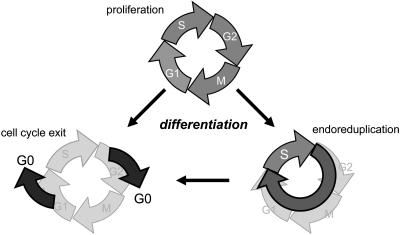
During the development of multicellular organisms, many different cell types are created. These display characteristic cell cycle programs that can radically change during the organism's lifetime (Jakoby and Schnittger, 2004; Fig. 1). Usually, in younger and less differentiated tissues, a proliferative cell cycle mode occurs. In this mitotic cell cycle program, DNA replication (also known as the synthesis or S phase) is followed by the segregation of the duplicated genetic material to two daughter cells in mitosis (M phase). Both the S and M phases are usually preceded by a preparative gap phase (G1 and G2 phase, respectively) during which cells monitor whether all conditions are favorable to continue a new round of DNA replication or mitosis. As cells differentiate, the rate of cell proliferation decreases. However, before complete withdrawal from the cell cycle and concomitant with differentiation, many plant cells switch from a mitotic cell cycle to an endoreduplication cycle (also called endocycle or endoreplication). During this endocycle the DNA is replicated without subsequent mitosis leading to polyploidy. Eventually, cells exit the cell cycle completely.
Figure 1.
Cell cycle modes during development. During the growth of a multicellular organism, different cell cycle programs are executed. Typically, in young and less differentiated tissues, a proliferative cell cycle mode occurs by which more cells are generated. As cells differentiate, the rate of cell proliferation decreases and eventually, cells exit the cell cycle. However, before complete withdrawal from the cell cycle, and along with differentiation, many plant and animal cells switch from a mitotic cell cycle to an endoreduplication cycle, in which the nuclear DNA becomes replicated without subsequent nuclear and cellular division, leading to polyploid cells.
Cell cycle progression is controlled by an evolutionarily conserved molecular mechanism. A central role is played by kinase complexes, which in their minimal configuration consist of a Ser/Thr kinase (the cyclin-dependent kinase [CDK]) and a regulatory cyclin subunit. CDKs phosphorylate a plethora of substrates, thereby triggering the transition from one cell cycle phase into the next one. The sequential and transient activation of different CDK-cyclin complexes dictates the unidirectional progression through the cell cycle.
Because of its importance in growth and development, the CDK-cyclin activity must be strictly controlled. In addition, mechanisms must be operational to ensure a correct exit of the cell cycle in response to antimitogenic stimuli. In yeast and mammals, one of the major regulators of CDK activity are CDK inhibitory molecules (CKIs) that bind and inhibit or sequester CDKs. Recently, putative orthologs of CKI proteins have been identified in plants as well. In this Update, we review the current knowledge on the biochemical properties of plant CKIs and discuss their physiological relevance during plant growth and development.
THE PLANT CELL CYCLE. A BASIC INTRODUCTION
Similar to animals, progression through the cell cycle in plants is regulated by CDKs (De Veylder et al., 2003; Inzé, 2005). In Arabidopsis (Arabidopsis thaliana), at least two classes of CDKs are involved in cell cycle regulation: the A-type CDKs that are represented by only one gene in the model species Arabidopsis (designated Arath;CDKA;1) and the B-type CDK family that has four members, grouped into the B1 (Arath;CDKB1;1 and Arath;CDKB1;2) and B2 (Arath;CDKB2;1 and Arath;CDKB2;2) subclasses (Vandepoele et al., 2002). A-type CDKs display kinase activity from late G1 phase until the end of mitosis, suggesting a role for this particular CDK at both the G1-to-S and G2-to-M transition points (Magyar et al., 1997; Porceddu et al., 2001; Sorrell et al., 2001). A central role for CDKA;1 in controlling cell number has been demonstrated using transgenic tobacco (Nicotiana tabacum) plants with reduced A-type CDK activity (Hemerly et al., 1995). The requirement for Arath;CKDA;1 at least for entry into mitosis has been demonstrated as well by cdka;1 null mutants that fail to progress through the second mitosis during male gametophytic development (Nowack et al., 2005). The group of B-type CDKs displays a peak of activity at the G2-to-M phase transition only (Magyar et al., 1997; Porceddu et al., 2001; Sorrell et al., 2001), suggesting that they play a role at the onset of, or progression through, mitosis. Correspondingly, cells of plants with reduced B-type CDK activity arrest in the G2 phase of the cell cycle (Porceddu et al., 2001; Boudolf et al., 2004a).
Although titration of CDK activity by the expression of dominant negative versions of both A- and B-type CDKs resulted in cell cycle defects, no extra cell divisions were stimulated by the overexpression of wild-type Arath;CDKA;1, Arath;CDKB1;1, and Arath;CDKB1;2 alleles in plants (Hemerly et al., 1995; Schnittger et al., 2003; Boudolf et al., 2004a). This observation is consistent with the view that a cofactor is required for full CDK activity, i.e. a cyclin. Cyclins are regulated both transcriptionally and posttranslationally, mainly by controlled protein degradation. By regulating the abundance of specific cyclins, the CDK activity is precisely tuned and targeted to substrates in a spatial and temporal manner.
The plant cyclin gene family is very complex. For instance, the Arabidopsis genome codes for at least 49 different cyclins (Vandepoele et al., 2002; Wang et al., 2004) that are classified into seven different subclasses (A, B, C, D, H, P, and T). To date, only a few members of the A-type, B-type, and D-type cyclins have been characterized. With a few exceptions, the expression patterns and activity profiles mimic those of their mammalian counterparts. A-type cyclins are important from S until M phase, while B-type cyclins primarily control the G2-to-M transition. D-type cyclins, whose expression is mainly correlated with the proliferative status of cells, are presumed to drive cells through the G1-to-S checkpoint in a mitogen-dependent manner (De Veylder et al., 2003; Inzé, 2005). In contrast to animals, some evidence points to an additional function of plant D-type cyclins at the G2-to-M transition (Schnittger et al., 2002; Kono et al., 2003; Koroleva et al., 2004).
PLANT CKIs. THE KIP-RELATED PROTEINS
In addition to binding of cyclins, CDK activity is regulated by docking of small proteins, generally known as CKIs, which have been found to induce cell cycle arrest or to delay cell cycle progression in response to intracellular or extracellular signals. CKIs have been identified in many different organisms, and, although all of them display CKI activity, they control a broad spectrum of often species-specific physiological processes. For example, in budding yeast (Saccharomyces cerevisiae), three CKIs have been described, Pho81, Far1, and Sic1 (Mendenhall, 1998). Pho81 inhibits a CDK-cyclin complex that controls gene expression under low-phosphate conditions; Far1 binds and inactivates G1 CDK complexes to mediate pheromone-dependent cell cycle blockage; and Sic1 plays a role in the timing of S-phase onset. In fission yeast (Schizosaccharomyces pombe), the CKI Rum1 is structurally and functionally related to Sic1, inhibits mitotic CDKs, and plays a central role in the regulation of the G1 phase.
In mammals, based on shared structural features and biochemical functions, CKIs have been divided into two major classes, the INK4 and the Kip/Cip class (Sherr and Roberts, 1999). Members of the INK4 family (p15INK4b, p16INK4a, p18INK4c, and p19INK4d) are structurally similar to the Pho81 inhibitor of budding yeast and are characterized by the presence of multiple ankyrin-type repeats for CDK binding. They bind and inhibit a small subset of CDKs (CDK4 and CDK6) that are primarily responsible for passage through G1. INK4 protein binding to monomeric CDKs or CDK-cyclin complexes causes allosteric changes that impair cyclin binding or lead to the dissociation of the CDK-cyclin complex, respectively. In contrast, inhibitors of the Kip/Cip family (p21Cip1, p27Kip1, and p57Kip2) bind and inhibit a broader range of CDKs and function in dimeric as well as heterotrimeric complexes with CDKs and cyclins; all share a conserved inhibitory domain at their N terminus. Kip/Cip binding does not dissociate the CDK-cyclin complex but distorts the catalytic ATP-binding center of the CDK subunit.
The first plant CKIs were detected in yeast two-hybrid screens performed to identify CDKA;1-associating proteins (Wang et al., 1997; Lui et al., 2000; De Veylder et al., 2001; Jasinski et al., 2002a). Additional plant CKIs have been discovered in silico through genome data mining (De Veylder et al., 2001; Coelho et al., 2005). Overall, the plant CKIs have only low sequence identity to each other and the nonplant CKIs. Interspecies sequence similarity is restricted to a short amino acid region shared between the plant CKIs and the mammalian Kip/Cip inhibitors. Because of this sequence similarity, which suggests that the plant proteins are homologous to the animal Kips, the name Kip-related proteins (KRPs) was suggested for the seven CKIs found in the Arabidopsis genome (De Veylder et al., 2001), but some family members are also known under the names ICK1 (KRP1) and ICK2 (KRP2; Wang et al., 1997; Lui et al., 2000). No Arabidopsis homologs to the INK4 or yeast inhibitors have been identified so far (Vandepoele et al., 2002).
Biochemical Properties
Despite their low sequence similarity with the mammalian CKIs, plant KRP genes encode functional CKIs, as demonstrated by their ability to inhibit CDK activity. In vitro, CKI activity was proven by adding recombinant KRP to partially purified CDK complexes (Wang et al., 1997; Lui et al., 2000; Jasinski et al., 2002a; Coelho et al., 2005). In vivo CKI activity was demonstrated by overexpressing diverse KRP genes in Arabidopsis (Wang et al., 2000; De Veylder et al., 2001; Jasinski et al., 2002a; Zhou et al., 2003a). The reduced CDK activity observed upon KRP overexpression correlates with a decrease in cell division rate, resulting in leaves whose cell number is substantially reduced. This decrease in cell number is accompanied by a change in leaf morphology (Fig. 2). Whereas KRPs have clearly been demonstrated to operate as inhibitors of CDK activity, the identity of the targeted CDK complexes remains unknown. Not all CDKs are KRP sensitive, as even application of a high dose of recombinant KRP to purified CDK complexes results in only a partial inhibition of total CDK activity (Wang et al., 1997). In accordance with this observation, yeast two-hybrid interaction analysis demonstrated that the KRPs bind A-type, but not B-type, CDKs (Lui et al., 2000; De Veylder et al., 2001; Jasinski et al., 2002a; Zhou et al., 2002). Recently strong biochemical evidence has been reported: In Arabidopsis plants overexpressing Arath;KRP2, only the A-type CDK complexes are targeted for inhibition (Verkest et al., 2005). Consistently, the phenotype of plants misexpressing Arath;KRP1 could be rescued by comisexpression of Arath;CDKA;1 but not Arath;CDKB1;2 (Schnittger et al., 2003).
Figure 2.
Phenotypes of KRP-misexpressing plants. A, Wild-type Columbia plants. B, Plants misexpressing the Arath;KPR2 gene under the control of the shoot meristem-specific STM promoter, resulting in smaller and more elongated leaves than observed for wild-type plants. Plants are at the same developmental stage and magnification as in A. C and D, Rosette leaves from a wild-type Columbia and a transgenic plant misexpressing the Arath;KRP1 gene under the control of the stomatal lineage-specific TMM promoter, respectively. Expression of Arath;KRP1 in TMM cells results in an altered leaf morphology because of reduced leaf cell numbers. E and F, Close-up of the leaves shown in C and D, respectively. Note the enlarged epidermal cells in F. Bars = 1 mm (C and E) and 100 μm (D and F). (C–F are from Weinl et al. [2005], reprinted with permission.)
Besides the CDK subunit, KRP binding is also directed by the cyclin subunit. Yeast two-hybrid assays have revealed interactions of Arabidopsis and tobacco KRPs with D-type cyclins, suggesting that KRPs are potential regulators of CDK-cyclinD complexes (Wang et al., 1998; Lui et al., 2000; De Veylder et al., 2001; Jasinski et al., 2002a; Zhou et al., 2002). Furthermore, in vivo binding specificity between plant CKIs and different D-type cyclins has been proven by the observation that the aberrant cell and leaf phenotypes seen upon KRP overexpression can be complemented by co-overexpression of D-type cyclins (Jasinski et al., 2002a; Schnittger et al., 2003; Zhou et al., 2003b). Recently, it was demonstrated that not only D-type but also A-type cyclin-harboring CDK complexes can be inhibited by KRPs in vitro (Coelho et al., 2005); so it seems that the plant CKIs resemble the mammalian Kip/Cip inhibitors, which bind and inhibit a broad range of CDKs, including both A- and D-type cyclin-containing CDK complexes.
Structural Organization
Like the mammalian Kip/Cip inhibitors, the plant CKIs have low sequence similarity to each other. Detailed analysis identified several sequence elements shared by different KRPs, but only three C-terminally located motifs are conserved in all plant inhibitors (De Veylder et al., 2001). This region of the KRPs shows partial homology with the Kip/Cip protein domain necessary for interaction with the CDK subunit, suggesting that the plant CKI function resides at their C terminus. Indeed, Wang et al. (1998) showed in a yeast two-hybrid interaction assay that the C-terminal domain of KRPs is sufficient for interaction of Arath;KRP1 with Arath;CDKA;1 and Arath;CYCD3;1. Moreover, the functionality of this domain for CDK binding and inhibition was proven in vitro and in vivo (Schnittger et al., 2003; Zhou et al., 2003a).
The role of the highly diverse N-terminal plant CKI sequences remains unclear. In Arath;KRP1, the N-terminal region was suggested to negatively regulate CKI function; deletion of this region increased the yeast two-hybrid physical interaction of Arath;KRP1 with CDKs and cyclins, and enhanced the phenotype of Arath;KRP1 overexpression in Arabidopsis (Wang et al., 1998; Schnittger et al., 2003). One possible function of the N terminus could be the regulation of the KRP stability. Arath;KRP2 protein is highly unstable and its degradation depends on the proteasome (Verkest et al., 2005). Indeed, removal of the N-terminal region increased the Arath;KRP1 protein level. However, the mechanism regulating this protein stability remains unknown (Zhou et al., 2003a; Weinl et al. 2005).
Regulation of KRP Activity at the Transcript Level
Yeast and mammalian CKIs are regulated at the transcriptional, translational, and posttranslational levels through mechanisms that affect their abundance rather than their intrinsic activity. Most plant tissues coexpress various KRP genes but at different mRNA levels, suggesting different transcriptionally regulatory mechanisms and possibly distinct roles for the plant CKIs within a single tissue (Wang et al., 1998; De Veylder et al., 2001; Jasinski et al., 2002b; Ormenese et al., 2004). A detailed spatial expression analysis by mRNA in situ hybridizations in the Arabidopsis shoot apex revealed different groups of KRP genes with similar expression patterns. Whereas Arath;KRP4 and Arath;KRP5 expression was confined to mitotically dividing tissues within the shoot apex, other KRP genes could be detected in both dividing and maturing cells (Arath;KRP3, Arath;KRP6, and Arath;KRP7) or exclusively in maturing cells (Arath;KRP1 and Arath;KRP2; Ormenese et al., 2004). These data hint at a function of Arath;KRP1, Arath;KRP2, Arath;KRP3, Arath;KRP6, and Arath;KRP7 during the process of cell cycle exit and onset of differentiation, whereas Arath;KRP4 and Arath;KRP5 might direct specific aspects of the mitotic cell cycle, such as functioning of the checkpoints that control the correct timing of S- and M-phase onset.
A role for the KRPs during the regular cell cycle is also suggested by their observed cell cycle phase-dependent temporal regulation (Menges et al., 2005). Transcript levels peak during S-phase for Arath;KRP3 and Arath;KRP5, in G2-phase for Arath;KRP4, during late G2-to-M for Arath;KRP1, and at M-to-G1 for Arath;KRP6. The expression of Arath;KRP2 and Arath;KRP7 is constitutive during the cell cycle.
Furthermore, transcript levels of the tobacco NtKIS1a and the Arath;KRP1 accumulated with flower bud and leaf aging, respectively (Wang et al., 1998; Jasinski et al., 2002b). This temporal increase in transcripts during the course of cell cycle arrest and cellular differentiation suggests possible functions for these KRPs in development.
KRP mRNA is controlled not only in a spatial and temporal manner, but also through the generation of alternative splicing variants, as illustrated by the NtKIS1 locus that generates two splice variants, NtKIS1a or NtKIS1b (Jasinski et al., 2002b). The splice variant NtKIS1b lacks the most C-terminal motif found in NtKIS1a and other plant CKIs. Consistently, NtKIS1b does not interact with A-type CDKs and D-type cyclins and is unable to inhibit CDK activity in vitro and in vivo.
Regulation of KRP Activity at the Posttranslational Level
Currently, little is known about the regulation of plant CKIs at the protein level. In mammals, regulation of CKI activity is complex and is accomplished through several mechanisms. Kip/Cip inhibitors can be inactivated through out-titration by CDK-cyclinD complexes. Other mechanisms control the subcellular localization. Kip/Cip proteins have distinct nuclear and cytoplasmic functions, and their cytoplasmatic compartmentalization releases and activates nuclear CDK-cyclin complexes (Coqueret, 2003). However, the best studied posttranslational regulatory mechanism of the mammalian CKIs affects their abundance through ubiquitin-dependent proteolysis. Two alternative proteolytic pathways control p27Kip1 stability (Hengst, 2004). One pathway acts in the nucleus and requires p27Kip1 phosphorylation at Thr-187 by CDK2-cyclinE complexes and subsequent recognition and degradation at the S-phase by the SCFSkp2 ubiquitin-ligase complex. The other one acts at the G1-phase, is independent of Skp2 and Thr-187 phosphorylation, and involves cytoplasmic sequestration of p27Kip1 and its degradation through the recently identified Kip ubiquitination-promoting complex (Hengst, 2004).
There is evidence that at least some plant KRPs are regulated through proteolysis. As described above, functional analysis of the Arath;KRP1 domains indicated the presence of a regulatory motif for protein instability in its N-terminal domain (Zhou et al., 2003a; Weinl et al., 2005). Additionally, both Zeama;KRP2 and Arath;KRP2 are regulated at the posttranslational level during maize (Zea mays) endosperm and Arabidopsis leaf development, respectively, demonstrated by their alteration in protein levels while their transcript levels remain constant (Coelho et al., 2005; Verkest et al., 2005). In the case of Arath;KRP2, protein stability is regulated by the proteasome. Moreover, in vitro analysis illustrated that Arath;KRP2 is a CDK-cyclin substrate and that its phosphorylation is at least in part responsible for Arath;KRP2 proteolysis. Although both Arath;CDKA;1 and Arath;CDKB1;1 complexes phosphorylate Arath;KRP2, neither the cell cycle phase when this event occurs nor the specific ubiquitin-ligase that is responsible for Arath;KRP2 degradation is currently known.
Another mechanism of posttranslational regulation has been identified through comparative analysis of the NtKIS1a and NtKIS1b splice variants (Jasinski et al., 2002b). Even though the spliced form NtKIS1b does not interact with Nicta;CDKA;1 and D-type cyclins, NtKIS1b counteracts the capacity of NtKIS1a to inhibit CDK activity in vitro. The two splice variants have a different transcriptional expression pattern: Whereas NtKIS1a is constitutively present during the cell cycle, NtKIS1b transcript levels peak at G2-to-M. These data, together with their cooperative subcellular localization, suggest that NtKIS1b antagonizes NtKIS1a inhibition of CDK activity at the G2-to-M transition. However, the mechanism by which this occurs remains to be elucidated.
Intercellular and Intracellular Localization of KRPs
In animals, CKI function depends on its intracellular localization. CKI p27Kip1 exerts its inhibitory function in the nucleus and entry into the nucleus appears to be used as a control mechanism. In addition, p27Kip1 degradation is precisely regulated and is seemingly also connected, at least to some degree, with its intracellular localization pattern (see above).
Fusions of the green fluorescent protein or the yellow fluorescent protein (YFP) and the Arath;KRP1 or the NtKIS1 have revealed a strict nuclear localization (Jasinski et al., 2002b; Zhou et al., 2003a; Weinl et al., 2005; Fig. 3). In addition, a putative nuclear localization signal has been identified in the protein sequences of Arath;KRP2, Arath;KRP5, and Arath;KRP7 (De Veylder et al., 2001).
Figure 3.
Intercellular and subcellular localization of Arath;KRP1. A, Trichome-specific expression of the GLABRA 2 promoter as revealed by green fluorescent protein fluorescence. B, Protein fusions of Arath;KRP1 with YFP expressed from the GLABRA 2 promoter spread from the trichome into the neighboring cells. C and D, Close-up of trichomes; the full-length Arath;KRP1-YFP fusion protein is found exclusively in the nuclei (C); the N-terminally truncated Arath;KRP1D2-108 localizes to the nucleus and the cytoplasm (D). Bars =50 μm.
In animals, regulation of nuclear import and export of p27Kip1 is complex and involves phosphorylation at Ser-10, Thr-157, Thr-187, and Thr-198. None of these phosphorylation sites is conserved in plant KRPs. However, the stabilized N-terminally truncated Arath;KRP1 shows, in addition to a nuclear, also a cytosolic localization (Fig. 3D). Thus, one likely possibility is that Arath;KRP1 becomes degraded in the cytoplasm and that a motif in the N terminus of the protein is required for this degradation. It will be interesting to see to what degree the intracellular localization of CKIs is an important regulatory mechanism in plants as well.
Surprisingly, plant CKIs function in a noncell-autonomous manner. Fusion proteins between YFP and Arath;KRP1 were found at least two to three cells away from their site of expression (Weinl et al., 2005; Fig. 3, B and C). Other plant cell cycle regulators have also been observed to travel between cells (M. Jakoby and A. Schnittger, unpublished data). Currently, it is still unclear whether it is the mRNA or the protein that moves, or whether this movement is based on a targeted versus a nontargeted mechanism. So far, the functional relevance of this movement is unknown. On the one hand, the noncell-autonomous behavior could simply be a consequence of overproduction of these proteins. On the other hand, the noncell-autonomous action of Arath;KRP1 offers the possibility to link decisions on a cellular level with the supracellular division and growth pattern in tissues and organs. For instance, during leaf development, epidermal cells have been observed to exit the cell cycle before palisade cells do (Donnelly et al., 1999). A mechanism can be envisaged in which the diffusion of synthesized KRPs in the epidermis coordinates the initial cell cycle exit of the dermal layer with that of the palisade parenchyma layer later during development. In any case, the molecular nature of the noncell-autonomous action of KRPs remains to be analyzed in detail.
KRPS AND PLANT DEVELOPMENT
KRPs as Integrators of Developmental Signals
The observation that several plant CKIs are transcriptionally regulated during development indicates that they share with the mammalian Cip/Kips the potential to integrate developmental signals into the core cell cycle machinery. Indeed, Arath;KRP1 transcripts are induced by cold treatment, which correlates with a decrease in CDK activity. Furthermore, Arath;KRP1 expression is activated by the phytohormone abscisic acid (Wang et al., 1998), suggesting that this particular KRP might be in part responsible for the growth inhibitory effect triggered upon abscisic acid treatment. By contrast, the mitogenic hormone auxin repressed Arath;KRP2 transcription, both in cell cultures and in planta (Richard et al., 2001; Himanen et al., 2002). Downregulation of Arath;KRP2 precedes the auxin-induced reentry of quiescent root pericycle cells into the cell cycle. Interestingly, Arath;KRP2 transcripts have been detected in young roots at the phloem but not the protoxylem poles of the pericycle, i.e. the sites at which lateral roots can initiate. In older root tissues, Arath;KRP2 expression has been observed at both the phloem and protoxylem poles, corresponding with the observation that new laterals mainly initiate at the young parts of the root. Curiously, upon the initiation of a lateral root primordium, Arath;KRP2 expression is induced in cells opposite the developing new lateral root, implying a mechanism by which Arath;KRP2 prevents the formation of two opposing lateral roots. A role for KRPs in controlling root architecture has been confirmed by overexpression analysis, as illustrated by the observation that ectopic Arath;KRP2 overexpression in Arabidopsis results in a dramatic decrease in the number of lateral roots (Himanen et al., 2002).
Control of Endocycle Onset
Mammalian Kip/Cip inhibitor gene expression has been found to correlate with the onset of endoreduplication (Bates et al., 1998; Hattori et al., 2000). The endocycle is an alternative cell cycle during which DNA replication is not followed by mitosis and cytokinesis, and often marks the onset of cell differentiation. Endoreduplication represents the most common mechanisms to increase the cellular DNA ploidy in plants, and, although the physiological relevance of the endoreduplication process is still unresolved, there are several indirect reasons to believe that an increase in the DNA ploidy level supports cell growth and high metabolic activity (Schnittger et al., 2003; Sugimoto-Shirasu and Roberts, 2003).
In yeast and fruitfly (Drosophila melanogaster), the onset of endoreduplication corresponds with a decrease in CDK activity (Edgar and Orr-Weaver, 2001; Larkins et al., 2001). A similar mechanism is probably operational in plants because the start of endoreduplication in Arabidopsis leaves, maize endosperm, and the fruit of tomato (Lycopersicon esculentum) is accompanied by a decline in extractable CDK activity (Grafi and Larkins, 1995; Joubès et al., 1999; Verkest et al., 2005). Recently, KRPs have been demonstrated to participate in control of this decrease in CDK activity. In strong Arath;KRP2-overexpressing lines, CDK activity is inhibited in both mitotically dividing and endoreduplicating leaf tissues. By contrast, in weak overexpressing lines, only the mitotic CDK-cyclin complexes are affected, blocking entry into mitosis but still allowing onset and progression through S-phase, resulting into an increase in DNA ploidy levels. The two apparently contradictory effects seen upon strong or weak KRP overexpression can be explained by assuming that KRPs have a binding preference toward the CDK-cyclin complexes that control the G2-M checkpoint or that higher levels of CDK-cyclin activity are required for entry into mitosis than for entry into S-phase. The capacity of KRPs to trigger the onset of the endocycle in dividing tissues was confirmed by the specific overexpression of Arath;KRP2 in proliferating tissues, causing an inhibition of mitotic CDK activity and a premature onset of endoreduplication (Verkest et al., 2005). Likewise, low levels of Arath;KRP1 in trichome socket cells or its specific expression in the mitotically dividing stomatal precursor cells triggered increased ploidy levels (Weinl et al., 2005).
Interestingly, the Arath;KRP2 protein is negatively regulated at the posttranslational level by B-type CDK activity as seen by the increase in Arath;KRP2 abundance in transgenic plants with reduced Arath;CDKB1;1 activity (Verkest et al., 2005). B-type CDKs phosphorylate KRPs, marking them for protein destruction (see above). Previously, Arath;CDKB1;1 activity has been demonstrated to play an important role in the decision process of the cell to divide or to endoreduplicate: Plants with reduced B-type CDK activity exit the mitotic cell cycle and enter the endocycle prematurely (Boudolf et al., 2004b). Because KRPs specifically inhibit A-type CDK activity, the controlled destruction of KRPs by B-type CDK complexes suggests a mechanism by which the entry into the endocycle is controlled by a sequential decrease of first B-type and then A-type CDK activities. In this model, A-type CDKs are protected from KRP2-mediated inhibition as long as cells possess a high level of B-type CDK activity because CDKB1;1 marks the KRP proteins for destruction (Fig. 4A). However, as cells enter the endocycle program, they lose B-type CDK activity, with a stabilization of KRPs and a subsequent inhibition of A-type CDK activity as a result (Verkest et al., 2005; Fig. 4, B and C).
Figure 4.
Model of KRPs controlling the switch between the different cell cycle programs. A, In proliferating cells, B-type CDKs phosphorylate KRPs, triggering their destruction. In addition, phosphorylation might change the conformation of KRPs, interfering with their binding to A-type CDKs. B, In cells triggered to endoreduplicate, B-type CDK activity ceases, resulting in a stabilization of the KRPs, which now bind and inhibit A-type CDK-cyclin complexes with a role in mitosis. The KRP concentration, however, is probably not high enough to inhibit as well the CDK-cyclin complexes driving S-phase entry, allowing cells to reenter the S-phase. C, During cell cycle exit, KRP expression is up-regulated. Now, in addition to blocking entry into mitosis, CDK-cyclin complexes controlling the entry into S-phase become inhibited, resulting in a complete cell cycle arrest.
Do KRPs Function Outside the Cell Cycle?
In animals, CKIs might also have functions outside the cell cycle, such as in differentiation, morphogenesis, and programmed cell death. So far, there is no clear evidence that plant CKIs also function outside the cell cycle. Strong and constitutive overexpression of Arath;KRP2 did not alter cellular differentiation processes as illustrated by the unaltered timing of stomatal differentiation patterns with respect to leaf development (De Veylder et al., 2001). Also, premature onset of endoreduplication caused by Arath;KRP1 expression in trichome-neighboring cells does not apparently interfere with the adoption of trichome socket cell fate (Weinl et al., 2005). However, misexpression of Arath;KRP1 in Arabidopsis trichomes induces cell death (Schnittger et al., 2003). This phenotype seems to be linked to the developmental program of trichomes because for other cell types no cell death phenotypes have been observed upon KRP overexpression. At the moment, it is not clear whether the observed trichome cell death phenotype is linked with a compromised endoreduplication program, with cell death being indirectly initiated as a consequence of a discrepancy between DNA content and cell size. Clearly, additional experiments are required to decide whether KRPs control cell survival.
CONCLUSION
Recent years have brought about a tremendous increase in our understanding of CKIs in plants. Some regulatory pathways are now emerging with KRPs functioning as dose-dependent cell cycle regulators. KRPs might be important for adjusting CDK activity within dividing cells, as well as in facilitating the transition between different cell cycle programs, such as entering endoreduplication cycles or executing cell cycle exit (Fig. 4). KRPs could possibly play a central role in connecting cell cycle progression with developmental as well as environmental cues. Many questions, however, still need to be resolved. For instance, KRPs are very poorly conserved among species; thus, do these proteins still share a common structure outside the CDK and cyclin-binding motif? Is there a developmental or physiological need for the many different KRP genes within an organism (seven in Arabidopsis)? How is the KRP abundance and localization regulated? Many tools have recently been developed that allow us to address these questions at a cellular, biochemical, and genetic level. In addition, KRPs from different plant species have been identified, setting the path for a comparative approach and leading to the anticipation of general principles in KRP function in plants.
Acknowledgments
L.D.V. and A.S. thank the members of their laboratories and John Larkin (Louisiana State University) for critical reading and helpful comments on the manuscript, and Martine De Cock for help in preparing it.
This work was supported by grants from the European Union (SY-STEM) and Interuniversity Poles of Attraction Program-Belgian Science Policy (P5/13), the Institute for the Promotion of Innovation by Science and Technology in Flanders (predoctoral fellowship to A.V.), and the Fund for Scientific Research (Flanders; postdoctoral fellowship to L.D.V.). Support by the Volkswagen-Stiftung (VW Foundation) and by the Deutsche Forschungsgemeinschaft to A.S. is kindly acknowledged.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Lieven De Veylder (lieven.deveylder@psb.ugent.be) and Arp Schnittger (schnitt@mpiz-koeln.mpg.de).
References
- Bates S, Ryan KM, Phillips AC, Vousden KH (1998) Cell cycle arrest and DNA endoreduplication following p21Waf1/Cip1 expression. Oncogene 17: 1691–1703 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Barrôco R, de Almeida Engler J, Verkest A, Beeckman T, Naudts M, Inzé D, De Veylder L (2004. a) B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inzé D, De Veylder L (2004. b) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho CM, Dande RA, Sabelli PA, Sun Y, Dilkes BP, Gordon-Kamm WJ, Larkins BA (2005) Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiol 138: 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coqueret O (2003) New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol 13: 65–70 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Joubès J, Inzé D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6: 536–543 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL (2001) Endoreplication cell cycles: more for less. Cell 105: 297–306 [DOI] [PubMed] [Google Scholar]
- Grafi G, Larkins BA (1995) Endoreduplication in maize endosperm: involvement of M phase-promoting factor inhibition and induction of S phase-related kinases. Science 269: 1262–1264 [DOI] [PubMed] [Google Scholar]
- Hattori N, Davies TC, Anson-Cartwright L, Cross JC (2000) Periodic expression of the cyclin-dependent kinase inhibitor p57Kip2 in trophoblast giant cells defines a G2-like gap phase of the endocycle. Mol Biol Cell 11: 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A, de Almeida Engler J, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst L (2004) A second RING to destroy p27Kip1. Nat Cell Biol 6: 1153–1155 [DOI] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D (2005) Green light for the cell cycle. EMBO J 24: 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Schnittger A (2004) Cell cycle and differentiation. Curr Opin Plant Biol 7: 661–669 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Perennes C, Bergounioux C, Glab N (2002. b) Comparative molecular and functional analyses of the tobacco cyclin-dependent kinase inhibitor NtKIS1a and its spliced variant NtKIS1b. Plant Physiol 130: 1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Riou-Khamlichi C, Roche O, Perennes C, Bergounioux C, Glab N (2002. a) The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3;1-overexpressing plants. J Cell Sci 115: 973–982 [DOI] [PubMed] [Google Scholar]
- Joubès J, Phan T-H, Just D, Rothan C, Bergounioux C, Raymond P, Chevalier C (1999) Molecular and biochemical characterization of the involvement of cyclin-dependent kinase A during the early development of tomato fruit. Plant Physiol 121: 857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono A, Umeda-Hara C, Lee J, Ito M, Ichimiya H, Umeda M (2003) Arabidopsis D-type cyclin CYCD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol 132: 1315–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Tomlinson M, Parinyapong P, Sakvarelidze L, Leader D, Shaw P, Doonan JH (2004) CycD1, a putative G1 cyclin from Antirrhinum majus, accelerates the cell cycle in cultured tobacco BY-2 cells by enhancing both G1/S entry and progression through S and G2 phases. Plant Cell 16: 2364–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins BA, Dilkes BP, Dante RA, Coelho CM, Woo Y-m, Liu Y (2001) Investigating the hows and whys of DNA endoreduplication. J Exp Bot 52: 183–192 [PubMed] [Google Scholar]
- Lui H, Wang H, DeLong C, Fowke LC, Crosby WL, Fobert PR (2000) The Arabidopsis Cdc2a-interacting protein ICK2 is structurally related to ICK1 and is a potent inhibitor of cyclin-dependent kinase activity in vitro. Plant J 21: 379–385 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Mészáros T, Miskolczi P, Deák M, Fehér A, Brown S, Kondorosi E, Athanasiadis A, Pongor S, Bilgin M, et al (1997) Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9: 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD (1998) Cyclin-dependent kinase inhibitors of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Curr Top Microbiol Immunol 227: 1–24 [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Nowack MK, Grini PE, Jakoby MJ, Lafos M, Koncz C, Schnittger A (2005) A novel positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat Genet (in press) [DOI] [PubMed]
- Ormenese S, de Almeida Engler J, De Groodt R, De Veylder L, Inzé D, Jacqmard A (2004) Analysis of the spatial expression pattern of seven Kip related proteins (KRPs) in the shoot apex of Arabidopsis thaliana. Ann Bot (Lond) 93: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheld J-P, Segers G, De Veylder L, De Pinho Barrôco R, Casteels P, Van Montagu M, Inzé D, Mironov V (2001) A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J Biol Chem 276: 36354–36360 [DOI] [PubMed] [Google Scholar]
- Richard C, Granier C, Inzé D, De Veylder L (2001) Analysis of cell division parameters and cell cycle gene expression during the cultivation of Arabidopsis thaliana cell suspensions. J Exp Bot 52: 1625–1633 [PubMed] [Google Scholar]
- Schnittger A, Schöbinger U, Bouyer D, Weinl C, Stierhof Y-D, Hülskamp M (2002) Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc Natl Acad Sci USA 99: 6410–6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Weinl C, Bouyer D, Schöbinger U, Hülskamp M (2003) Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15: 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512 [DOI] [PubMed] [Google Scholar]
- Sorrell DA, Menges M, Healy JMS, Deveaux Y, Amano X, Su Y, Nakagami H, Shinmyo A, Doonan JH, Sekine M, et al (2001) Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar Bright Yellow-2 cells. Plant Physiol 126: 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, de O. Manes C-L, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Inzé D, De Veylder L (2005) The cyclin-dependent kinase inhibitor KRP2 controls the mitosis-to-endocycle transition during Arabidopsis leaf development through a specific inhibition of the mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, dePamphilis CW, Ma H (2004) Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 135: 1084–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Fowke LC, Crosby WL (1997) A plant cyclin-dependent kinase inhibitor gene. Nature 386: 451–452 [DOI] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- Weinl C, Marquardt S, Kuijt SJH, Nowack MK, Jakoby MJ, Hülskamp M, Schnittger A (2005) Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17: 1704–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Fowke LC, Wang H (2002) Plant CDK inhibitors: studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparisons in transgenic Arabidopsis plants. Plant Cell Rep 20: 967–975 [Google Scholar]
- Zhou Y, Li G, Brandizzi F, Fowke LC, Wang H (2003. a) The plant cyclin-dependent kinase inhibitor ICK1 has distinct functional domains for in vivo kinase inhibition, protein instability and nuclear localization. Plant J 35: 476–489 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wang H, Gilmer S, Whitwill S, Fowke LC (2003. b) Effects of co-expressing the plant CDK inhibitor ICK1 and D-type cyclin genes on plant growth, cell size and ploidy in Arabidopsis thaliana. Planta 216: 604–613 [DOI] [PubMed] [Google Scholar]