was a significant induction of in vivo ICL1 expression in C. neoformans as infection progressed. The elevated in vivo expression of ICL1 detected at day 7 of infection suggested that an increasing number of yeast cells in the CSF are contained inside macrophages which have been activated during progression of infection. From previous studies on the dynamic immunological reactions occurring in this experimental model of cryptococcal meningitis (42, 45), Perfect et al. observed induced ICL1 expression which correlated with a time (day 7 of infection) when the host had developed its maximum numbers of CSF cells and CSF macrophages had acquired their activation state for fungicidal or tumoricidal activity (43). Thus, there may be several factors which induce ICL expression at 1 week of infection. First, exposure to the intracellular lipids or acetate of activated macrophages has been shown to induce isocitrate lyase gene expression in other organisms studied (26, 37). Second, there is a changing of CSF glucose concentrations in the subarachnoid space of the rabbit during infection, with some animals developing CSF hypoglycorrachia (45), and this three- to fivefold drop of CSF glucose concentrations might induce ICL1 expression. Third, there are magnetic nuclear resonance data for mice with cryptococcomas which include reports of detection of the metabolites ethanol and acetate in brain tissue and thus, these carbon molecules might be available in the subarachnoid space for induction of ICL1 (T. E. Dzendrowski, V. Himmelreich, S. Dowd, C. Allen, C. Mountford, and T. C. Sorrell, 4th Int. Conf. on Cryptococcus and Cryptococcosis, Paris, France, Abstr. PC18, 1999).
Using this inducible expression data, it was exciting to hypothesize that the glyoxylate pathway is crucial for C. neoformans energy needs when the yeast is within a hostile environment such as the central nervous system. There has been animal model support for the importance of the pathway in pathogenesis. For example, an intracellular pathogen, Mycobacterium tuberculosis, was found to have had its expression of isocitrate lyase induced during infection of activated macrophages (26, 37). In elegant pathogenesis experiments, McKinney et al. showed the importance of isocitrate lyase and the glyoxylate shunt in vivo for the persistence of Mycobacterium tuberculosis infection (37). Using comparisons of icl mutant strains with the wild-type strain, those investigators showed that in immunocompetent mice, acute infection was not impacted by the presence of isocitrate lyase but that this enzyme was needed for persistence in the host tissue. Furthermore, it was demonstrated that isocitrate lyase was important only for growth in activated macrophages and that its negative impact on the virulence composite was lost in immunodeficient mice. These observations illustrate that the potential importance of isocitrate lyase for M. tuberculosis is profoundly influenced by the host's response to infection. Similarly, in the rabbit model, ICL1 induction appeared to correlate with accelerated host immune responses in that the induction took place during times when CSF macrophage activation occured (43).
The glyoxylate pathway has also been linked to fungal pathogenesis. With the use of microarray technology, Lorenz and Fink examined the transcriptional profile of the model yeast S. cerevisiae. A population of S. cerevisiae cells which had been phagocytosed by macrophages was isolated, and a whole-genome microarray analysis was performed to compare transcripts from intracellular yeasts to those from extracellular yeasts. Many of the most highly induced S. cerevisiae genes intracellularly encoded proteins related to the glyoxylate cycle. Those investigators then examined a common fungal pathogen, Candida albicans, by creating an icl1 null mutant to determine whether the intact glyoxylate pathway was necessary to complete the virulence composite. In a disseminated murine candidiasis model, the icl1 null mutant was found to be attenuated, exhibiting a prolonged survival compared to a wild-type strain, but eventually the icl1 mutant was able to kill the mice (33).
Given our in vivo gene expression studies and the previous work investigating Mycobacterium and Candida, our findings on virulence impact with C. neoformans icl1 mutants were surprising. First, two independent icl1 mutants showed a slight initial decrease in CSF yeast counts compared to those of the wild-type strain in early infection but yeast counts of mutants rose to wild-type levels as the infection progressed and at times during infection when ICL1 expression was induced. The yeast counts in the CSF continued to rise later in infection, with no suggestion that ICL1 was uniquely important to the growth and survival of this yeast within the central nervous system site. More than a dozen null mutants for different C. neoformans genes have been studied in the rabbit model; it is clear that various levels of attenuation can be detected (1, 2, 13, 14, 16, 17, 41, 47, 57). However, in relationship to these other attenuated strains, the icl1 mutants possessed excellent survival abilities in the CSF and generally possess wild-type growth characteristics in vivo.
For a separate check on isocitrate lyase's impact on pathogenesis, the icl1 mutants were introduced into an inhalation murine model. In this model, yeasts travel into the lungs and through macrophages until dissemination into the central nervous system occurs. This model puts different demands on the yeast's ability to survive and produce disease in another animal species. It was clear that in the murine model, an intact glyoxylate pathway was not necessary for the production of disease at the site at which the inoculum was administered. A significant component of survival in this model is the yeast's ability to survive and grow intracellularly within macrophages (20). Consistent with their ability to produce disease compared to that of wild-type cells, the icl1 mutants showed no growth defect in either resting or activated macrophages.
The results of our investigations of ICL1 impact on a central nervous system infection with C. neoformans are consistent with previous in vivo studies using S. cerevisiae. Goldstein and McCusker showed that despite induction of the isocitrate lyase gene after macrophage phagocytosis of S. cerevisiae, this yeast can grow and compete well in the brains of mice, even though its ICL1 gene and glyoxylate pathway are not functional (22). These data support the C. neoformans finding that there is a disconnection between the importance of the glyoxylate pathway in in vivo gene expression profiling and the observed phenotypes of null mutants for pathogenesis in this pathway. It is hypothesized that despite the block in the glyoxylate pathway, host glucose levels in these models at the site of infection still provide a consistent energy source for the yeast to support growth and produce disease. Alternatively, there may be a second pathway in C. neoformans which can utilize two- and three-carbon products of fatty acid breakdown for energy when the glyoxylate pathway is blocked by the absence of isocitrate lyase. For instance, S. cerevisiae has an ICL2 gene (2-methyl isocitrate lyase) which is involved in propionyl-coenzyme A metabolism (34), but in our search of the cryptococcal genomic databases there was no apparent homolog in C. neoformans. Further studies will be needed to determine if another pathway exists.
The discovery of molecular weak points in fungal pathogens may help us develop targets to test and identify new antifungal drugs. Isocitrate lyase and the glyoxylate pathway have been potential targets for several reasons. First, they exist in prokaryotic and eucaryotic pathogens but not in mammals and this meets an important criteria for selectivity. Second, this pathway has a focus on inhibition of nutrient availability which has been used successfully with herbicides. Third, most conventional drugs target processes for cell growth and division but this focus could target slow-growing yeast at a metabolic stage. Fourth, compounds or inhibitors of this target have already been shown to possess antifungal activity in vitro (23), and finally, the structure of the isocitrate lyase enzyme for some pathogens has been elucidated to help in drug design (53). Despite the attractive features of this target for drug development, it is clear from our studies that the impact on blocking the glyoxylate pathway in C. neoformans at isocitrate lyase is not sufficient to prevent the yeast from producing disease in certain animal models. However, the importance of isocitrate lyase for the virulence composite of C. neoformans might be found under other conditions or models. Further investigation of how isocitrate lyase and the glyoxylate pathway (i) affect tissue-specific yeast growth, (ii) produce different pathogenesis outcomes with two different fungal pathogens, and (ii) interact with other specific energy inhibitors or other fatty acid degradation pathways is needed to fully appreciate this enzyme's potential as an antifungal target.
Acknowledgments
This work was supported by NIAID-sponsored grants (AI28388 and AI44975) and as part of the Duke University Mycology Research Unit.
Editor: T. R. Kozel
REFERENCES
- 1.Alspaugh, J. A., L. M. Cavallo, J. R. Perfect, and J. Heitman. 2000. RAS1 regulates filamentation, mating, and growth at high temperature of Cryptococcus neoformans. Mol. Cell. Biol. 36:352-365. [DOI] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein gamma subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alspaugh, J. A., R. Pukilla-Worley, T. Harashima, L. M. Cavallo, D. Funnel, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002.. Adenylylcyclase functions downstream of the G-alpha protein GPA1 and controls mating and pathogenicity. Eukaryot. Cell 1:75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atomi, U. K., T. Kanai, S. Takeshita, N. Kanayama, M. Veda, and A. Tanaka. 1997. Derepression of gene expression mediated by the 5′ upstream region of the isocitrate lyase gene of Candida tropicalis is controlled by two distinct regulatory pathways in Saccharomyces cerevisiae. Eur. J. Biochem. 243:748-752. [DOI] [PubMed] [Google Scholar]
- 5.Barth, G., and T. Scheuber. 1993. Cloning of the isocitrate lyase gene (ICL1) from Yarrowia lipolytica and characterization of the deduced protein. Genetics 241:422-430. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall, A. 1995. Antibody immunity and invasive fungal infections. Infect. Immun. 63:4211-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadevall, A., and J. R. Perfect. 1998. Therapy of cryptococcosis, p. 457-518. In A. Casadevall and J. R. Perfect (ed.), Cryptococcus neoformans. ASM Press, Washington, D.C.
- 8.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficiency mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., and K. J. Kwon-Chung. 1998. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 66:2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. C., and K. J. Kwon-Chung. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, Cap64, is essential for virulence. Infect. Immun 64:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, Y. C., B. L. Wickes, G. F. Miller, L. A. Penoyer, and K. J. Kwon-Chung. 2000. Cryptococcus neoformans STE12 alpha regulates virulence but is not essential for mating. J. Exp. Med. 191:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox, G. M., H. C. McDade, S. C. Chen, S. C. Tucker, M. Gottfredsson, L. C. Wright, T. C. Sorrell, S. D. Leidich, A. Casadevall, M. A. Ghannoum, and J. R. Perfect. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39:166-175. [DOI] [PubMed] [Google Scholar]
- 14.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox, G. M., T. H. Rude, C. C. Dykstra, and J. R. Perfect. 1995. The actin gene from Cryptococcus neoformans: structure and phylogenetic analysis. J. Med. Vet. Mycol. 33:261-266. [DOI] [PubMed] [Google Scholar]
- 16.Del Poeta, M., D. L. Toffaletti, T. H. Rude, C. C. Dykstra, J. Heitman, and J. R. Perfect. 1999. Topoisomerase 1 is essential in Cryptococcus neoformans: role in pathobiology and as an antifungal target. Genetics 152:167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Poeta, M., D. L. Toffaletti, T. H. Rude, S. D. Sparks, J. Heitman, and J. R. Perfect. 1999. Cryptococcus neoformans differential gene expression detected in vitro and in vivo with green fluorescent protein. Infect. Immun. 67:1812-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, and J. R. Perfect. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen C. neoformans. Mol. Cell. Biol. 21:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erickson, T., L. Liu, A. Gueylkian, X. Zhu, J. Gibbons, and P. R. Williamson. 2001. Multiple virulence factors of Cryptococcus neoformans are dependent on UPH1. Mol. Microbiol. 42:1121-1131. [DOI] [PubMed] [Google Scholar]
- 20.Feldmesser, M., S. C. Tucker, and A. Casadevall. 2001. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 9:273-278. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez, E., F. Moreno, and R. Rodicio. 1992. The ICL1 gene from Saccharomyces cerevisiae. Eur. J. Biochem. 204:983-990. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein, A. L., and J. H. McCusker. 2001. Development of Saccharomyces cerevisiae as a model pathogen: a system for the genetic identification of gene products required for survival in the mammalian host environment. Genetics 159:499-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hautzel, R., H. Anke, and W. S. Sheldrick. 1990. Mycenon, a new metabolite from a Mycena sp. TA87202 (basidiomycetes) as an inhibitor of isocitrate lyase. J. Antibiot. 43:1240-1244. [DOI] [PubMed] [Google Scholar]
- 24.Heinisch, J., E. Valdes, J. Alvarez, and R. Rodicio. 1996. Molecular genetics of ICl2 encoding a non-functional isocitrate lyase in Saccharomyces cerevisiae. Yeast 12:1285-1295. [DOI] [PubMed] [Google Scholar]
- 25.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 26.Honer, Z. U., K. Bentrup, A. Miczak, D. L. Swenson, and D. G. Russell. 1999. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 181:7161-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the o-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42:453-467. [DOI] [PubMed] [Google Scholar]
- 28.Kanai, T., S. Takeshita, H. Atomi, K. Unemura, M. Veda, and A. Tanaka. 1998. A regulatory factor, FIL1p, involved in derepression of the isocitrate lyase gene in Saccharomyces cerevisiae—a possible mitochondrial protein necessary for synthesis in mitochondria. Eur. J. Biochem. 256:212-220. [DOI] [PubMed] [Google Scholar]
- 29.Ko, Y. H., C. R. Cremo, and B. A. McFadden. 1992. Vanadate-dependent photomodification of serine 319 and 321 in the active site of isocitrate lyase from Escherichia coli. J. Biol. Chem. 267:91-95. [PubMed] [Google Scholar]
- 30.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon-Chung, K. J., I. Polacheck, and T. J. Popkin. 1982. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J. Bacteriol. 150:1414-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodge, J. K., E. Jackson-McChelski, D. L. Toffaletti, J. R. Perfect, and J. I. Gordon. 1994. Targeted gene replacement demonstrates that myristoyl-CoA: protein N-myristoyl transferase is essential for the viability of Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 91:12008-12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cell is required for fungal virulence. Nature 412:83-86. [DOI] [PubMed] [Google Scholar]
- 34.Luttik, M. A., P. Kotter, F. A. Salomons, I. J. Van Der Klel, J. P. Van Dijken, and J. T. Pronk. 2000. The Saccharomyces cerevisiae ICL2 gene encodes a mitochondrial 2-methylisocitrate lyase involved in propionyl-coenzyme A metabolism. J. Bacteriol. 182:7007-7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCammon, M. T. 1996. Mutants of Saccharomyces cerevisiae with defects in acetate metabolism: isolation and characterization of Acn-mutants. Genetics 144:567-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCusker, J. H., K. V. Clemons, D. A. Stevens, and R. W. Davis. 1994. Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42°C and form pseudohyphae. Infect. Immun. 62:5447-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKinney, J. D., Z. U. Hoener, K. Bentrup, E. J. Munoz-Elias, B. Chen, W. T. Chan, D. Swensen, J. C. Sacchettini, W. R. Jacobs, and D. G. Russell. 2000. Resistance of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS 100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy, J. W. 1998. Protective cell-mediated immunity against Cryptococcus neoformans. Res. Immunol. 149:373-386. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, R. T., J. Hua, B. Pryor, and J. K. Lodge. 2001. Identification of virulence mutants of the fungal pathogen Cryptococcus neoformans using signature-tagged mutagenesis. Genetics 157:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odom, A., S. Muir, E. Lim, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perfect, J. R., and D. T. Durack. 1985. Chemotactic activity of cerebrospinal fluid in experimental cryptococcal meningitis. Sabouraudia 23:37-46. [DOI] [PubMed] [Google Scholar]
- 43.Perfect, J. R., M. M. Hobbs, D. L. Granger, and D. T. Durack. 1988. Cerebrospinal fluid macrophage response to experimental cryptococcal meningitis: relationship between in vivo and in vitro measurements of cytotoxicity. Infect. Immun. 56:849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perfect, J. R., N. Ketabchi, G. M. Cox, C. I. Ingram, and C. Beiser. 1993. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J. Clin. Microbiol. 31:3305-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perfect, J. R., S. D. R. Lang, and D. T. Durack. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101:177-194. [PMC free article] [PubMed] [Google Scholar]
- 46.Perfect, J. R., B. B. Magee, and P. T. Magee. 1989. Separation of chromosomes of Cryptococcus neoformans by pulsed field gel electrophoresis. Infect. Immun. 57:2624-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perfect, J. R., D. L. Toffaletti, and T. H. Rude. 1993. The gene encoding for phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect. Immun. 61:4446-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhodes, J. C., I. Polacheck, and K. J. Kwon-Chung. 1982. Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect. Immun. 36:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salas, S. D., J. E. Bennett, K. J. Kwon-Chung, J. R. Perfect, and P. R. Williamson. 1996. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J. Exp. Med. 184:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 51.Sanger, F., A. R. Coulson, B. G. Barrell, A. J. H. Smith, and B. A. Roe. 1980. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J. Mol. Biol. 143:161-178. [DOI] [PubMed] [Google Scholar]
- 52.Schoeler, A., and H. J. Schueller. 1993. Structure and regulation of the isocitrate lyase gene (ICL1) from the yeast Saccharomyces cerevisiae. Curr. Genet. 23:375-381. [DOI] [PubMed] [Google Scholar]
- 53.Sharma, V., S. Sharma, Z. U. Hoener, K. Bentrup, J. D. McKinney, D. G. Russell, W. R. Jacobs, and J. C. Sacchettini. 2000. Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat. Struct. Biol. 7:663-668. [DOI] [PubMed] [Google Scholar]
- 54.Shen, L. L., J. Baranowski, J. Fostel, D. A. Montgomery, and P. A. Lartey. 1992. DNA topoisomerases from pathogenic fungi: targets for the discovery of antifungal drugs. Antimicrob. Agents Chemother. 36:2778-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, P., M. E. Cardenas, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Two cyclophilin A homologs with shared and distinct functions are expressed in Cryptococcus neoformans. EMBO Rep. 21:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waugh, M. S., C. B. Nichols, C. M. DeCesare, G. M. Cox, J. Heifman, and J. A. Alspaugh. 2002. RAS-1 and RAS2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191-201. [DOI] [PubMed] [Google Scholar]
- 57.Wayne, L. G., and K. Y. Lin. 1982. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect. Immun. 37:1042-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wills, E. A., I. S. Roberts, M. Del Poeta, J. Rivera, A. Casadevall, and G. M. Cox. 2001. Identification and characterization of the Cryptococcus neoformans phosphomannose isomerase-encoding gene, MAN1, and its impact on pathogenicity. Mol. Microbiol. 40:610-620. [DOI] [PubMed] [Google Scholar]
- 59.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed]