Abstract
The mechanisms underlying the adherence of Escherichia coli O157:H7 and other enterohemorrhagic E. coli (EHEC) strains to intestinal epithelial cells are poorly understood. We have identified a chromosomal region (designated lpfABCC′DE) in EHEC O157:H7 containing six putative open reading frames that was found to be closely related to the long polar (LP) fimbria operon (lpf) of Salmonella enterica serovar Typhimurium, both in gene order and in conservation of the deduced amino acid sequences. We show that lpfABCC′DE is organized as an operon and that its expression is induced during the exponential growth phase. The lpf genes from EHEC strain EDL933 were introduced into a nonfimbriated (Fim−) E. coli K-12 strain, and the transformed strain produced fimbriae as visualized by electron microscopy and adhered to tissue culture cells. Anti-LpfA antiserum recognized a ca. 16-kDa LpfA protein when expressed under regulation of the T7 promoter system. The antiserum also cross-reacted with the LP fimbriae in immunogold electron microscopy and Western blot experiments. Isogenic E. coli O157:H7 lpf mutants derived from strains 86-24 and AGT300 showed slight reductions in adherence to tissue culture cells and formed fewer microcolonies compared with their wild-type parent strains. The adherence and microcolony formation phenotypes were restored when the lpf operon was introduced on a plasmid. We propose that LP fimbriae participate in the interaction of E. coli O157:H7 with eukaryotic cells by assisting in microcolony formation.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is recognized as a significant enteric pathogen that has been implicated in numerous outbreaks worldwide (reviewed in reference 24). This organism colonizes the intestine and can cause bloody or nonbloody diarrhea and hemolytic uremic syndrome. A potent cytotoxin, Shiga toxin (Stx), is the best-characterized virulence factor, but many aspects of the pathogenesis of the disease associated with E. coli O157:H7 are poorly characterized. In particular, the mechanisms underlying the adherence of EHEC to intestinal epithelial cells are not well understood (24, 27). Colonization of the gastrointestinal tract, which is presumably mediated by specific adherence factors, is a key aspect of enteric infection caused by EHEC. Several potential virulence determinants of EHEC have been described, but the only adherence factor that has been demonstrated to play a role in intestinal colonization in vivo in an animal model is the outer membrane protein intimin (8, 22, 43). Most EHEC and all enteropathogenic E. coli (EPEC) strains produce this adhesin (17). Intimin, encoded by the eae gene, is located within the locus for enterocyte effacement (LEE) pathogenicity island, which is required for the classic attaching and effacing intestinal lesion produced by these organisms (14, 15, 21).
The presence of a second adherence factor has been described in EPEC but not in EHEC strains. The type IV bundle-forming pilus (BFP) encoded by the plasmid of EPEC strains is involved in bacterium-to-bacterium adherence in the localized adherence pattern (4, 12) and potentially in direct interaction with the host epithelial cells (41, 42). Neither BFP nor analogous adhesins have been described in EHEC, but the existence of intestinal adherence factors distinct from intimin is suggested by the isolation of human EHEC strains of serotypes other than O157:H7 that lack the eae gene but are still associated with bloody diarrhea or hemolytic uremic syndrome (10).
Several research groups have explored this hypothesis and proposed potential novel adhesin factors. Tarr et al. (37) identified Iha (Vibrio cholerae IrgA-homologue adhesin) in E. coli O157:H7. This protein was associated with adherence to HeLa cells while expressed in a nonfimbriated E. coli strain, but no difference in adherence was observed with an iha mutant of O157:H7. Nicholls et al. (25) characterized a chromosomal genetic locus termed efa1 (EHEC factor for adherence) in an EHEC strain serotype O111:H− and observed that this locus is associated with high levels of adherence to cultured Chinese hamster ovary (CHO) cells. They demonstrated that the efa1 locus was necessary for the in vitro adhesion to CHO cells and that the efa1 isogenic mutant strain lost its ability to adhere and also was defective in its hemagglutination and autoaggregation phenotypes. Tatsuno et al. (39) performed a transposon mutagenesis in the EHEC O157:H7 strain (O157Sakai), and the insertion mutants were screened for their ability to adhere to Caco-2 cells. Almost half of the insertion mutants were found within the LEE pathogenicity island, while the other half were mapped to open reading frames (ORFs) with unknown functions or with functions not directly associated with adherence. Their results suggested that EHEC might contain additional adherence-associated loci which are not contained within the LEE pathogenicity island. Recently, Brunder et al. (6) characterized a gene cluster in the large plasmid of a sorbitol-fermenting EHEC O157:H− strain which is required for the expression of fimbriae with homology to the P fimbriae of uropathogenic E. coli. The Sfp (for sorbitol-fermenting EHEC O157 fimbriae, plasmid encoded) fimbriae mediate mannose-resistant hemagglutination, but this sfp gene cluster is restricted to sorbitol-fermenting EHEC O157:H− strains and is absent in other EHEC serotypes, including O157:H7.
In this work, we characterized a chromosomal fimbrial operon of E. coli O157:H7. Sequence analysis indicated that this operon showed high similarity to the long polar (LP) fimbria (lpf) operon of Salmonella enterica serovar Typhimurium and to a lesser degree to other well-characterized fimbrial operons. Introduction of the EHEC lpf operon into a nonfimbriated E. coli K-12 strain resulted in the expression of fimbriae and increased adhesion to tissue culture cells. We also provide evidence suggesting that LP fimbriae participate in the adherence of E. coli O157:H7 to eukaryotic cells and play a role in microcolony formation.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The strains were routinely grown in Luria-Bertani (LB) broth or on Luria agar at 37°C (20). Additional media used to grow bacteria were CFA agar (11) and MacConkey agar (Difco). When required, antibiotics were added to the media at the following concentrations: kanamacyn (Km) 50 μg/ml, ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 30 μg/ml; and streptomycin (Sm), 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| EDL933 | Prototype EHEC O157:H7 | 30 |
| 86-24 | EHEC O157:H7 strain 86-24; Smr Nalr | 38 |
| CVD468 | 86-24; lpfA::cat Smr Cmr | This study |
| AGT300 | Derivative of wild-type EHEC O157:H7 strain 87-23 (stx mutant); Smr | This study |
| AGT301 | AGT300; lpfA::cat Smr Cmr | This study |
| BL21(DE3) | hsdS gal (λcIts857ind1Sam7 nin5 lacUV5-T7 gene 1) | 35 |
| ORN172 | E. coli Δfim Kmr | 46 |
| SM10 (λ Pir) | thi thr leuB tonA lacY supE recA::RP4-2-Tc::Mu-Km Kmr | 33 |
| Plasmids | ||
| pJOR5 | pACYC184 with 5,929-bp fragment containing lpfABCC′DE; Cmr | This study |
| pSK+ | pBluescript cloning vector; Apr | Stratagene |
| plpfA | pBluescript SK(+) with KpnI/BamHI; lpfA Apr | This study |
| plpfA::cat | cat cassette in SmaI of lpfA | This study |
| pBR322 | Cloning vector; Apr Cmr Tcr | New England BioLabs |
| pLPF100 | pBR322 with 5,929-bp BamHI; lpfABCC′DE Apr | This study |
| pACYC184 | Cloning vector; source of Cmr cassette; Cmr Tcr | New England BioLabs |
| pRS551 | Protein fusion vector; Apr Kmr | 34 |
| pPLPFA | pRS551 with EcoRI/BamHI;1,164-bp lpf promoter region | This study |
| pT7-5 | T7 expression system vector; Apr | 36 |
| pLPFA01 | pT7-5 with EcoRI/BamHI;537-bp lpfA ORF | This study |
| pCVD442 | Suicide vector; Apr Sucs | 7 |
| pLPF::cat | lpfA::cat in pCVD442 | This study |
Recombinant DNA techniques.
Plasmid DNA was isolated by the method of Kado and Liu (16). Alternatively, a Qiagen QIAprep plasmid preparation kit was used to isolate plasmid DNA from 3 ml of overnight bacterial culture. Reagents were used according to the manufacturers' protocols. Plasmids were introduced into clinical isolates of E. coli by electroporation as described by Dower et al. (9). Restriction endonuclease analyses, ligation and transformation of plasmid DNA, and isolation of chromosomal DNA from bacteria were performed following standard methods (20, 31).
Cloning of the lpf operon.
Total genomic DNA from strain EDL933 was isolated and used as a template for PCR amplification with Platinum Pfx DNA polymerase (Gibco BRL, Life Technologies) with the primer pair 5LPF (5′-CGGGATCCGTATTGCGTGAGGCGCATATTTAGCCAGAAA-3′) and 3LPF (5′-CGGGATCCGTGCAAGTCCGGAATAGACCATTTTAACGGA-3′). The PCR product was purified and digested with BamHI, and the 5,929-bp product was isolated from an agarose gel and ligated into the BamHI site of the plasmid pACYC184 to create pJOR5. The ligated products were transformed into E. coli strain ORN172, and recombinant clones containing the lpf operon were selected by PCR and plasmid screening and confirmed by DNA sequencing. The nucleotide sequence of the recombinant lpf operon showed 100% identity to the chromosomal lpf operon of strain EDL933. The lpf operon was also cloned into the BamHI site of pBR322 to obtain pLPF100. ORN172 containing either pJOR5 or pLPF100 was selected for further analysis. Electron microscopic analysis confirmed that pJOR5 and pLPF100 encode the EHEC LP fimbriae (see Results).
RNA extraction and RT-PCR technique.
EHEC strain EDL933 was grown to exponential phase (optical density at 600 nm [OD600], 0.5), and whole-cell RNA was isolated using the Trizol reagent (Gibco-BRL) according to the manufacturer's instructions. The RNA was treated with DNase to eliminate contaminating DNA, and cDNA was synthesized using the random primers provided in the Superscript First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer's instructions. The primers used for the reverse transcriptase (RT)-PCR analysis were A (5′-CGATTTCAACCTGTCTTACGAG-3′), B (5′-GCTATGCAGCAATCGTTTGAAC-3′), C (5′-GGGAATTCAATTTTTTAAATGGAGTTTTTC-3′), and D (5′-CGGTGGCTTCCTGGAAGGT-3′). PCRs on the RT product and no RT control were performed with Taq DNA polymerase using standard procedures.
Construction of lpfA isogenic mutants.
EHEC strains defective in expression of LP fimbriae were constructed in the chromosome of strains 86-24 and AGT300 (Table 1) by marker exchange as follows. The fimbrial subunit gene lpfA was amplified by PCR with the primer pairs 5KLPF (5′-CGGGTACCATGCCTGCTTT-3′) plus 3SMLPF (5′-GGCCCGGGCAAAACCTTTCGAAATCAAA-3′) and 3BILPF (5′-CCGGATCCGTGTTATCACCATTGGT-3′) plus 5SMLPF (5′-GGCCCGGGATTTGTCACCAACCGC-3′) and cloned as a KpnI/SmaI 5′ fragment and an SmaI/BamHI 3′ fragment, respectively, in pBluescript SK(+) (Stratagene). The cloned lpfA gene was digested with SmaI, and the cat cassette (obtained by PCR from pACYC184) was introduced. The disrupted gene was amplified by PCR using Pwo polymerase (Boehringer Mannheim) and cloned into the suicide vector pCVD442 (7). The resulting plasmid, pLPF::cat, was introduced into 86-24 and AGT300 by conjugation using the donor strain SM10 (λ Pir). Colonies resistant to chloramphenicol and sucrose were tested for ampicillin sensitivity. The presence of the cat cassette within the chromosomal lpfA gene of CVD468 and AGT301 was confirmed by PCR with the primers listed above.
Bacterial adhesion to epithelial cells.
The ability of E. coli lpf+[supi]+ and lpf mutant strains to adhere to HeLa and Madin-Darby bovine kidney (MDBK) cell monolayers was assessed as previously described (37, 44). The cells were grown to semiconfluence at 37°C in 5% CO2 in 24-well plates (Corning) in Dulbecco's minimal essential medium (DMEM) with 10% (vol/vol) heat-inactivated fetal bovine serum, 2 mM l-glutamine, penicillin (100,000 IU/liter), and streptomycin (100 mg/liter). Before use, the cells were washed with sterile phosphate-buffered saline (PBS; pH 7.4) and replenished with DMEM containing 1% d-mannose. The strains were grown in LB broth overnight at 37°C, and for both qualitative and quantitative assays, tissue culture cells were incubated with 107 bacteria per well for 3 or 6 h at 37°C. The monolayers were washed, fixed, and stained with Giemsa solution for microscopic evaluation. To quantify E. coli lpf+[supi]+ and lpf mutant adherence, the infected monolayers were washed two times with PBS, and the adherent bacteria were recovered with 200 μl of 0.1% Triton X-100 in PBS buffer and plated on Luria agar plates containing the proper antibiotic. Data are expressed as the percentage of the bacterial inoculum recovered from triplicate wells and are the mean of at least two separate experiments. Statistical difference was expressed as the P value determined by a t test analysis.
Construction of the lpf-lacZ promoter fusion.
An operon fusion with lacZ was constructed by amplifying the regulatory region of the lpf operon in strain 86-24 by PCR using Pwo polymerase with the primer pairs 5RPLPFA (5′-CCGAATTCGCTATCGGTTCTTATG-3′) and 3BPLPFA (5′-GCGGATCCGGCAAAAACGACCTTTTTC-3′). The PCR product was digested and cloned into the EcoRI and BamHI sites of plasmid pRS551, which contains a promoterless lac operon (34), to create plasmid pPLPFA.
β-Galactosidase assay.
The E. coli strains containing the lpf promoter::lacZ fusion were grown with shaking at 250 rpm for 18 h at 37°C in LB broth, diluted 1:100 in fresh DMEM or LB broth, and grown at 37°C to early, mid-, and late exponential phase (OD600 = 0.3, 0.6, and 0.9, respectively). The cultures were diluted 1:10 in Z buffer (Na2HPO4 [0.06 M], NaH2PO4 [0.04 M], KCl [0.01 M], MgSO4 [0.001 M], and β-mercaptoethanol [0.05 M]) and were assayed for β-galactosidase activity using o-nitrophenyl-β-d-galactopyranoside as the substrate as previously described (23).
Preparation of antiserum.
Anti-LpfA (α-LpfA) rabbit immune serum was obtained from Zymed Laboratories, Inc., using purified peptide with the sequence TGYGNAQVDFNLSY-COOH, corresponding to the C-terminal region of LpfA conjugated to the immunogen keyhole limpet hemocyanin. LpfA peptide antiserum was prepared by multiple absorption with nonfimbriated E. coli strain ORN172 grown at 37°C. LpfA antiserum from S. enterica serovar Typhimurium was kindly provided by A. J. Bäumler (Texas A&M University).
Cloning the lpfA gene in the T7 promoter system.
The lpfA ORF was amplified by PCR with primer pairs 5LPFA (5′-GGGAATTCAGGAGGTTAAATGGAGTTTTTC-3′) and 3LPFA (5′-CGGGATCCGATTACTCGTAAGACAG-3′), digested and cloned into the EcoRI and BamHI sites of pT7-5, a vector in which transcription is directed by the T7 RNA polymerase promoter (36). The recombinant plasmid pLPFA01 was transformed into E. coli strain BL21(DE3), and the expression of LpfA was induced with 0.1 M IPTG (isopropyl-β-d-thiogalactopyranoside) as previously described (36).
Preparation of fimbrial crude extracts.
In brief, bacteria from 40 MacConkey or CFA agar plates were harvested and suspended in 100 ml of PBS buffer. The fimbriae were mechanically shredded by vortexing and harvested by centrifugation (8,000 × g for 20 min). The supernatant was transferred to a fresh tube, and the remaining bacteria were separated by centrifugation (12,000 × g for 30 min). The crude fimbrial extracts were harvested by centrifugation (40,000 × g for 30 min) and resuspended in PBS buffer. For Western blot assays, overnight cultures grown in DMEM were diluted to an OD600 of 1.0, and the bacteria were recovered by centrifugation (8,000 × g for 20 min). The bacterial pellet was resuspended in distilled water acidified with concentrated HCl to pH 1.8. The bacterial suspensions were boiled for 5 min, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) solubilization buffer was added. The samples were neutralized with NaOH and then separated in SDS-PAGE gels.
Western blot assay.
Crude cell lysates and fimbrial extracts were separated by SDS-12% PAGE minigels according to the method of Laemmli (19). Proteins were either stained with Coomassie brilliant blue or transferred to Immobilon-P (polyvinylidene difluoride) membranes (Millipore) using a Trans-Blot SD transfer cell (Bio-Rad) at 15 V for 22 min. The transfer of proteins was verified by staining the membrane with Ponceau S. The membrane was blocked with a PBS (pH 7.4)-0.5% Triton X-100 solution containing 5% nonfat milk. Incubations with primary (1:10,000) and secondary (1:30,000) antibodies were carried out for 1 h at room temperature. The blot was developed with ECL detection reagents (Amersham Pharmacia Biotech).
Electron microscopy.
Bacteria were grown overnight on MacConkey or CFA agar plates at 37°C. The bacteria were recovered and allowed to adhere to a carbon-Formvar coated 300-mesh copper grid (Electron Sciences). The fimbriae were visualized by negative staining with 0.1% phosphotungstic acid, pH 7.4, and the grids were analyzed by electron microscopy. For immunogold labeling of LP fimbriae, bacteria were reacted with α-LpfA antiserum from S. enterica serovar Typhimurium (1:500 dilution) and gold (12-nm-diameter)-labeled anti-rabbit immunoglobulin G and negatively stained as before. Specimens were examined in a Philips CM 120 electron microscope (FEI Co.). The samples were prepared at the microscopy facility in the Department of Cell Biology at The Johns Hopkins University.
Computerized sequence analysis.
Comparison of different subunit proteins with their related proteins was performed by the multiple sequence alignment program Clustal W (40) at the Institute for Chemical Research, Kyoto University (http://www.clustalw.genome.ad.jp/). A sequence similarity search was performed using the sequence manipulation site of the University of Alberta, Edmonton, Canada (http://www.ualberta.ca/∼stothard/javascript/ident_sim.html).
RESULTS
Identification of the lpfABCC′DE operon and homology with other fimbrial operons.
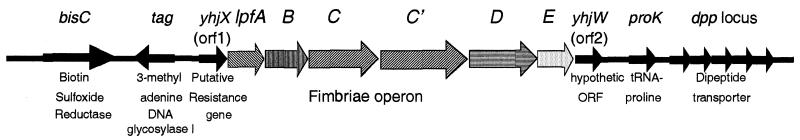
The entire genome of enterohemorrhagic E. coli O157:H7 strain EDL933 has been sequenced (28). Analysis of the strain-specific DNA sequences of EDL933 that correspond to the 76- to 81.5-min region on the E. coli K-12 chromosome led to the identification of a 6.0-kb DNA segment whose predicted protein products are similar to several fimbria-associated proteins (see below). This DNA segment was inserted in a region that maps to minute 78 on the E. coli K-12 chromosome (Fig. 1).
FIG. 1.
Schematic representation of the organization of the lpf operon in the chromosome of E. coli O157:H7 strain EDL933. lpfABCC′DE genes are shown as arrows with different patterns (see Fig. 3), and the genes with homology to E. coli K-12 genes are represented by solid arrows. The region shown corresponds to the minute 78 region in the E. coli K-12 chromosome.
This region contains six ORFs whose predicted protein sequences have considerable similarity to those included in the S. enterica serovar Typhimurium lpf operon, encoding the LP fimbriae (2). To retain the same nomenclature, they were designated lpfABCC′DE. The S. enterica serovar Typhimurium lpf operon consists of five ORFs in comparison to the six found in the EHEC lpf operon. Outside the lpf genes, two ORFs found in the S. enterica serovar Typhimurium sequence (orf1 and orf2), with homology to orf103 and orf102 of E. coli K-12, were also found flanking lpfABCC′DE in EHEC EDL933. Only genes with homology to E. coli K-12 genes and no insertion elements were found on either side of the EHEC lpf region (Fig. 1). These data indicate that the lpf regions in S. enterica serovar Typhimurium and E. coli O157:H7 are inserted in similar chromosomal locations and suggest common patterns of horizontal gene acquisition. The order and orientation of the ORFs lpfABCC′DE and the masses of the predicted polypeptides are given in Fig. 1 and Table 2.
TABLE 2.
Proteins encoded by the lpf operon of EHEC O157:H7
| Protein | Amino acids | Calculated, mass (kDa) | % Identity (% similarity) to fimbria-specific proteinsa
|
Proposed function | ||
|---|---|---|---|---|---|---|
| LpfS.t. | FimE.c. | FimS.t. | ||||
| LpfA | 179 | 18.5 | 73.0 (81.5) to LpfA | 32.3 (43.8) to FimA | 33.7 (45.5) to FimA | Major fimbrial subunit |
| LpfB | 233 | 25.5 | 67.4 (80.3) to LpfB | 40.2 (53.7) to FimC | 37.8 (55.4) to FimC | Chaperone |
| LpfC | 368 | 40.2 | 64.3 (74.2) to LpfC | 44.4 (55.1) to FimD | 37.2 (47.7) to FimD | Outer membrane usher chaperone |
| LpfC′ | 443b | 48.3b | 66.7 (76.8) to LpfC | 37.7 (50.5) to FimD | 37.0 (50.7) to FimD | Outer membrane usher protein |
| LpfD | 352 | 37.2 | 39.8 (54.6) to LpfD | 22.4 (37.0) to FimH | 22.3 (35.8) to FimH | Minor fimbrial subunit |
| LpfE | 177 | 18.4 | 48 (56.5) to LpfE | 28.4 (39.3) to FimI | 28.2 (42) to FimA | Fimbrial subunit |
LpfS.t., S. enterica serovar Typhimurium LP fimbriae; FimE.c., E. coli type I fimbriae; FimS.t., S. enterica serovar Typhimurium type I fimbriae.
The calculated size and amino acid sequence of lpfC′ are obtained from the first start codon directly downstream of lpfC. The first start codon with a good Shine-Dalgarno sequence is further downstream and will produce an LpfC′ protein of 166 amino acids with a calculated mass of 17.8 kDa.
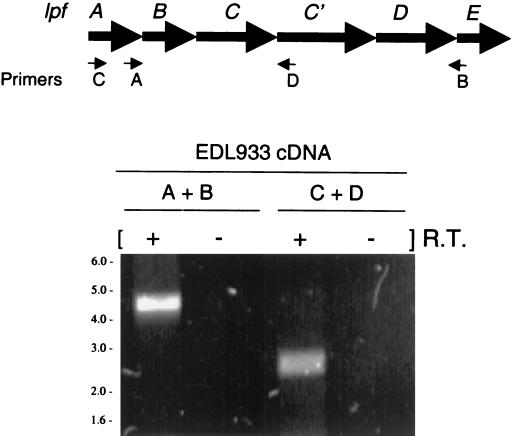
Analysis of the gene organization and the deduced amino acid sequences of all six lpf gene products indicated that lpfABCC′DE is organized in an operon. To confirm this prediction, we determined the operon structure of lpfABCC′DE by RT-PCR analysis (Fig. 2). Whole-cell RNA was isolated from EHEC strain EDL933, and cDNA was synthesized as a template. Because the putative operon is at least 5.5 kb in length, PCR was used to amplify shorter amplicons corresponding to segments within lpfABCC′DE. As expected from the predicted operon structure, RT-PCR analysis demonstrated that lpfA and lpfE were transcriptionally coupled (primers A and B) (Fig. 2). Similarly, lpfA was found to be transcriptionally linked to lpfC′ by using primers C and D. Because lpfA lies 5′ to lpfB and lpfE is 3′ from lpfD, we concluded that lpfABCC′DE is transcribed as a single polycistronic message.
FIG. 2.
RT-PCR analysis. RNA was extracted from EHEC EDL933 and subjected to RT-PCR amplification. The locations of the primer pairs (A+B and C+D) used in the RT-PCR analysis are indicated in the schematic lpf operon. The presence (+) or absence (−) of RT in the reaction is indicated. PCR products were not visualized in the control lanes (−). The sizes (in kilobases) and positions of DNA markers are indicated on the left.
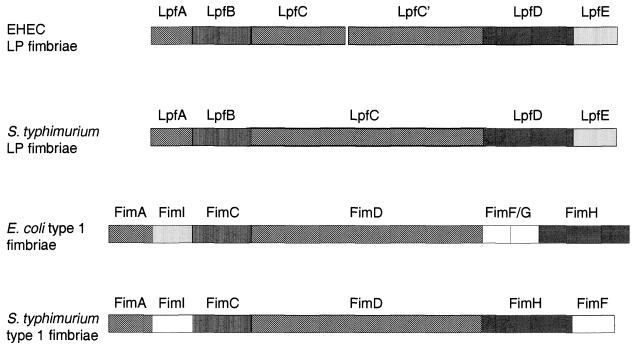
In addition to the similarity of the predicted Lpf protein products to proteins encoded by the lpf operon in S. enterica serovar Typhimurium (lpfABCDS.t.), these proteins were also similar to the products of the fim operons in E. coli (fimAICDFGHE.c.) and S. enterica serovar Typhimurium (fimAICDHFS.t.) (Table 2 and Fig. 3). LpfAEHEC, the putative major fimbrial subunit, is the first gene product in the operon, which shows homology to LpfAS.t. and FimAE.c./S.t. The second deduced protein product, LpfBEHEC, shows substantial identity to LpfBS.t. and FimCE.c./S.t. These proteins are proposed to function as chaperones in the other fimbrial systems. LpfCEHEC and LpfC′EHEC are homologous to the outer membrane usher proteins LpfCS.t. and FimDE.c./S.t.. All the genes in the operon are intact compared with genes in other fimbrial operons, with the exception of lpfC, which is disrupted in EHEC (Fig. 3). Two EHEC ORFs showed significant homology with the S. enterica serovar Typhimurium lpfC, with one ORF showing homology with the 5′ region of S. enterica serovar Typhimurium lpfC and the other with the 3′ region. In order to confirm that the disrupted ORFs, which we will refer to as lpfC and lpfC′, were not sequencing artifacts, primers flanking the disrupting region were used to amplify a segment from the 3′ end of lpfC to the putative 5′ end of lpfC′ in EHEC O157:H7 strains EDL933 and 86-24. Sequence analysis of the amplified fragments from both strains confirmed that EHEC lpfC is truncated and a new putative start codon for lpfC′ is located 144 nucleotides downstream of lpfC, but this start site lacks a good Shine-Dalgarno sequence (data not shown). The next possible start codon for lpfC′ with a good ribosome-binding site is located 972 nucleotides downstream of lpfC. The last two proteins encoded by the operon, LpfDEHEC and LpfEEHEC, show lower identity to other fimbrial proteins. LpfDEHEC shows homology to LpfDS.t. and FimHE.c./S.t., which are proposed to function as minor fimbrial subunits. Finally, LpfEEHEC is another putative fimbrial subunit with homology to LpfES.t., FimIE.c., and FimAS.t..
FIG. 3.
Comparison of the deduced amino acid sequences and the gene order of related fimbrial operons. The EHEC O157:H7 lpf operon (top) is compared with (from top down) the S. enterica serovar Typhimurium lpf operon and the fim operons in E. coli and S. enterica serovar Typhimurium. The position and length of each gene is indicated by bars. Homologous genes are shown with identical patterns, and genes with no homology are shown as open bars.
Transcription of the lpf operon is stimulated during exponential growth.
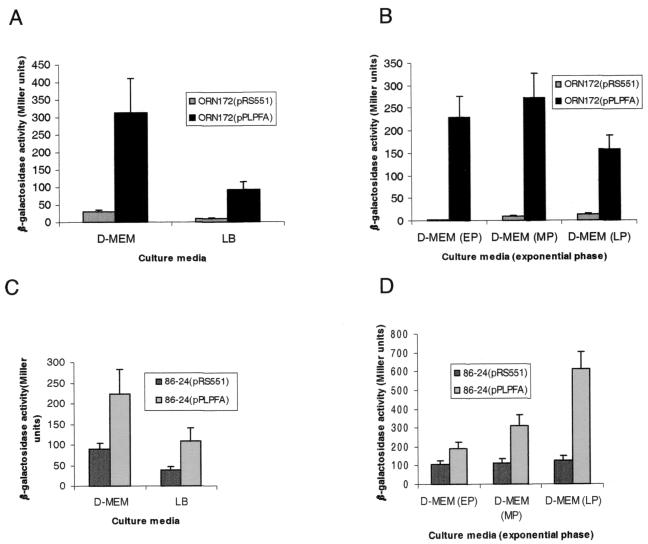
The visualization and purification of fimbriae from EHEC strains was very difficult because of inconsistent expression of the fimbriae, perhaps due to unknown stringent regulatory mechanisms. To approach this problem, we generated an operon fusion of the lpfA promoter (the first gene in the operon) with a reporter lacZ gene (plasmid pPLPFA) and examined the expression of this fusion under different medium and growth conditions. Initially, pPLPFA and its parent plasmid containing the promoterless lacZ gene, pRS551, were transformed into E. coli strain ORN172. We studied the transcription of the lpf operon in this strain because it was used in subsequent experiments with the LP fimbriae. ORN172 is an E. coli Δfim strain shown by electron microscopy not to express fimbriae and commonly used to study fimbrial expression (46). The expression of β-galactosidase was increased 9.5-fold in LB broth and 10.1-fold in DMEM in strain ORN172(pPLPFA) compared to that in ORN172(pRS551) during the mid-exponential growth phase (Fig. 4A). To determine the effect of the growth phase on lpfAp::lacZ expression, cultures of ORN172 strains containing pRS551 or pPLPFA were tested at early, mid-, and late exponential phase in DMEM. Maximal expression of β-galactosidase was observed during mid-exponential phase in strain ORN172 (Fig. 4B).
FIG. 4.
β-Galactosidase assays of plasmid pRS551 (vector control) and pPLPFA (lpfAp::lacZ) in E. coli strains ORN172 and 86-24 in LB broth and DMEM at different times point during the exponential growth phase. Strain ORN172(pPLPFA) was compared with its parent strain in LB broth and DMEM at an OD600 of 0.6 (A) or in DMEM at early (EP), mid- (MP), and late (LP) exponential phases (OD600, 0.3, 0.6, and 0.9, respectively) (B). β-Galactosidase activity was determined in strains 86-24(pRS551) and 86-24(pPLPFA) growing in LB broth and DMEM during mid-exponential phase (OD600, 0.6) (C) or at different time points during the exponential phase (OD600, 0.3, 0.6, and 0.9) (D). The error bars indicate standard deviations.
In order to determine the expression of the lpfp::lacZ gene fusion in the wild-type strain, pPLPFA and its parent plasmid were transformed into EHEC strain 86-24 (Fig. 4C and D). At mid-exponential phase, β-galactosidase expression was induced 2.8-fold in LB broth and 2.5-fold in DMEM in strain 86-24(pPLPFA) compared with that in 86-24(pRS551) (Fig. 4C). This induction was similar to that observed in strain ORN172, although the induction was more moderate. In contrast, EHEC 86-24 showed a different pattern of β-galactosidase expression throughout the exponential phase (Fig. 4D). Strain 86-24(pPLPFA) went from a 1.8-fold induction in early exponential phase to a 4.8-fold induction in late exponential phase compared with 86-24(pRS551) (Fig. 4D).
These data indicate that transcription of the lpf operon is induced under in vitro culture conditions throughout the exponential growth phase.
Cloning and expression of the lpfABCC′DE operon in E. coli and identification of fimbriae.
The lpf gene cluster of EHEC strain EDL933 was amplified by PCR using specific primers (see Materials and Methods) The 5,929-bp amplicon was cloned into pACYC184 to yield pJOR5, which was transformed into E. coli strain ORN172 (Table 1).
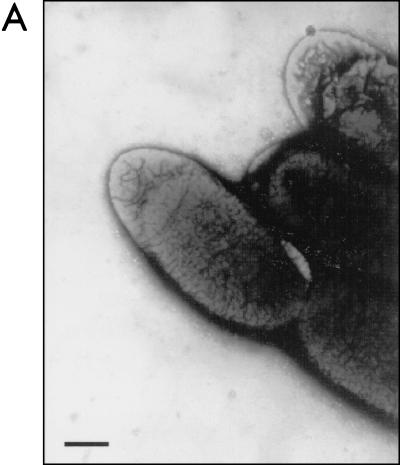
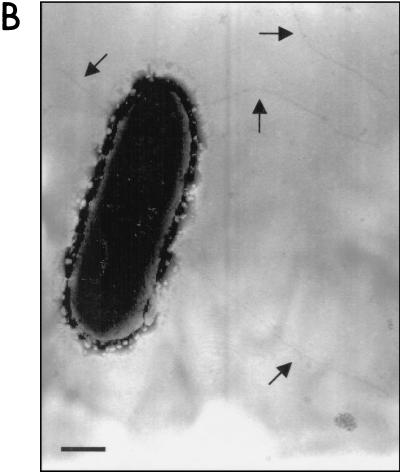
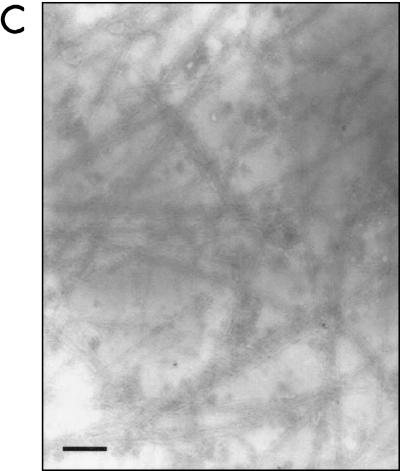
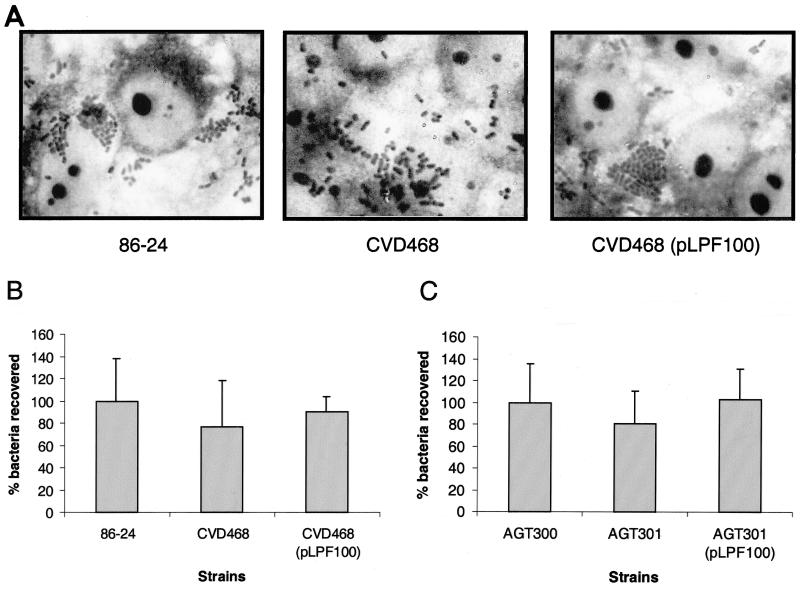
To determine whether pJOR5 encodes fimbrial structures, the strains ORN172 and ORN172(pJOR5) were analyzed by electron microscopy (Fig. 5). Fimbriae exhibiting a long rod-like appearance were detected in strain ORN172(pJOR5) (Fig. 5B) but not in strain ORN172 (Fig. 5A). The morphology of the recombinant EHEC fimbriae was structurally similar to that of the E. coli type I fimbriae and exhibited the approximate length observed when the S. enterica serovar Typhimurium LP fimbriae are expressed in a nonfimbriated E. coli strain (2). In the recombinant Salmonella LP fimbriae, the 2- to 10-μm-long fimbriae showed a polar distribution. However, unlike the Salmonella LP fimbriae, the fimbriae detected on strain ORN172(pJOR5) did not show the polar pattern previously reported. Instead, the recombinant EHEC fimbriae appear to be peritrichously distributed.
FIG. 5.
Expression of fimbriae in an Lpf+ E. coli strain as visualized in transmission electron micrographs. (A) ORN172 (E. coli Δfim strain); (B) ORN172(pJOR5) (lpfABCC′DE); (C) crude preparation of LP fimbriae. Bars, 0.35 μm.
We then verified whether pJOR5 encodes EHEC LP fimbriae. Culture supernatants of strains ORN172 and ORN172(pJOR5) were recovered after centrifugation and analyzed by electron microscopy (see Materials and Methods). The crude fimbrial preparation of strain ORN172(pJOR5) contained rod-like structures that were not visualized in the preparation of strain ORN172 (Fig. 5C and data not shown).
Adhesion to tissue culture cells and construction of an EHEC lpfA mutant.
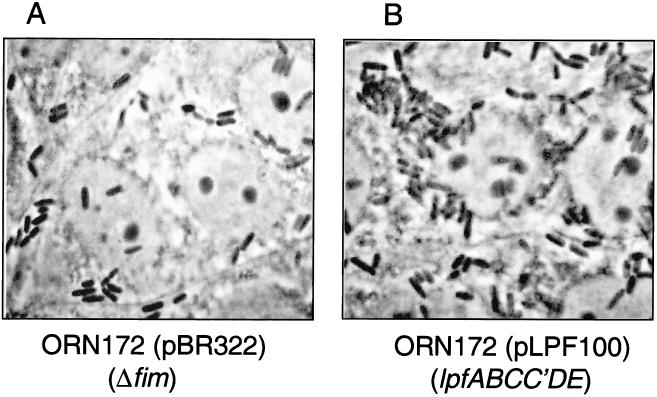
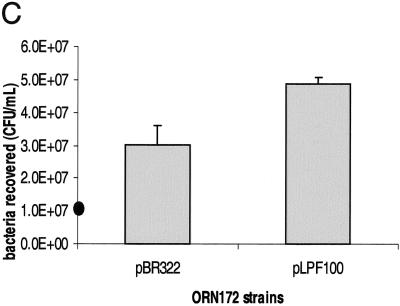
To determine the role of the lpf operon in adhesion, we selected two tissue culture cell lines that had been previously used to test adhesion factors in EHEC (5, 37). HeLa and MDBK cells were incubated with ORN172(pBR322) (Fig. 6A) or ORN172(pLPF100) expressing LP fimbriae (Fig. 6B and data not shown). After 3 h of infection, the cells were fixed, stained with Giemsa solution, and visualized by phase-contrast microscopy. E. coli ORN172 containing pLPF100 was able to adhere to both cell lines in a clustered pattern compared with the poorly adherent ORN172(pBR322). The percentage of bacteria recovered from infected cultured cells increased 60.3% in HeLa cells (P = 0.044) [from 3.0 × 107 CFU in ORN172(pBR322) to 4.9 × 107 CFU in ORN172(pLPF100)] and 62.6% in MDBK cells (P = 0.031) (from 1.6 × 107 to 2.5 × 107 CFU) when ORN172 carried the lpf operon (Fig. 6C and data not shown).
FIG. 6.
(A and B) Adhesion assays in HeLa cells showing adherence patterns of E. coli strains ORN172(pBR322) (A) and ORN172(pLPF100) (B) after 3 h of incubation and staining with Giemsa solution. (C) E. coli strains ORN172(pBR322) and ORN172(pLPF100) recovered after 3 h of incubation on cultured HeLa cells (P = 0.044). The solid oval on the y axis indicates the initial inoculum.
To further characterize the role of the lpf operon in EHEC adherence, an isogenic mutation was created in the proposed major fimbrial subunit. The lpfA gene, disrupted with a chloramphenicol resistance cassette, was introduced by allelic exchange into EHEC strain 86-24 (38) to create strain CVD468 (see Materials and Methods). Wild-type 86-24, CVD468 (an 86-24 lpfA mutant strain), and CVD468(pLPF100) were assayed for the ability to adhere to tissue culture cells (Fig. 7A), and the percentages of bacteria recovered after 6 h of incubation were calculated (Fig. 7B) (see Materials and Methods). We observed only a modest reduction in the adherence of strain CVD468 compared with that of EHEC strain 86-24. CVD468 showed a 23.4% reduction in the number of bacteria recovered. The adherence was restored to levels similar to that of the wild-type strain when the lpfA mutant was complemented with pLPF100 (90.4% recovery compared with the wild type) (Fig. 7B). Strain 86-24 showed typical localized adherence clusters on HeLa cells 6 h after incubation (Fig. 7A). Although CVD468 did not show a significant reduction in adherence relative to 86-24, the bacteria adhered to the tissue culture cells in a diffuse rather than a localized adherence pattern and the presence of microcolonies was rarely observed. The formation of stable microcolonies was restored when pLPF100 was introduced into CVD468. A similar phenotype was observed when EHEC strain 86-24 and its isogenic mutant were used to infect monolayers of MDBK cells (data not shown).
FIG. 7.
(A) Adherence patterns of EHEC O157:H7 strains 86-24 and AGT300 and their corresponding lpfA isogenic mutants (CVD468 and AGT301, respectively) to cultured HeLa cells after 6 h of incubation and staining with Giemsa solution. (B and C) Percentages of EHEC O157:H7 strains 86-24 (B) and AGT300 (C) and their corresponding isogenic lpfA mutants adherent to cultured HeLa cells after 6 h of incubation. The error bars indicate standard deviations.
We have observed that the cytotoxin (Stx) produced by EHEC strain 86-24 hinders the study of the adherence phenotype, since it is difficult to determine adherence in a situation in which the tissue culture cells may be sustaining a lethal toxic injury. To address this problem, we tested a set of isogenic strains derived from EHEC O157:H7 strain 87-23 (Table 1) for their adherence phenotypes. 87-23 was selected because it is an stx mutant EHEC strain isolated from the same outbreak in Washington state as strain 86-24 (13). HeLa cells were incubated with strains AGT300 (a streptomycin-resistant derivative of strain 87-23), AGT301 (lpfA::cat in AGT300), and AGT301(pLPF100) for 6 h, and the percentages of adherent bacteria were quantified as described above. AGT301 was observed to adhere less to HeLa cells (19.4% reduction) than did AGT300, but the difference was not significant (P = 0.468) (Fig. 7C). Complementation of AGT301 with pLPF100 restored adherence to wild-type levels (102.6% of bacteria recovered). Microscopic analysis of the Giemsa-stained infected cells indicated that microcolony formation was rarely observed on cells infected with strain AGT301. Like CVD468, strain AGT301 exhibited a diffuse pattern of adherence to HeLa cells compared with those of AGT300 and AGT301(pLPF100), where the formation of microcolonies was more often observed (data not shown). Taken together, these results suggest that LP fimbriae are expressed during in vitro infection and participate in the adherence phenotype in particular during the formation of microcolonies.
Detection of LpfA expression by Western blotting.
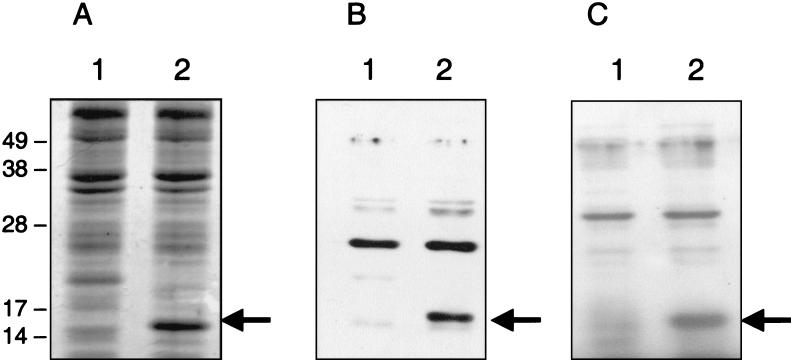
To provide evidence for the expression of the EHEC lpf operon in vitro, rabbit polyclonal antiserum was raised against a peptide that was designed from a region of the EHEC major fimbrial subunit, LpfA. We first tested the specificity of the LpfA peptide antiserum by Western blotting using an E. coli strain hyperexpressing the LpfA subunit. Crude cell lysates of E. coli strain BL21(DE3) containing either plasmid pT7-5 or pLPFA01 were prepared after overnight induction with IPTG and then separated by SDS-PAGE gels (Fig. 8). Coomassie blue staining revealed a strong band of ca. 16.0 kDa present in the IPTG-induced crude extract of strain BL21(pLPFA01) compared with BL21(pT7-5) (Fig. 8A). Western blot analysis showed that the LpfA peptide antiserum reacted with the 16-kDa protein band of strain BL21(pLPFA01) (Fig. 8B). [The antiserum also reacted with an unidentified ca. 26-kDa protein in strain BL21(pT7-5) lacking the lpf genes.] To confirm that the 16-kDa band corresponds to the EHEC LpfA protein, rabbit polyclonal antiserum raised against the S. enterica serovar Typhimurium LpfA protein was used in Western blots. The 16.0-kDa protein expressed in strain BL21(pLPFA01) cross-reacted with the Salmonella LpfA antiserum, indicating that this band corresponded to the EHEC LpfA protein and suggesting common epitopes in the two proteins (Fig. 8C).
FIG. 8.
SDS-PAGE analysis of crude cell lysates from E. coli strains BL21(pT7-5) (lanes 1) and BL21(pLPFA01) (lanes 2) after overnight incubation with 0.1 M IPTG. The corresponding lysates were separated and either stained with Coomassie brilliant blue (A) or transferred to polyvinylidene difluoride membranes for Western blotting with LpfA peptide antiserum (B) or S. enterica serovar Typhimurium LpfA antiserum (C). The molecular mass markers (in kilodaltons) are indicated on the left, and the 16-kDa EHEC LpfA protein in each gel is identified with an arrow.
Detection of LP fimbriae in EHEC and ORN172 strains.
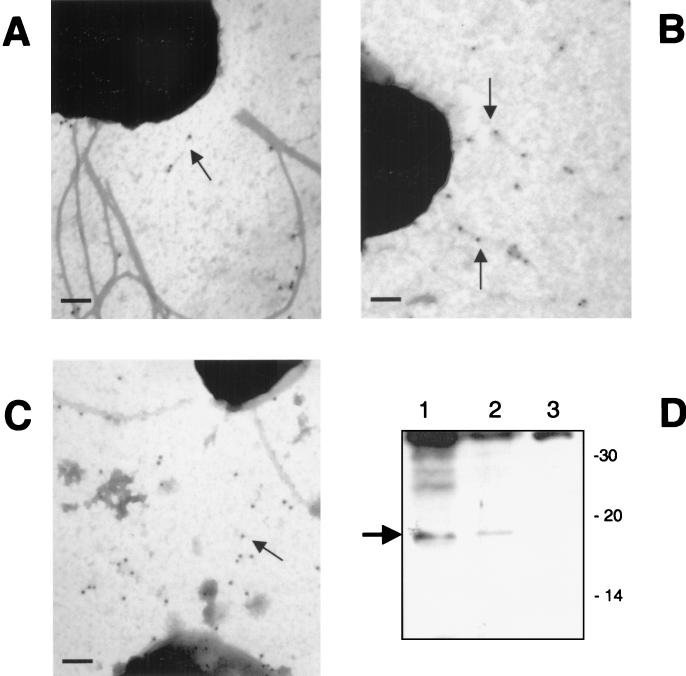
We then attempted to visualize the LP fimbriae by electron microscopy using immunogold and negative-staining techniques. As previously indicated, EHEC strains such as 86-24 express several fimbria-like structures in their surfaces. We were unable to identify fimbriae on the surface of the wild-type strain that was specifically labeled with gold particles (data not shown). Therefore, we tried to detect the LP fimbriae in the recombinant ORN172 strains. Bacterial fimbriation was highly dependent on culture conditions. Thus, after growth at 37°C on MacConkey agar, the LP fimbriae were visualized (Fig. 5B), while no fimbriae were seen with bacteria grown on CFA or LB agar at 37 or 30°C. When grown at 37°C, ORN172(pJOR5) and ORN172(pLPF100) produce LP fimbriae (Fig. 9). Immunogold-labeling electron microscopy of these ORN172 recombinant strains grown at 37°C on MacConkey agar showed that α-Salmonella LpfA antiserum bound to the fimbrial structures (Fig. 9A to C). Attempts were made to detect the LP fimbriae on the surface of EHEC strain 86-24 by a similar approach, but we were unable to specifically gold label a fimbrial structure with the α-LpfA antiserum (data not shown). Therefore, we tested crude fimbrial extracts for the presence of the LP fimbriae by Western blotting. As shown in Fig. 9D, we detected a protein band of ca. 18-kDa mass that cross-reacted with the α-Salmonella LpfA antiserum in the crude fimbrial extracts of strain ORN172(pJOR5) (Fig. 9D, lane 1). In a similar way, crude fimbrial extracts were prepared from EHEC strains 86-24 and CVD468 and tested by Western blotting. A protein band was detected in the crude extract of strain 86-24 with an electrophoretic mobility similar to that of the protein band identified in strain ORN172(pJOR5). A similar protein band was absent in the crude extract of strain CVD468. These data indicate that the protein band corresponded to the major fimbrial subunit, LpfA. Attempts to determine the N-terminal sequence of the LpfA protein in these extracts were unsuccessful due to the low levels of protein.
FIG. 9.
Electron micrographs showing strains ORN172(pJOR5) (A and B) and ORN172(pLPF100) (C) labeled with immunogold conjugated to S. enterica serovar Typhimurium LpfA antiserum. Bars, 0.2 μm. (D) Western blot analysis of fimbrial crude lysates from E. coli strains ORN172(pJOR5) (lane 1), 86-24 (lane 2), and CVD468 (lane 3) reacted with S. enterica serovar Typhimurium LpfA antiserum. LpfA is identified with an arrow, and the molecular mass markers (in kilodaltons) are indicated on the right.
DISCUSSION
The mechanism of infection of EHEC O157:H7, like those of other pathogens, is known to depend on a variety of bacterial properties that enable organisms to cause disease. During infection, EHEC must encounter and attach to one or more cell types found in the intestinal mucosa, evade host defenses, and compete with other bacterial species for nutrients. The intestinal tropism may involve several types of adherence factors, in addition to intimin, to assist in colonization of the gastrointestinal tract. In spite of the efforts of several researchers to demonstrate the production of adherence fimbriae in EHEC, the results have been inconsistent. We, as well as other groups, have observed fimbrial structures on the surface of E. coli O157:H7 (references 1, 32, and 45 and data not shown) and have been investigating the nature of these structures. In this study, we have characterized a chromosomal fimbrial operon in EHEC O157:H7 strain EDL933 that shows homology to the LP fimbria operon of S. enterica serovar Typhimurium and to other well-characterized fimbrial operons. The lpf operon in S. enterica serovar Typhimurium was initially identified as a locus not present in related members of the family Enterobacteriaceae (2). We found that this operon is indeed present in EHEC O157:H7 and O55:H7 but not in other EHEC strains tested (data not shown). The lpf operon in EHEC O157:H7 mapped at the same chromosomal location as the lpf operon in S. enterica serovar Typhimurium (78 min). Both loci are flanked by sequences homologous to those in E. coli K-12, supporting the idea that the lpf loci have been acquired by horizontal transfer during the evolution of S. enterica serovar Typhimurium and E. coli O157:H7.
Although EHEC LP fimbriae in E. coli strain ORN172 carrying the cloned lpf operon were visualized by electron microscopy, the polar distribution previously observed with the S. enterica serovar Typhimurium LP fimbriae was not observed (2). Instead, the EHEC LP fimbriae, which resemble the E. coli type 1 fimbriae, appear to be distributed peritrichously on the bacteria. Our results suggest that the LP fimbriae were synthesized by the lpf operon and that increased adherence to tissue culture cells occurs upon introduction of the lpf operon. We cannot absolutely rule out the possibility that a cryptic fimbrial operon in E. coli is induced upon introduction of the lpf operon and could explain the increase in the adherence pattern, but such a possibility is unlikely, since these fimbriae reacted with antisera prepared against a synthetic peptide derived from the predicted lpfA gene product. lacZ fusion analysis with the lpf promoter region indicates that this operon is transcribed in strain ORN172, supporting the idea that expression of LP fimbriae is responsible for the increased adherence to tissue culture cells, and we have shown that the ORN172 strain carrying the lpf operon expresses the LP fimbriae in its surface. The slight, albeit not statistically significant, reduction in the adherence to tissue culture cells observed with the EHEC lpfA mutants (CVD468 and AGT301) further suggests the role of these fimbriae in adherence. The possibility also exists that LP fimbriae facilitate adherence to epithelial cells in other biological niches not tested here, such as the gastrointestinal tracts of animals. Indeed, we found that E. coli strain ORN172 expressing LP fimbriae had the ability to adhere to cultured bovine kidney cells, namely, MDBK cells. We were also able to detect the LpfA protein by Western blotting under in vitro culture conditions, suggesting that the fimbriae are composed of this protein.
Another unusual characteristic of the lpf operon is the presence of two ORFs (lpfC and lpfC′) that are predicted to encode putative outer membrane components of the fimbriae. DNA sequence comparison with other related fimbrial outer membrane proteins indicated that lpfC is disrupted in EHEC O157:H7. In S. enterica serovar Typhimurium, there is one lpfC gene that encodes a protein of 94.4 kDa, whereas the EHEC lpf operon contains lpfC and lpfC′, which code for predicted proteins of 40.2 and 17.8 kDa, respectively. However, we have not yet directly demonstrated the production of these proteins, and we cannot rule out an unusual translation mechanism that could produce a single larger protein from the lpfC and lpfC′ ORFs. It is also possible that the assembly of LP fimbriae utilizes a native outer membrane component synthesized elsewhere in the E. coli chromosome.
S. enterica serovar Typhimurium LP fimbriae have been shown to mediate adhesion to murine Peyer's patch cells of the small intestine (3), and it has recently been proposed that phase variation of the major fimbrial subunit gene (lpfA) is a mechanism to evade cross-immunity between Salmonella serotypes (3, 26). In the case of E. coli O157:H7, it is believed that the site of colonization is the human large-bowel mucosa (24). Association of EHEC to human tissue in vivo in an attaching and effacing pattern had not been previously demonstrated, but recently Phillips et al., using in vitro organ cultures of human intestine, showed that EHEC O157:H7 adhered to human intestinal mucosa in this characteristic pattern (29). Furthermore, the attaching and effacing lesion formation was found to be restricted to follicle-associated epithelium of the Peyer's patches. Together with the tropism of Salmonella LP fimbriae for murine Peyer's patches, these data suggest that EHEC LP fimbriae might be an important surface-exposed factor that promotes binding to this specific intestinal location.
The data presented in this paper contribute to a better understanding of the pathogenesis of EHEC and add one more element to the existing model of EHEC intestinal colonization. The model is based on the attaching and effacing intestinal histopathology shown in vitro and in humans by EPEC and in vitro and in animal models by EHEC. This phenotype is characterized by intimate adherence of the bacteria, effacement of intestinal epithelial cell microvilli, and marked changes in the host cell cytoskeleton (17). The adhesin intimin is important in the final stage of adherence, but other factors that mediate initial adherence in EHEC strains are not known. Our in vitro results suggest that LP fimbriae might participate during adherence at some stage of the process. While the bacteria are intimately attached to eukaryotic cells, the expression of LP fimbriae seems to favor the formation of microcolonies, but it is not known if the expression of fimbriae participates in bacteria-to-bacteria interactions or if their presence enhances adherence to tissue epithelial cells.
The potential function of LP fimbriae in EHEC adherence resembles the one recently described for BFP in EPEC strains, where the pilus is proposed to alter its structure associated with bacterial adherence, aggregation, and dispersal of microcolonies (18). This raises the possibility that LP fimbriae mediate the preferential binding of EHEC O157:H7 to epithelial cells and help to regulate the transition between initial binding and formation of a more complex tridimensional bacterial cluster structure.
The diversity and partial redundancy of these fimbrial and nonfimbrial adherence factors, coupled with the small but reproducible effect seen for the LP fimbriae, illustrate the need to characterize adherence as a complex trait and point to ample opportunities for subtle phenotypic variation in adherence profiles in addition to gross differences in colonization strategies.
Acknowledgments
This work was supported by grants AI41325 and DK58957 to J.B.K. from the National Institutes of Health (NIH) and grants from NIH (NIAID and NCHGR) and RMHC to F.R.B. A.G.T. was supported by research supplements for underrepresented minorities from the NIAID and NIDDK, NIH. J.A.G. thanks Conacyt, México (grant 32777-M).
We thank Vanessa Sperandio and Jane Michalski for critical reading of the manuscript and Bradley Harris for his valuable technical assistance.
Editor: A. D. O'Brien
REFERENCES
- 1.Ashkenazi, S., L. May, M. LaRocco, E. L. Lopez, and T. G. Cleary. 1991. The effect of postnatal age on the adherence of enterohemorrhagic Escherichia coli to rabbit intestinal cells. Pediatr. Res. 29:14-19. [DOI] [PubMed] [Google Scholar]
- 2.Bäumler, A. J., and F. Heffron. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäumler, A. J., R. M. Tsolis, and F. Heffron. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc. Natl. Acad. Sci. USA 93:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieber, D., S. W. Ramer, C.-Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 5.Bilge, S. S., J. C. Vary, S. F. Dowell, and P. I. Tarr. 1996. Role of the Escherichia coli O157:H7 O-side chain in adherence and analysis of an rfb locus. Infect. Immun. 64:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunder, W., A. S. Khan, J. Hacker, and H. Karch. 2001. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H−. Infect. Immun. 69:4447-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of a eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnenberg, M. S., C. O. Tacket, S. P. James, G. Losonsky, J. P. Nataro, S. S. Wasserman, J. B. Kaper, and M. M. Levine. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J. Clin. Investig. 92:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dytoc, M. T., A. Ismaili, D. J. Philpott, R. Soni, J. L. Brunton, and P. M. Sherman. 1994. Distinct binding properties of eae-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect. Immun. 62:3494-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, D. J., Jr., D. G. Evans, and H. L. DuPont. 1979. Hemagglutination patterns of enterotoxigenic and enteropathogenic Escherichia coli determined with human, bovine, chicken, and guinea pig erythrocytes in the presence and absence of mannose. Infect. Immun. 23:336-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girón, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 13.Griffin, P. M., S. M. Ostroff, R. V. Tauxe, K. D. Greene, J. G. Wells, J. H. Lewis, and P. A. Blake. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann. Intern. Med. 109:705-712. [DOI] [PubMed] [Google Scholar]
- 14.Jerse, A. E., K. G. Gicquelais, and J. B. Kaper. 1991. Plasmid and chromosomal elements involved in the pathogenesis of attaching and effacing Escherichia coli. Infect. Immun. 59:3869-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kado, D. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaper, J. B., L. J. Gansheroff, M. R. Wachtel, and A. D. O'Brien. 1998. Intimin-mediated adherence of Shiga toxin-producing Escherichia coli and attaching-and-effacing pathogens, p. 148-156. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 18.Knutton, S., R. K. Shaw, R. P. Anantha, M. S. Donnenberg, and A. A. Zorgani. 1999. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol. Microbiol. 33:499-509. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 22.McKee, M. L., A. R. Melton-Celsa, R. A. Moxley, D. H. Francis, and A. D. O'Brien. 1995. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to Hep-2 cells. Infect. Immun. 63:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 26.Norris, T. L., and A. J. Bäumler. 1999. Phase variation of the lpf operon is a mechanism to evade cross-immunity between Salmonella serotypes. Proc. Natl. Acad. Sci. USA 96:13393-13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-532. [DOI] [PubMed] [Google Scholar]
- 29.Phillips, A. D., S. Navabpour, S. Hicks, G. Dougan, T. Wallis, and G. Frankel. 2000. Enterohaemorrhagic Escherichia coli O157:H7 target Peyer's patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut 47:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sherman, P., R. Soni, M. Petric, and M. Karmali. 1987. Surface proteins of the vero cytotoxin-producing Escherichia coli O157:H7. Infect. Immun. 55:1824-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 34.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 35.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 36.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarr, P. I., M. A. Neill, C. R. Clausen, J. W. Newland, R. J. Neill, and S. L. Moseley. 1989. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984-1987. J. Infect. Dis 159:344-347. [DOI] [PubMed] [Google Scholar]
- 39.Tatsuno, I., H. Kimura, A. Okutani, K. Kanamaru, H. Abe, S. Nagai, K. Makino, H. Shinagawa, M. Yoshida, K. Sato, J. Nakamoto, T. Tobe, and C. Sasakawa. 2000. Isolation and characterization of mini-Tn5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect. Immun. 68:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tobe, T., and C. Sasakawa. 2001. Role of the bundle-forming pilus of enteropathogenic Escherichia coli in host cell adherence and in microcolony development. Cell Microbiol. 3:579-585. [DOI] [PubMed] [Google Scholar]
- 42.Tobe, T., and C. Sasakawa. 2002. Species-specific cell adhesion of enteropathogenic Escherichia coli is mediated by the bundle-forming pili. Cell Microbiol. 4:29-42. [DOI] [PubMed] [Google Scholar]
- 43.Tzipori, S., F. Gunzer, M. S. Donnenberg, L. de Montigny, J. B. Kaper, and A. Donohue-Rolfe. 1995. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect. Immun. 63:3621-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vial, P. A., J. J. Mathewson, L. Guers, M. M. Levine, and H. L. DuPont. 1990. Comparison of two assay methods for patterns of adherence to Hep-2 cells of Escherichia coli from patients with diarrhea. J. Clin. Microbiol. 28:882-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winsor, D. K., Jr., S. Ashkenazi, R. Chiovetti, and T. G. Cleary. 1992. Adherence of enterohemorrhagic Escherichia coli strains to a human colonic epithelial cell line (T84). Infect. Immun. 60:1613-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woodall, L. A., P. W. Russell, S. L. Harris, and P. E. Orndorff. 1993. Rapid, synchronous, and stable induction of type 1 piliation in Escherichia coli by using a chromosomal lacUV5 promoter. J. Bacteriol. 175:2770-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]