Abstract
The distinctive properties of Brucella outer membrane have been considered to be critical for Brucella sp. virulence. Among the outer membrane molecules possibly related to these properties, Omp10 and Omp19 are immunoreactive outer membrane lipoproteins. Moreover, these proteins of Brucella could constitute a new family of outer membrane proteins specifically encountered in the family Rhizobiaceae. We evaluated the impact of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. The omp10 mutant was dramatically attenuated for survival in mice and was defective for growth in minimal medium but was not impaired in intracellular growth in vitro, nor does it display clear modification of the outer membrane properties. Significantly fewer brucellae were recovered from the spleens of mice infected with the omp19 mutant than from those of mice infected with the parent strain at 4 and 8 weeks postinfection. The omp19 mutant exhibited an increase in sensitivity to the polycation polymyxin B and to sodium deoxycholate. These results indicate that inactivation of the omp19 gene alters the outer membrane properties of B. abortus.
Brucellae are gram-negative bacteria that cause human disease and significant worldwide economic losses due to infection of livestock. These bacteria are able to multiply within professional and nonprofessional phagocytes, but the exact mechanisms whereby Brucella spp. intracellularly parasitize the host are still to be defined (4, 5, 7, 15, 30, 36, 37).
The Brucella outer membrane has been proposed to be involved in virulence (i.e., resistance to bactericidal cationic peptides and polycations, permeability to hydrophobic agents, resistance to divalent cation chelators, and poor activation of bactericidal mechanisms by lipopolysaccharide [LPS]) (for a review, see reference 35). Correlation between these properties and specific surface molecules can be studied by genetically engineering mutations in the genes and determining the resultant phenotype.
The analysis of genetically defined rough mutants of Brucella melitensis and Brucella abortus confirmed the involvement of lipopolysaccharide O side chain in Brucella in vivo survival (1, 24, 25, 34, 45, 46). The recently identified BvrR-BvrS two-component regulatory system is involved in the control of outer membrane properties such as resistance to bactericidal polycations and is highly relevant for the virulence of B. abortus (40).
The molecular characterization of several Brucella outer membrane proteins (Omps) has been reported over the past years. The genes omp25, omp31, and omp2b (which encode the major 25-, 31-, and 36-kDa Omps, respectively) and the genes pal, omp10, omp19, and omp1 (which encode the 16-, 10-, 19-, and 89-kDa minor Omps, respectively) have been cloned and sequenced (10, 16, 18, 28, 31, 43, 44, 50; Bearden and Ficht, GenBank accession number U51683). Omp2b functions as a porin, and the 16-kDa Omp shows significant similarity to the peptidoglycan-associated lipoproteins Pal of gram-negative bacteria (32, 44). An omp25 mutant of B. melitensis is attenuated in mice and in the natural host (16a and 16b), indicating that Omp25 is critical for the maintenance of a B. melitensis infection. The 10- and 19-kDa Omps (Omp10 and Omp19, respectively) are surface-exposed lipoproteins (i.e., covalently linked to fatty acids) expressed in all six Brucella species and all their biovars (8, 42, 43). Omp10 and Omp19 share antigenic determinants with bacteria of the family Rhizobiaceae (9), and the only homologs present in the current sequence databases are two protein sequences deduced from the Mesorhizobium loti genome (27).
Antibody is elicited to Omp10 and Omp19. By using purified recombinant Omp10 and Omp19, a significant antibody response specific for these Omps could be detected in a large fraction of sera from sheep naturally infected by B. melitensis. However, there was no serologic response to these recombinant Omps in cattle naturally infected by B. abortus (29, 43). Kovach et al. also reported antibodies to Omp19 in infected mice, goats, dogs, and humans (28).
Notwithstanding the conservation of Omp10 and Omp19 among Brucella species, their possibly unique association with the Rhizobiaceae, and their immunoreactivity, no biological function has been assigned to these Omps. To define the function and the pathogenic significance of Omp10 and Omp19, we constructed deletion mutants of the corresponding genes by allelic replacement. The omp mutants were used to examine the role for both Omps in bacterial growth, outer membrane properties, and virulence, as assessed in appropriate in vitro (cell culture) and animal models.
(A portion of this work was presented at the 96th General Meeting of the American Society for Microbiology, Washington D.C., 1996.).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A variant of B. abortus strain 544, CO2 independent and resistant to nalidixic acid (Nalr), was obtained from J.-M. Verger, Laboratoire de Pathologie Infectieuse et Immunologie, Institut National de la Recherche Agronomique, Nouzilly, France. B. melitensis rough mutant B3B2 results from mini-Tn5 transposon insertion in the perosamine synthetase gene (25). Escherichia coli strain S17-1 (39) was kindly provided by G. Cornelis, Microbial Pathogenesis Unit, Christain de Duve Institute of Cellular Pathology, Brussels, Belgium. Nalidixic acid, kanamycin, ampicillin, and/or chloramphenicol was added when appropriate at the following concentrations: 25, 25, 25, and 30 μg/ml, respectively. Brucella organisms were grown on tryptic soy agar supplemented with 0.1% (wt/vol) yeast extract (TSAYE), on 2× YT (2YT) agar (38), in tryptic soy broth with 0.1% yeast extract, or in 2YT broth. A minimal medium slightly modified from that of Gerhardt et al. was used (nitrogen and energy were supplied by glutamic acid, lactic acid, and glycerol) (23). The Brucella growth rate in liquid media was monitored by recording a culture's optical density (OD) at 590 nm. For carbon substrate utilization patterns, tests were performed using 96-well GN MicroPlates (Biolog, Hayward, Calif.) mainly as directed by the manufacturer. Briefly, Brucella organisms freshly grown on TSAYE were recovered, suspended in 0.85% NaCl, and spectrophotometrically standardized to 2 × 109 cells/ml. These suspensions were then added to individual wells (150 μl) and incubated for 24 h at 37°C in 5% CO2. Color reactions were determined visually (51).
Construction and characterization of omp mutants.
In order to allow our constructs to be mobilized from a donor strain expressing RP4-conjugative function, like E. coli S17-1, the f1 origin of replication of pBluescript SK(−) (Stratagene, La Jolla, Calif.) was excised by SspI restriction and replaced by a 0.76-kb filled-in SmaI-SalI fragment encoding the RK2 origin of transfer (excised from pTJS82, kindly provided by G. Cornelis). This generated the plasmid pSK-oriT.
The plasmid pD192 was designed for the replacement of the first 320 bp of the omp19 coding sequence by the kanamycin resistance cassette (kan). A 479-bp BsmI-NruI fragment was excised from plasmid p191 (43) and replaced by the filled-in 1.3-kb BamHI kan cassette from pUC4K (Pharmacia P-L Biochemicals, Uppsala, Sweden). The entire insert containing 261 bp located upstream of omp19 and 223 bp of the end of omp19 separated by the kan marker was recovered by EcoRI digestion and subcloned into the EcoRI site of pSK-oriT, producing plasmid pD192. This plasmid was transferred to B. abortus 544 Nalr by conjugation as described by Verger et al. except that the mating time was shortened to 1 h (49). All the transconjugants isolated resulted from a single crossover in the 5′ flanking region of omp19 (5′ integrant) or in the 3′ end of omp19 (3′ integrant), as demonstrated by Southern blot analysis. No omp19 deletion mutant was isolated using pD192. The use of larger flanking regions should increase recombination frequency and, accordingly, mutation efficacy. In order to isolate a larger DNA fragment containing omp19, we took advantage of the pD192 integrant clones.
Genomic DNA was prepared from 5′ and 3′ integrant clones, digested with HindIII and BamHI, respectively, and ligated. After transformation into E. coli XL1-blue and selection on ampicillin plates, plasmid DNA was extracted from some colonies and digested by HindIII and BamHI, for clones recovered from a 5′ and a 3′ integrant, respectively. Plasmid pIa19.16 was selected, the 1.8-kb insert of which contained in addition to omp19 about 1 kb of sequence downstream of this coding sequence. Plasmid pIb19.22 was also selected and was characterized by a longer insert containing omp19 and about 10 kb upstream of this sequence. Both clones expressed Omp19 as shown by immunoblot analysis with anti-Omp19 monoclonal antibody (MAb) A76/02A04/A07. A 2.4-kb HindIII-SmaI fragment of pIb19.22 located immediately upstream of omp19 was subcloned into pSK-oriT as well as a 1-kb BglI-ClaI fragment of pIa19.16 encompassing the end of omp19. Plasmid pD193 was obtained by cloning the kan cassette into the EcoRI site located between these two segments. In pD193, omp19 coding sequence is deleted from the first 396 bp.
The plasmid pDS10 was designed for the replacement of part of the omp10 coding sequence by the kan cassette. Two Brucella DNA sequences, a 1.62-kb EcoRI-SmaI fragment and a 1.4-kb SacI-EagI fragment, both excised from p101.2 (43), and located upstream and downstream of omp10, respectively, were cloned into the pSK-oriT corresponding sites. The kan cassette was inserted as a 1.3-kb BamHI fragment from vector pUC4K between these two elements. In the resulting pD103 construct, the omp10 coding sequence was deleted from a 266-bp internal fragment. The sacB gene of Bacillus subtilis along with its regulator sacR sequence was excised from plasmid pUCD800 (kindly provided by G. Cornelis) (22) as a 2.6-kb BamHI-PstI fragment and cloned into the corresponding sites of pD103, leading to plasmid pDS10.
The constructs pD193 and pDS10, both unable to replicate in B. abortus, were conjugated from E. coli S17-1 into B. abortus 544 Nalr. A double crossover due to homologous recombination events in each of the omp flanking arms resulted in replacement of the omp sequence by the kan marker and loss of the delivery vector sequences. Brucella transconjugants were selected in the presence of nalidixic acid, kanamycin, and 5% sucrose when needed. Transconjugants were then tested for ampicillin sensitivity to screen for or to confirm (in the case of pDS10 transconjugants) loss of suicide vector sequences.
To provide genetic evidence in the transconjugants of omp replacement by the kan cassette, DNA isolated from the mutant and wild-type strains was digested with HindIII and hybridized to omp, kan, and sacB probes. A chromosomal DNA miniprep procedure was performed as described previously (48). An omp19-specific probe was prepared from the 0.96-kb EcoRI insert from plasmid p191 (43), and an omp10-specific probe was prepared from a 1.65-kb EcoRI-SmaI fragment containing the 5′ end of omp10 excised from p101.2 (43). A kan-specific probe was prepared from the BamHI fragment containing the kan cassette from pUC4K, and a sacB-specific probe was prepared from a 2.6-kb BamHI-PstI fragment from pUCD800 subcloned into pSK-oriT. Chemiluminescent detection of biotinylated probes was performed according to the Phototope-Star detection protocol (New England Biolabs, Schwalbach, Germany).
The authenticity of the mutants was also verified by immunoblot analysis on whole-cell extracts with the anti-Omp10 MAb A68/07G11/C10 and anti-Omp19 MAb.
All Brucella transconjugants analyzed were checked for purity, species, and biovar characterization by standard procedures by J.-M. Verger and M. Grayon (3).
Indirect enzyme-linked immunosorbent assay on whole Brucella cells was performed as described previously (6) with anti-Omp10 MAb to detect Omp10 and with anti-peptidoglycan MAb 3D6 to assess outer membrane integrity (11).
For electron scanning microscopy, freshly grown Brucella cells were suspended in phosphate-buffered saline (PBS) to an OD of 1 at 590 nm and killed by addition of gentamicin (50 μg/ml) and sodium azide (0.1%). Cells were fixed with glutaraldehyde (2%) for 15 min, and 1.5 ml was transferred on a 0.2-μm-pore-size cellulose acetate filter (Sartorius). Samples were dried, coated to a thickness of 20 nm with a Balzers DCM010 gold sputter coater, and viewed with a Philips XL20 scanning electron microscope.
Susceptibility assays.
Brucella strains were tested for sensitivity to bovine serum essentially as described by Corbeil et al. (12). Sera were collected from blood of two naive cattle and stored at −80°C. The log of the number of CFU per milliliter of fresh serum was subtracted from the log number of CFU per milliliter of heated serum. Results were expressed as the log of the number of bacteria killed by 1 ml of serum (referred to herein as the “log killed”). Bacterial survival after a controlled exposure to polymyxin B (7,870 U/mg; Sigma-Aldrich Chemie, Steinheim, Germany) was assayed essentially as described by Sola-Landa et al. (40). B. melitensis rough mutant B3B2 was used as a control strain sensitive to polymyxin B (25). Briefly, serial dilutions of polymyxin B prepared in 1 mM HEPES (pH 8.0) were made in 96-well microtiter-type plates. Bacteria resuspended at around 2 × 104 CFU/ml were dispensed into triplicate rows, and plates were incubated for 1 h at 37°C. Viable counts were performed by spreading 20 μl from each well onto 2YT agar. Results were expressed as the percentage of survival with respect to that of controls incubated without the peptide. Data presented are means of triplicate rows and are representative of four experiments. MICs of detergents (sodium dodecyl sulfate, sodium deoxycholate, and N-lauroyl-N-methylglycine [Sarkosyl] [all from Sigma-Aldrich]) were determined in 2YT plates. The sensitivities of Brucella strains to killing by H2O2 as well as by antibiotics (penicillin G, ampicillin, tetracycline, erythromycin, and rifampin [bioDiscs], Bio-Mérieux, Marcy l'Etoile, France) were evaluated by a disk sensitivity assay as described by Elzer et al. (17).
Cell culture and bacterial infection.
Survival of Brucella strains was evaluated in an immortalized cell line of bovine peritoneal macrophages (41) and in epitheloid human HeLa cells by the procedure described by Delrue et al. (14). Briefly, Brucella strains were grown to log phase for 18 h in 2YT broth in the presence of the appropriate antibiotics and added to cells at a multiplicity of infection of 200 to 300 in cell culture medium. Culture plates were centrifuged for 5 min (for macrophages) or 10 min (for HeLa cells) at 1,000 rpm in a Jouan centrifuge at room temperature and placed in a 5% CO2 atmosphere at 37°C. After 1 h, wells were washed and further incubated with culture medium supplemented with 50 μg of gentamicin (Life Technologies) per ml to kill the remaining extracellular brucellae. The number of intracellular viable brucellae was determined at 2, 24, and 48 h postinfection. The data presented are means of four to six culture wells and representative of three experiments.
Survival in the mouse model.
Groups of 8 (first assay)- or 10 (second assay)-week-old female BALB/c mice were inoculated intraperitoneally with 0.2 ml of a suspension containing around 104 CFU of each bacterial strain harvested with PBS from a 24-h TSAYE slope (exact doses were retrospectively assessed). At appropriate intervals postinoculation (p.i.), five mice from each treatment group were sacrificed for blood and spleen collection. The presence of O-antigen antibody in the mouse sera was determined by enzyme-linked immunosorbent assay. Spleens were weighed, and bacterial survival was determined following homogenization of the mouse spleens in 5 ml of PBS (first assay) or 2 ml of distilled water (second assay) with a stomacher 80 homogenizer. Serial dilutions of the spleen homogenates were plated in triplicate on TSAYE to determine bacterial counts. For the first assay, the detection limit was 50 CFU per spleen. For the second assay, the whole-spleen homogenates from mice infected with strain 544D10.1 and recovered at 8 and 14 weeks p.i. were plated in order to reduce the detection limit from 20 to 1 CFU per spleen. The number of CFU per spleen was expressed as the log number of CFU per spleen. The maintenance of the phenotypic and genetic markers was checked throughout these experiments. The 544D19 and 544D10 isolates tested showed the same phenotypic properties and immunoblot patterns as the inoculum, indicating that they were stable.
Statistical analysis.
To determine the significance of differences observed in our experiments, pairwise comparisons were performed by Scheffé tests, after a two-way analysis of variance providing the residual mean square estimate with the highest available degree of freedom number (13).
RESULTS
Construction of omp10 and omp19 mutants.
The majority of the omp19 coding sequence was removed from the chromosome of virulent B. abortus 544 Nalr by the gene replacement strategy originally described by Halling et al. (26). Among 278 Nalr Kanr colonies isolated following introduction of the pD193 deletion vector into B. abortus 544, only six were Amps. The failure of these six isolates to express Omp19 was confirmed by immunoblotting with an anti-Omp19 MAb. Two Kanr Amps colonies were chosen for further study and given the designation 544D19.1 and 544D19.2. Southern blot analysis of these two putative omp19 mutants confirmed gene replacement (data not shown). Following HindIII digestion, the 3.8-kb band characteristic of the presence of omp19 in the 544 parent DNA (48) was replaced by two bands of 3.1 and 1.6 kb in both 544D19 mutant DNAs, consistent with omp19 replacement by kan containing an HindIII site. The kan probe did not hybridize to the 544 DNA but did hybridize to the same-size fragments recognized by the omp19 probe from both 544D19 mutants.
Cloning of larger fragments upstream and downstream of omp19 into pD193 allowed us to isolate omp19 mutants (probably by increasing the frequency of crossovers), although with a low efficiency. To increase the efficiency of deletion mutant isolation upon conjugal transfer of replacement vector, we used the B. subtilis sacB gene as a counterselectable marker in B. abortus 544. Therefore, plasmid pDS10 was constructed by introducing the sacB marker into plasmid pD103, designed for omp10 gene replacement. All the Nalr Kanr transconjugants selected in the presence of sucrose were Amps. Wild-type 544 and two Amps Sucr clones were analyzed by Southern blotting. Due to an HindIII internal site in the omp10 probe, it hybridized with two bands of 4.8 and 4.5 kb in the 544 DNA (48). The 4.5-kb band was replaced by a 1-kb band in the Amps Sucr clones, consistent with replacement of omp10 coding sequence by kan. The kan probe did not hybridize to 544 DNA but did hybridize to 4.5- and 1-kb fragments in Amps Sucr clones. These data confirm that the allelic exchange had occurred, resulting in the omp10 gene being replaced by the kan cassette. Western blot analysis with an anti-Omp10 MAb confirmed that Omp10 was not expressed in the Amps Sucr clones. Two Nalr Kanr Amps Sucr clones were chosen for further study and named 544D10.1 and 544D10.2. Counterselection with sacB was also used successfully for the deletion of omp10 in other Brucella strains or species: B. abortus vaccine strain S19, B melitensis 16 M, and Brucella ovis Reo 198.
omp10 and omp19 are not essential genes for Brucella survival in vitro. This was shown by the deletion of the genes from several brucellae. In addition, deletion of the omp10 gene or omp19 gene had no detectable effect on conventional species and biovar phenotypic properties, and oxidative metabolic patterns (data not shown) (3). Omp10 was not shown to be involved in Tb, Iz, Wb, or R/C brucella phage adhesion or entry, since phage susceptibility patterns of the parent B. abortus, B. melitensis, Brucella suis, B. ovis, and the omp10 mutants are identical. Omp19 is also not necessary to infection by the first three of these brucella phages since the B. abortus omp19 mutant remains susceptible to these phages.
Survival of the omp mutants in the mouse model.
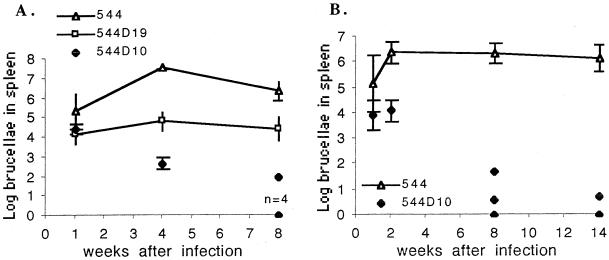
We compared the numbers of bacteria in the spleens of mice infected with 104 CFU of 544D10.1, 544D19.1, or 544 Nalr. Growth of 544D19.1 and that of 544 were parallel over the 8-week period, but mean numbers of bacteria in the spleens of 544D19.1-infected mice were always significantly lower (P = 0.05 at week 1 and P = 0.001 at weeks 4 and 8) than the number of bacteria in the spleens of 544-infected mice (Fig. 1A). In mice infected with 544D10.1, the number of bacteria decreased at 4 weeks p.i. At 8 weeks p.i., no bacteria (detection limit, 50 CFU) were detected in the spleens of four of the five mice infected with 544D10.1, and only 102 CFU per spleen was detected in the spleen of the remaining mouse. In contrast, mean counts in spleens from mice infected with 544 were higher than 105 CFU during the entire 8-week period. The 544D10 and 544D19 mutants colonized the liver (detection limit, 50 CFU) but almost disappeared from this organ by 4 weeks, in contrast to 544, which persisted at about 4 logs for the 8-week period. These results show a slight attenuation of the omp19 mutant in the mouse.
FIG. 1.
Virulence of B. abortus 544 Nalr and omp mutant strains in BALB/c mice. Mice were infected by intraperitoneal injection with 104 brucellae. Values are means (log number of CFU per spleen) ± standard deviations (error bars) (n = 5). For infection with strain 554D10.1 at 8 and 14 weeks p.i., individual results are shown. (A) First infection assay with 544 Nalr, 544D19.1, and 544D10.1. (B) Second infection assay with 544 Nalr and 554D10.1 (n = 4; no bacteria detected in four mice).
In order to precisely determine omp10 mutant clearance from BALB/c mice, we performed a second infection assay with strain 544D10.1 or 544 Nalr, with a lowered detection limit of 1 organism per spleen (Fig. 1B). The 544 Nalr parent strain quickly grew in spleens and reached high numbers that persisted over the 14-week period. Eight weeks after infection with 544D10.1, one of the mice was free of brucellae, and only between 1 and 47 CFU per spleen was detected in the other four mice. At 14 weeks p.i., five bacteria were detected in the spleen of only one of five mice infected with this strain. Serologic data confirmed that the mice had effectively been infected by Brucella even though no brucellae were recovered from their spleens. These data confirmed the results of the first assay and established that 544D10.1 was attenuated in the mouse.
The experiments described in the following sections were designed to determine the effect of the omp mutations to several host killing mechanisms.
Survival of the omp mutants in cells.
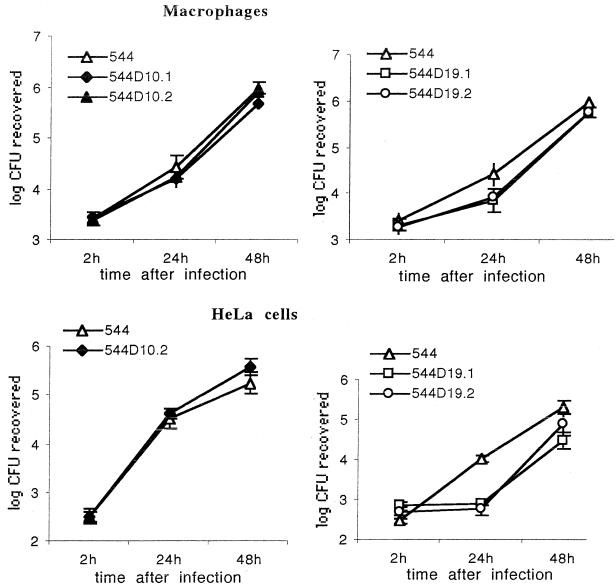
The ability of omp mutants to enter cells and to replicate intracellularly was studied in professional and nonprofessional phagocytes (Fig. 2). The parent B. abortus 544 strain replicated within bovine macrophages and HeLa cells. No significant difference in uptake and intracellular growth in both cell types between the omp10 mutants and their parent strain was seen.
FIG. 2.
Intracellular replication of B. abortus 544 Nalr, 544D19.1, 544D19.2, 544D10.1, and 544D10.2 in bovine macrophages or HeLa cells. The data presented are the results of a representative experiment and are means ± standard deviations (error bars) of plate counts from four to six culture wells.
The omp19 mutants replicate to levels close to those of 544 Nalr in bovine macrophages. However, in HeLa cells omp19 mutants replicate at a lower rate than do 544 cells during the first 24 h (P < 0.001). After 24 h, the replication rate increased and paralleled that of 544.
Physiological and morphological characterization of omp mutants.
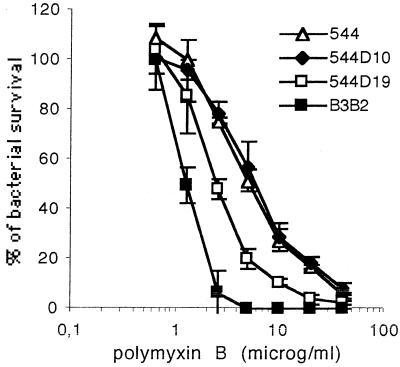
Compared to other gram-negative bacteria, Brucella outer membrane is resistant to bactericidal polycations such as polymyxin B. To evaluate the effect of the loss of Omp10 or Omp19 on B. abortus outer membrane properties, we first tested omp mutant viability after controlled exposure to polymyxin B. Omp10 mutants showed no increase in sensitivity to polymyxin B (Fig. 3). However, the survival of the omp19 mutant 544D19.1 was reduced relative to that of 544 Nalr over a polymyxin B concentration range of 2.5 to 20 μg/ml (P < 0.001). In addition, MIC assays showed that omp10 and omp19 mutants were more sensitive to sodium deoxycholate (1.5 times and 3 times more, respectively) than the parent strain, suggesting altered outer membrane properties. Sensitivities of mutant and parent strains to sodium dodecyl sulfate, Sarkosyl, and the five tested antibiotics were not significantly different.
FIG. 3.
Bactericidal effect of polymyxin B on B. abortus 544 Nalr, 544D19.1, 544D10.1, and B3B2. The data presented are the results of a representative experiment and are means ± standard deviations (error bars) of plate counts from three wells. Results are expressed as percentages of the brucellae surviving in wells incubated in the absence of the peptide.
In vitro exposure revealed no appreciable increase in the sensitivity of the omp mutants to H2O2.
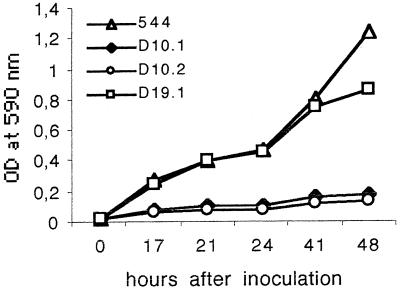
Mutant and parent strains have similar carbon substrate utilization patterns. The mutants showed no defect in the ability to grow in rich media (tryptic soy broth or 2YT broth). In contrast omp10 mutants were unable to grow in minimal medium independently of the addition of amino acids or nitrogenous bases (Fig. 4).
FIG. 4.
Growth of B. abortus 544 Nalr, 544D19.1, 544D10.1, and 544D10.2 in minimal medium. Cultures were inoculated to identical ODs from fresh cultures (0.03 at 590 nm), and growth was monitored by recording the ODs of the cultures at 590 nm at different time points during 48 h. These data are representative of three independent assays.
Mutation of Omp10 or Omp19 had no dramatic effect on the morphology of B. abortus 544 cells as determined by scanning electron microscopy, except that a very small proportion of 544D19 cells have a branched aspect or an irregular shape with swelling (data not shown).
DISCUSSION
The outer membrane lipoproteins Omp10 and Omp19 could constitute a new family of Omps specifically encountered in the Rhizobiaceae. No function has been assigned to these proteins, and analysis of the B. melitensis and B. suis genomic sequences surrounding the omp10 and omp19 genes has not provided more information about the function of these two lipoproteins.
Using a murine model, we found that deletion of the omp10 gene from B. abortus 544 dramatically reduces bacterial virulence. Over the 14-week study period, mice clear the mutant 544D10 more rapidly than the parent strain and display almost no splenomegaly. Since significant attenuation is observed by 2 weeks, omp10 mutant replication is probably hampered during the intracellular stage in phagocytes. The omp10 mutants showed no reduced resistance to polycationic peptide or to the reactive oxidative agent H2O2, two in vitro mimics of host intracellular defenses.
Since brucellae enter and replicate within professional and nonprofessional phagocytic cells, growth in both types of cells is used as a model system for measuring the attenuation of Brucella mutant strains. By using a bovine macrophage cell line and HeLa cells, no in vitro intracellular attenuation of the omp10 mutant was detected. Whether these discrepancies between in vivo and in vitro results are due to differences between mice and human or bovine cells remains to be determined. In addition, reduced omp10 mutant survival in mice is probably not attributable to enhanced extracellular killing by complement, since no increase in complement-mediated lysis was observed for this mutant (data not shown). The only detected effect of omp10 mutation on outer membrane properties was a 1.5-fold increase in sensitivity to sodium deoxycholate.
The omp10 mutant exhibited a marked growth defect in minimal medium, indicating dependence for growth on an unidentified compound absent from this medium (that does not seem to be secreted by the parent strain and made available by diffusion). If this dependence is relevant to the mutant attenuation in the mouse, availability of this compound in the Brucella replication niche would be a limiting factor for Brucella infection in mice.
The results obtained with mice suggest that omp10 mutant virulence should be further evaluated in the natural bovine host. In addition, the efficacy of omp10 mutant as a live vaccine against B. abortus should be investigated.
The sequence of the omp10 3′ region was determined. It contains, 138 bp downstream of the omp10 stop codon, a coding sequence for a homologue to the HemH ferrochelatase, the final enzyme of the heme biosynthetic pathway. Although we cannot definitely preclude a polar effect of omp10 mutation on hemH expression, omp10 mutants exhibited none of the phenotypes associated with hemH mutants in B. abortus and in other gram-negative bacteria: auxotrophy for hemin and brownish red color of colonies (2, 20, 21, 52). In addition, a putative transcription terminator is present 18 bases downstream from the omp10 stop codon (43). Attempts to complement the omp10 mutant with a copy of the omp10 gene cloned on the pBBR1MCS plasmid vector under the control of its own promoter failed, probably because of the overexpression of Omp10 in this strain.
Mice remained infected, but at a significantly lower level, with the omp19 mutant 544D19 over an 8-week period and exhibited almost no splenomegaly. The omp19 mutants showed a lower growth rate, at least in HeLa cells, during the first 24 h of infection, suggesting that they are more sensitive to the conditions within phagosomes before adapting to intracellular conditions and residency in an intracellular replication compartment. Disruption of the gene encoding Omp19 (referred to as an 18-kDa lipoprotein by these researchers) in vaccine strain B. abortus RB51 had no influence on the survival of this strain in mice (47), indicating that loss of Omp19 does not aggravate the attenuation of this rough strain.
Omp19 mutants exhibited altered outer membrane properties, as indicated by their reduced resistance to deoxycholate and polymyxin B. The low number of negatively charged groups in the Brucella LPS would account for the reduced affinity of the Brucella outer membrane for cationic peptides such as polymyxin B (33). The results of Freer et al. indicate that the involvement of Brucella outer membrane proteins in polymyxin B sensitivity “cannot be excluded, although their direct participation is unlikely” (19). Moreover, brucellae are sensitive to bovine complement-mediated lysis via the classical pathway (12). This sensitivity is a factor of extracellular killing within the host. Preliminary results suggest that the omp19 mutant exhibited an increased sensitivity to bovine serum complement compared to that of the parent 544 strain (1.8 log killed compared to 1.1 log killed).
All together, these results suggest an indirect effect of Omp19 deletion on interaction between polymyxin B and Brucella outer membrane, either at the initial interaction step or beyond this step. Freer et al. (19) indeed also showed that non-LPS outer membrane molecules participate in the arrangement of the LPS lattice. It is possible that the absence of Omp19 modifies interactions between outer membrane components which could disorganize this lattice. Enhanced sensitivities to deoxycholate and bovine complement-mediated lysis are also consistent with this hypothetical fragility of the omp19 mutant outer membrane. It would be interesting to evaluate the effect of omp19 mutation on the naturally rough B. ovis strain outer membrane properties to eliminate the contribution of the O-chain.
The slight but significant attenuation of 544D19 mutant in mice could be attributed to altered outer membrane properties of this mutant which would affect both its extracellular and intracellular survival.
Further studies are necessary to establish Omp10 and Omp19 function in Brucella growth and pathogenesis.
Acknowledgments
We gratefully acknowledge the electron microscopy department staff of the University of Namur, K. de Fays and E. Depiereux for their help in the statistical analysis of results, K. Kaniga for helpful discussion about the use of sacB, S. Halling and I. Lopez-Goni for their advice, and M. Grayon and J.-M. Verger for their valuable collaboration.
This work was supported by the Commission of the European Communities, contract Eclair AGRE-CT90-0049-C(EDB).
Editor: R. N. Moore
REFERENCES
- 1.Allen, C., G. Adams, and T. Ficht. 1998. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect. Immun. 66:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almiron, M., M. Martinez, N. Sanjuan, and R. A. Ugalde. 2001. Ferrochelatase is present in Brucella abortus and is critical for its intracellular survival and virulence. Infect. Immun. 69:6225-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alton, G., L. Jones, R. Angus, and J.-M. Verger. 1988. Techniques for the brucellosis laboratory. Institut National de la Recherche Agronomique, Paris.
- 4.Anderson, T. D., and N. F. Cheville. 1986. Ultastructural morphometric analysis of Brucella abortus-infected trophoblasts in experimental placentitis. Am. J. Pathol. 124:226-237. [PMC free article] [PubMed] [Google Scholar]
- 5.Arenas, G., A. S. Staskevich, A. Aballay, and L. Mayorga. 2000. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect. Immun. 68:4255-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowden, R., A. Cloeckaert, M. Zygmunt, S. Bernard, and G. Dubray. 1995. Surface exposure of outer membrane protein and lipopolysaccharide epitopes in Brucella species studied by enzyme-linked immunosorbant assay and flow cytometry. Infect. Immun. 63:3945-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canning, P. 1990. Phagocyte function in resistance to brucellosis, p. 151-163. In L. G. Adams (ed.), Advances in brucellosis research. Texas A&M University Press, College Station.
- 8.Cloeckaert, A., P. de Wergifosse, G. Dubray, and J. N. Limet. 1990. Identification of seven surface exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labelling for electron microscopy and enzyme-linked immunosorbent assay. Infect. Immun. 58:3980-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cloeckaert, A., A. Tibor, and M. Zygmunt. 1999. Brucella outer membrane lipoproteins share antigenic determinants with bacteria of the family Rhizobiaceae. Clin. Diagn. Lab. Immunol. 6:627-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloeckaert, A., J. M. Verger, M. Grayon, M. S. Zygmunt, and O. Grepinet. 1996. Nucleotide sequence and expression of the gene encoding the major 25-Kilodalton outer membrane protein of Brucella ovis: evidence for antigenic shift, compared with other Brucella species, due to a deletion in the gene. Infect. Immun. 64:2047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cloeckaert, A., M. Zygmunt, P. de Wergifosse, G. Dubray, and J. Limet. 1992. Demonstration of peptidoglycan-associated Brucella outer-membrane proteins by use of monoclonal antibodies. J. Gen. Microbiol. 138:1543-1550. [DOI] [PubMed] [Google Scholar]
- 12.Corbeil, L. B., K. Blau, T. I. Inzana, K. H. Neilsen, R. H. Jacobson, C. R. R., and A. J. Winter. 1988. Killing of Brucella abortus by bovine serum. Infect. Immun. 56:3251-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dagnelie, P. 1998. Statistique théorique et appliquée. De Boek et Larcier, Brussels, Belgium.
- 14.Delrue R.-M., M. J. Martinez-Lorenzo, P. Lestrate, I. Danese, V. Bielarz, P. Mertens, X. De Bolle, A. Tibor, J.-P. Gorvel, and J.-J. Letesson. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol. 3:487-497. [DOI] [PubMed] [Google Scholar]
- 15.Detilleux, P. G., B. L. Deyoe, and N. F. Cheville. 1990. Penetration and intracellular growth of Brucella abortus in nonphagocytic cells in vitro. Infect. Immun. 58:2320-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Wergifosse, P., P. Lintermans, J. N. Limet, and A. Cloeckaert. 1995. Cloning and nucleotide sequence of the gene coding for the major 25-kilodalton outer membrane protein of Brucella abortus. J. Bacteriol. 177:1911-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Edmonds, M., A. Cloeckaert, and P. Elzer. 2002. Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet. Microbiol. 88:205-221. [DOI] [PubMed] [Google Scholar]
- 16b.Edmonds, M., A. Cloeckaert, S. Hagius, L. Samartino, W. Fulton, J. Walker, F. Enright, N. Booth, and P. Elzer. 2002. Pathogenicity and protective activity in pregnant goats of a Brucella melitensis Δomp25 deletion mutant. Res. Vet. Sci. 72:235-239. [DOI] [PubMed] [Google Scholar]
- 17.Elzer, P., R. Phillips, M. Kovach, K. Peterson, and R. Roop II. 1994. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect. Immun. 62:4135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ficht, T., S. Bearden, B. Sowa, and L. Adams. 1989. DNA sequence and expression of the 36-kilodalton outer membrane protein gene of Brucella abortus. Infect. Immun. 57:3281-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freer, E., E. Moreno, I. Moriyon, J. Pizzaro-Cerda, A. Weintraub, and J.-P. Gorvel. 1996. Brucella-Salmonella lipopolysaccharide chimeras are less permeable to hydrophobic probes and more sensitive to cationic peptides and EDTA than are their native Brucella sp. counterparts. J. Bacteriol. 178:5867-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frustaci, J., and M. O'Brian. 1992. Characterization of a Bradyrhizobium japonicum ferrochelatase mutant and isolation of the hemH gene. J. Bacteriol. 174:4223-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frustaci, J., and M. O'Brian. 1993. The Escherichia coli visA gene encodes ferrochelatase, the final enzyme of the heme biosynthetic pathway. J. Bacteriol. 175:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerhardt, P., L. A. Tucker, and J. B. Wilson. 1950. The nutrition of Brucellae: utilization of single amino acids for growth. J. Bacteriol. 59:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godfroid, F., A. Cloeckaert, B. Taminiau, I. Danese, A. Tibor, X. De Bolle, P. Mertens, and J.-J. Letesson. 2000. Genetic organization of the lipopolysaccharide O-antigen biosynthesis region of Brucella melitensis 16M (wbk). Res. Microbiol. 151:655-668. [DOI] [PubMed] [Google Scholar]
- 25.Godfroid, F., B. Taminiau, I. Danese, P. Denoel, A. Tibor, V. Weynants, A. Cloeckaert, J. Godfroid, and J.-J. Letesson. 1998. Identification of the perosamine synthetase gene of Brucella melitensis 16M and involvement of lipopolysaccaride O side chain in Brucella survival in mice and in macrophages. Infect. Immun. 66:5485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halling, S. M., P. G. Detilleux, F. M. Tatum, B. A. Judge, and J. E. Mayfield. 1991. Deletion of the BCSP31 gene of Brucella abortus by replacement. Infect. Immun. 59:3863-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 28.Kovach, M. E., P. H. Elzer, G. T. Robertson, R. L. Chirhart-Gilleland, M. A. Christensen, K. M. Peterson, and M. R. Roop II. 1997. Cloning and nucleotide sequence analysis of a Brucella abortus gene encoding an 18 kDa immunoreactive protein. Microb. Pathogen. 22:241-246. [DOI] [PubMed] [Google Scholar]
- 29.Letesson, J.-J., A. Tibor, G. van Eynde, V. Wansard, V. Weynants, P. A. Denoel, and E. Saman. 1997. Humoral immune response of Brucella infected cattle, sheep and goats to eight purified recombinant Brucella proteins in iELISA. Clin. Diagn. Lab. Immunol. 4:556-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liautard, J.-P., A. Gross, J. Dornand, and S. Köhler. 1996. Interactions between professional phagocytes and Brucella spp. Microbiologia 12:197-206. [PubMed] [Google Scholar]
- 31.Lichtfouse, B. 1997. Identification of immunogenic domains and peptides mimics of Brucella lipopolysaccharide and outer membrane protein, OMP89, potentially useful in the improvement of brucellosis control. Ph.D. thesis. University of Namur, Namur, Belgium.
- 32.Marquis, H., and T. A. Ficht. 1993. The omp2 gene locus of Brucella abortus encodes for two homologous outer membrane proteins with properties characteristic of bacterial porins. Infect. Immun. 61:3785-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez de Tejada, G., J. Pizzaro-Cerda, E. Moreno, and I. Moriyon. 1995. The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect. Immun. 63:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McQuiston, J. R., R. Vemulapalli, T. J. Inzana, G. G. Schurig, N. Sriranganathan, D. Fritzinger, T. L. Hadfield, R. A. Warren, L. E. Lindler, N. Snellings, D. Hoover, S. M. Halling, and S. M. Boyle. 1999. Genetic characterization of a Tn5-disrupted glycosyltransferase gene homolog in Brucella abortus and its effect on lipopolysaccharide composition and virulence. Infect. Immun. 67:3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriyon, I., and I. Lopez-Goni. 1998. Structure and properties of the outer membranes of Brucella abortus and Brucella melitensis. Int. Microbiol. 1:19-26. [PubMed] [Google Scholar]
- 36.Pizarro-Cerda, J., S. Meresse, R. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goni, E. Moreno, and J.-P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rittig, M., M.-T. Alvarez-Martinez, F. Porte, J.-P. Liautard, and B. Rouot. 2001. Intracellular survival of Brucella spp. in human monocytes involves conventional uptake but special phagosomes. Infect. Immun. 69:3995-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:783-791. [Google Scholar]
- 40.Sola-Landa, A., J. Pizzaro-Cerda, M.-J. Grillo, E. Moreno, I. Moriyon, J.-M. Blasco, J.-P. Gorvel, and I. Lopez-Goni. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125-138. [DOI] [PubMed] [Google Scholar]
- 41.Stabel, J. R., and T. J. Stabel. 1995. Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet. Immunol. Immunopathol. 45:211-220. [DOI] [PubMed] [Google Scholar]
- 42.Tibor, A., B. Decelle, and J.-J. Letesson. 1999. Outer membrane proteins Omp10, Omp16, and Omp19 of Brucella spp. are lipoproteins. Infect. Immun. 67:4960-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tibor, A., E. Saman, P. de Wergifosse, A. Cloeckaert, J. Limet, and J. J. Letesson. 1996. Molecular characterization, occurrence, and immunogenicity in infected sheep and cattle of two minor outer membrane proteins of Brucella abortus. Infect. Immun. 64:100-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tibor, A., V. Weynants, P. Denoel, B. Lichtfouse, X. De Bolle, E. Saman, J. Limet, and J. J. Letesson. 1994. Molecular cloning, nucleotide sequence, and occurrence of a 16.5-kilodalton outer membrane protein of Brucella abortus with similarity to PAL lipoproteins. Infect. Immun. 62:3633-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ugalde, J., C. Czibener, M. Feldman, and R. A. Ugalde. 2000. Identification and characterization of the Brucella abortus phosphoglucomutase gene: role of lipopolysaccharide in virulence and intracellular multiplication. Infect. Immun. 68:5716-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vemulapalli, R., Y. He, L. S. Buccolo, S. M. Boyle, N. Sriranganathan, and G. G. Schurig. 2000. Complementation of Brucella abortus RB51 with a functional wboA gene results in O-Antigen synthesis and enhanced vaccine efficacy but no change in rough phenotype and attenuation. Infect. Immun. 68:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vemulapalli, R., S. Cravero, C. Calvert, T. Toth, N. Sriranganathan, S. Boyle, O. Rossetti, and G. Schurig. 2000. Characterization of specific immune responses of mice inoculated with recombinant vaccinia virus expressing an 18-kilodalton outer membrane protein of Brucella abortus. Clin. Diagn. Lab. Immunol. 7:114-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verger, J.-M., M. Grayon, A. Tibor, V. Wansard, J.-J. Letesson, and A. Cloeckaert. 1998. Differentiation of Brucella melitensis, B. ovis and B. suis biovar 2 strains by use of membrane protein- or cytoplasmic protein-specific gene probes. Res. Microbiol. 149:509-517. [DOI] [PubMed] [Google Scholar]
- 49.Verger, J. M., M. Grayon, E. Chaslus-Dancla, M. Meurisse, and J. P. Lafont. 1993. Conjugative transfer and in vitro/in vivo stability of the broad-host-range IncP R751 plasmid in Brucella spp. Plasmid 29:142-146. [DOI] [PubMed] [Google Scholar]
- 50.Vizcaino, N., A. Cloeckaert, M. S. Zygmunt, and G. Dubray. 1996. Cloning, nucleotide sequence, and expression of the Brucella melitensis omp31 gene coding for an immunogenic major outer membrane protein. Infect. Immun. 64:3744-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, J., M. Janda, and P. Duffey. 1992. Preliminary studies on the use of carbon substrate utilization patterns for identification of Brucella species. Diagn. Microbiol. Infect. Dis. 15:109-113. [DOI] [PubMed] [Google Scholar]
- 52.Xu, K., J. Delling, and T. Elliott. 1992. The genes required for heme synthesis in Salmonella typhimurium include those encoding alternative functions for aerobic and anaerobic coproporphyrinogen oxidation. J. Bacteriol. 174:3953-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]