Abstract
The wall-associated kinase (WAK) gene family, one of the receptor-like kinase (RLK) gene families in plants, plays important roles in cell expansion, pathogen resistance, and heavy-metal stress tolerance in Arabidopsis (Arabidopsis thaliana). Through a reiterative database search and manual reannotation, we identified 125 OsWAK gene family members from rice (Oryza sativa) japonica cv Nipponbare; 37 (approximately 30%) OsWAKs were corrected/reannotated from earlier automated annotations. Of the 125 OsWAKs, 67 are receptor-like kinases, 28 receptor-like cytoplasmic kinases, 13 receptor-like proteins, 12 short genes, and five pseudogenes. The two-intron gene structure of the Arabidopsis WAK/WAK-Likes is generally conserved in OsWAKs; however, extra/missed introns were observed in some OsWAKs either in extracellular regions or in protein kinase domains. In addition to the 38 OsWAKs with full-length cDNA sequences and the 11 with rice expressed sequence tag sequences, gene expression analyses, using tiling-microarray analysis of the 20 OsWAKs on chromosome 10 and reverse transcription-PCR analysis for five OsWAKs, indicate that the majority of identified OsWAKs are likely expressed in rice. Phylogenetic analyses of OsWAKs, Arabidopsis WAK/WAK-Likes, and barley (Hordeum vulgare) HvWAKs show that the OsWAK gene family expanded in the rice genome due to lineage-specific expansion of the family in monocots. Localized gene duplications appear to be the primary genetic event in OsWAK gene family expansion and the 125 OsWAKs, present on all 12 chromosomes, are mostly clustered.
Efficient communication between the plant cell wall and the cytoplasm is important in plant development and in responding to biotic and abiotic stresses (Kohorn, 2000; Brownlee, 2002; Somerville et al., 2004). The wall-associated kinase (WAK) gene family, which belongs to the receptor-like kinase (RLK) superfamily in plants (Shiu and Bleecker, 2001, 2003), plays a critical role in this communication (He et al., 1996; Kohorn, 2001). WAK1, the first member of the WAK gene family identified in Arabidopsis (Arabidopsis thaliana), encodes a protein containing an intracellular Ser-Thr kinase domain and extracellular domains with similarities to vertebrate epidermal growth factor (EGF)-like domains (He et al., 1996). From the Arabidopsis genome (Arabidopsis Genome Initiative, 2000), 26 WAK and WAK-Like (WAKL) genes were identified and most of the 26 members are expressed in Arabidopsis (Verica and He, 2002; Verica et al., 2003).
Functional studies of the different WAK members in Arabidopsis demonstrated that they are involved in various functions in plants, including pathogen resistance (He et al., 1998), heavy-metal tolerance (Sivaguru et al., 2003), and plant development (Lally et al., 2001; Wagner and Kohorn, 2001). Biochemical studies demonstrated that WAK proteins are covalently bound to pectin in the cell wall (Wagner and Kohorn, 2001). They can also form an approximately 500-kD protein complex via interactions with a Gly-rich extracellular protein, AtGRP-3 (Park et al., 2001), and can interact with a cytoplasmic, type 2C kinase-associated protein phosphatase (Anderson et al., 2001). Based on these attributes, it was suggested that WAKs serve as physical links between the extracellular matrix and the cytoplasm and also as a signaling component between the cell wall and the cytoplasm (He et al., 1996; Kohorn, 2001). Recently, a study of seven (group II) WAKL members in Arabidopsis showed that they have tissue-specific and developmentally regulated expression patterns and are functionally similar to WAKs (Verica et al., 2003).
To further understand the functions and evolution of the WAK gene family in plants, we analyzed the WAK gene family in rice (Oryza sativa). Rice is one of the most important cereals and also considered a model for other cereal species, including maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), and sorghum (Sorghum bicolor). Rice genomic sequences are now available from both subspecies indica (Yu et al., 2002) and japonica (Feng et al., 2002; Goff et al., 2002; Sasaki et al., 2002; Rice Chromosome 10 Sequencing Consortium, 2003; International Rice Genome Sequencing Project, 2005). Previous analyses of the RLK superfamily from the indica genome sequence indicated that the WAK/WAKL gene family was one of the few RLK subfamilies that expanded in rice compared to Arabidopsis (Shiu et al., 2004).
Here we present a detailed analysis of the rice WAK/WAKL (OsWAK) gene family from japonica. First, through public database searches, we retrieved all genes annotated as putative OsWAKs, followed by reiterative database searches from which we obtained additional putative OsWAK gene family members. Manual reannotation was performed to correct or reannotate the misannotated putative OsWAK genes, such as split genes, fused genes, short genes, and pseudogenes. We determined expression characteristics for certain OsWAK members and, based on domain composition, we classified OsWAKs into five groups and performed comparative phylogenetic analyses of WAKs in Arabidopsis, rice, and barley to understand the possible mechanisms of gene family expansion.
RESULTS
Identification and Classification of OsWAKs from Genome Sequences of Rice Subsp. japonica cv Nipponbare
Four analytical steps were used to identify and classify OsWAKs from japonica subsp. cv Nipponbare. First, all genes annotated as putative OsWAKs were retrieved from three public databases: (1) The Institute for Genomic Research (TIGR) Rice Genome Annotation Database (Osa1, release 1 and 2; Yuan et al., 2005); (2) Rice Protein Database in GRAMENE (Ware et al., 2002); and (3) GenBank in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). There were different numbers of putative OsWAKs annotated in the different databases. Identical sequences from the same or overlapping contigs, but annotated with different putative OsWAK names in the various databases, were identified and removed. The second step, aimed at a more complete search for putative OsWAKs in rice, was performed using BLASTp and tBLASTn (E < 0.1) searches of Osa1 and the japonica cv Nipponbare genome sequences in GenBank. These analyses utilized 10 uniquely different putative OsWAK protein sequences (2768.t00013, 2541.t00004, 8323.t00010, 4577.t00017, 9637.t02571, 4967.t00013, 3135.t00009, gi|19881772, 9638.t00862, and 6563.t00002), which contained both the extracellular EGF-like domains (EGF-2, PS01186/PROSITE and/or EGF-Ca2+, PS01187/PROSITE) and an intracellular protein kinase domain. Most new genes obtained from this reiterative database search were originally annotated either as kinase domain-containing protein genes or receptor-like protein (RLP) genes.
In the third step, each putative OsWAK sequence was manually assessed for the EGF-like domains, kinase domain, or its sequence similarity to other putative OsWAKs. In our analyses, in order for a gene to be defined as a putative OsWAK, the gene had to fit into one of the five OsWAK types (see Table I). Based on these five criteria, a total of 129 putative OsWAKs were initially identified from japonica cv Nipponbare.
Table I.
Classification of 125 OsWAKs identified from rice genome of japonica cv Nipponbare
| Gene Type | Domain Content | No. in Category |
|---|---|---|
| OsWAK-RLK | Both extracellular EGF-like domains and a cytoplasmic protein kinase domain | 67 |
| OsWAK-RLCK | Only cytoplasmic protein kinase domain with its amino acid identity ≥40% to a kinase domain of an OsWAK-RLK member | 28 |
| OsWAK-RLP | Only extracellular EGF-like domains | 13 |
| OsWAK short gene | No domain, protein sequence <300 amino acids but its amino acid identity ≥40% to a longer OsWAK protein | 12 |
| OsWAK pseudogene | Stop codons or frame shifts in coding region | 5 |
As the last step, manual reannotation was performed to correct or reannotate the misannotated putative OsWAKs, as described below. This analysis included 10 putative split OsWAKs merged into five OsWAK-RLKs, four fused putative OsWAKs split into five different OsWAKs, and six non-OsWAKs, leaving a final total of 125 OsWAKs; the numbers of each type of OsWAK are shown in Table I. For the 125 OsWAKs, the detailed annotations (Supplemental Tables I and II), genomic and predicted coding/full-length (FL)-cDNA sequences (Supplemental Data 1), and predicted protein sequences (Supplemental Data 2) are presented.
Correction and Reannotation of Misannotated Putative OsWAKs
Automated annotations of rice genome sequences of japonica cv Nipponbare in TIGR and GenBank are useful as initial resources for gene annotation, but individual genes and gene families are generally not verified and corrected. Several types of annotation errors were found in the automated annotation of the Arabidopsis genome, including intron numbers/positions, merged/split genes, and missed short genes; approximately 35% of the initially annotated genes from Arabidopsis were corrected from FL-cDNA sequences (Haas et al., 2003). We therefore performed a similar assessment of the initially identified, 129 putative OsWAKs derived from automated annotation using the rice FL-cDNA sequences in the Knowledge-Based Oryza Molecular Biological Encyclopedia database (Kikuchi et al., 2003; http://cdna01.dna.affrc.go.jp/cDNA). For 31 putative OsWAKs, one corresponding FL-cDNA sequence was obtained; for eight other OsWAKs, more than one corresponding FL-cDNA sequence was obtained (Supplemental Table III). Of the 39 putative OsWAKs that matched with FL-cDNAs, only 20 had predicted coding sequences that perfectly or nearly perfectly matched the corresponding FL-cDNA sequences (identity ≥99.5%). For the other 19, the FL-cDNA sequences were either shorter (seven) or longer (12) than the predicted coding sequences. All of the FL-cDNA sequences, except one, have both 5′ untranslated region (UTR) and 3′ UTR, indicating they are most likely intact cDNA sequences. Therefore, these 19 putative OsWAKs were corrected using their corresponding FL-cDNA sequences, although it is possible that some or all of the shorter FL-cDNAs could derive from truncated sequences. Reannotation of 14 out of the 19, along with 13 additional putative OsWAKs, is described below.
Split/Fused OsWAK Genes
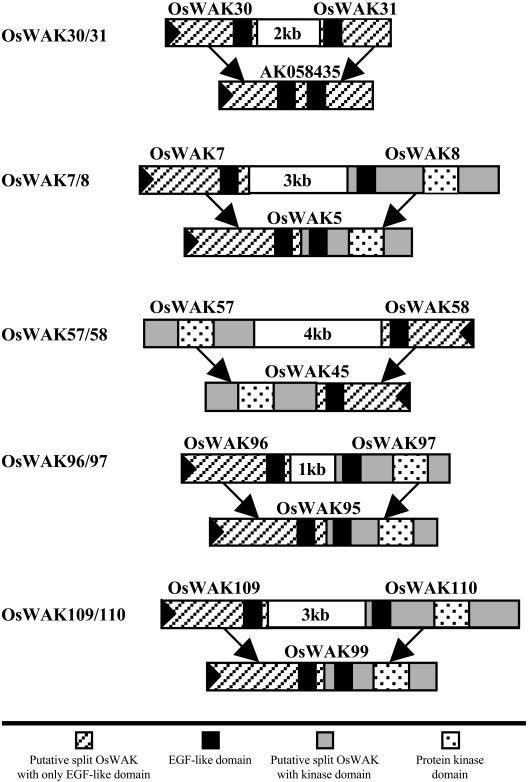
From the FL-cDNA sequence analysis described above, two putative OsWAK-RLPs, OsWAK30 and OsWAK31, matched to the same FL-cDNA sequence (AK058435). By examining matched regions, the two OsWAK-RLPs were found to match complementally to the FL-cDNA sequence (OsWAK30 on the 5′ side and OsWAK31 on the 3′ side). Also, both putative genes were located within an approximately 12-kb genomic region on chromosome 4 and no extra genes were predicted in the region between the two putative OsWAKs (Fig. 1). Therefore, these two putative, split OsWAK-RLPs were merged into a single OsWAK-RLP, OsWAK30/31. Based on similar analyses, eight more putative, split OsWAKs (four putative OsWAK-RLPs plus four putative OsWAK-receptor-like cytoplasmic kinases [RLCKs]) were merged into four OsWAK-RLKs (OsWAK7/8, OsWAK57/58, OsWAK96/97, OsWAK109/110). Merging was based on the fact that each pair matched complementally to the same OsWAK-RLK and different pairs matched to different OsWAK-RLKs (Fig. 1). They were split in the initial annotations at an intron between the two EGF-like domains within the extracellular region.
Figure 1.
Five OsWAKs merged from five pairs of putative split OsWAKs. Each pair of putative split OsWAKs is shown to be split at an intron in the original annotation; size of the intron is indicated in kilobase pairs. Each pair complementally matched either to a FL-cDNA or an OsWAK gene family member that is displayed below each pair; the corresponding matched regions are indicated by black arrows; large black triangles in boxes indicate direction of transcription.
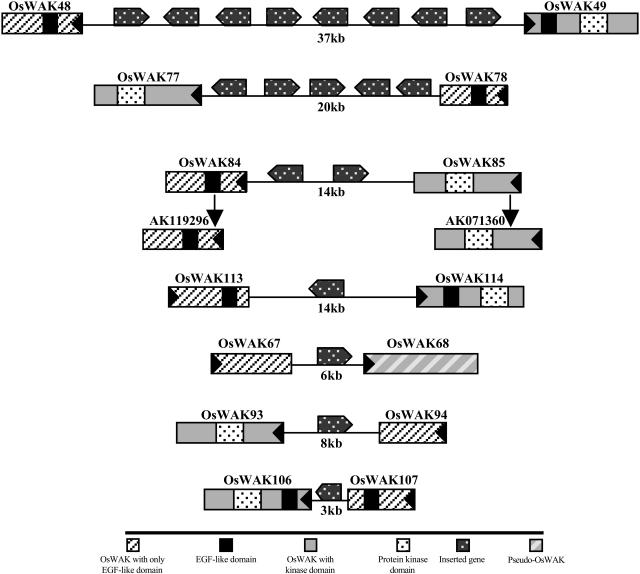
Fourteen other putative, split OsWAKs (seven putative OsWAK-RLPs and seven putative OsWAK-RLCKs) also matched complementally to various OsWAK-RLKs (data not shown). However, for each of these pairs, one to eight different genes interrupt the sequence (Fig. 2). These split OsWAKs appear to have already become independent genes; for example, the pair, OsWAK84 and OsWAK85, has two different FL-cDNA sequences. Therefore, these 14 putative, split OsWAKs remain annotated as independent OsWAKs.
Figure 2.
Genomic organization of 14 split OsWAKs. Each pair of OsWAKs is interrupted by one to eight different genes as indicated. The pair, OsWAK84 and OsWAK85, matched two different FL-cDNA sequences, as indicated by vertical arrows. Large black triangles in boxes indicate direction of transcription of OsWAKs; distance between each pair of OsWAKs is indicated in kilobase pairs.
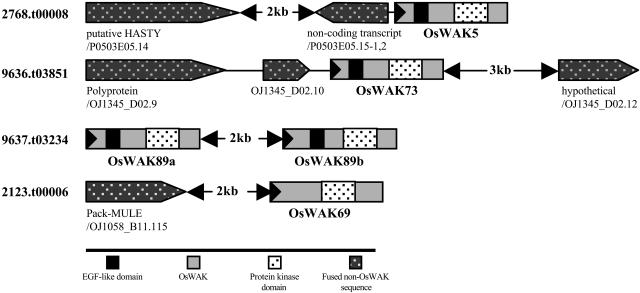
Three putative OsWAKs (OsWAK5/2768.t00008, OsWAK73/9636.t03851, OsWAK89/9637.t03234) from Osa1 in TIGR were found to be three fused genes. Putative OsWAK5 and OsWAK73 are fused with non-OsWAKs, and the third has two fused OsWAKs (Fig. 3). Therefore, the three fused genes were split, obtaining four OsWAKs (OsWAK5, OsWAK73, OsWAK89a, OsWAK89b). OsWAK89a and OsWAK89b have 57% identity in overall protein sequence and >93% identity in their kinase domains; both contain EGF-like domains in their extracellular regions, suggesting they are independent OsWAK-RLKs that likely derived from tandem duplication. Interestingly, putative OsWAK69 is fused to a Pack-Mu-like element (MULE; Jiang et al., 2004) on its 5′ side, and an approximately 2-kb space is found between the Pack-MULE and the kinase domain. The sequence of a FL-cDNA (AK064911) matched only to the Pack-MULE, not to the kinase domain (Fig. 4). Accordingly, the fusion is believed to be an annotation artifact and the kinase domain of the original OsWAK69 was split out as an OsWAK-RLCK.
Figure 3.
Five OsWAKs split from four fused putative OsWAKs. The five OsWAKs are illustrated with boxes; large black triangles in boxes indicate direction of transcription. Fused non-OsWAK sequences are indicated (see key). Annotations with the GenBank accession number are shown below each non-OsWAK sequence; distances between fused genes are indicated in kilobase pairs, except in cases where the distance is less than 1 kb.
Figure 4.
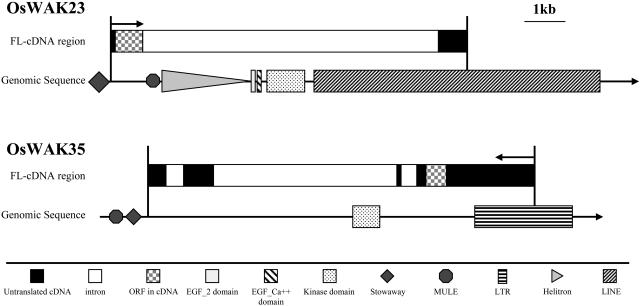
Two examples of OsWAK short genes resulting from TE insertions. MULE and Helitron elements inserted within OsWAK23 and LINE at the 3′ end of OsWAK23 provide a novel splicing site, splicing out the EGF-like domain and kinase domain in the FL-cDNA of OsWAK23. The LTR element at the 3′ end of OsWAK35 provides a transcription start site producing an antisense transcript of OsWAK35, which has only a short ORF.
OsWAK Short Genes
Five putative OsWAKs (OsWAK17, OsWAK19, OsWAK54, OsWAK62, OsWAK67) were initially identified as OsWAK short genes. This was due to the fact that their predicted protein sequences were <300 amino acids and did not encode an EGF-like or kinase domain, but they had >40% amino acid identity to the longer OsWAKs. From the FL-cDNA sequence analyses described above, seven more putative OsWAKs (OsWAK18, OsWAK23, OsWAK35, OsWAK37, OsWAK52, OsWAK101, OsWAK127) were reannotated as OsWAK short genes. This was also due to the fact that the deduced protein sequences from their longest open reading frames (ORFs) are <300 amino acids and do not encode an EGF-like or kinase domain. Of the seven OsWAK short genes, four (OsWAK18, OsWAK52, OsWAK101, OsWAK127) had antisense transcripts and the other three (OsWAK23, OsWAK35, OsWAK37) had sense transcripts. In order to further understand how the OsWAK short genes might have arisen in the rice genome, genomic sequences adjacent to the seven OsWAK short genes were analyzed. These analyses found various transposable elements (TEs) were inserted at either the 5′ or 3′ ends of the FL-cDNAs of five of the OsWAK short genes (Table II).
Table II.
TEs inserted within or adjacent to five OsWAK short genes
| Name | Accession No. of FL-cDNA | Type of TE | Insertion Position of TE |
|---|---|---|---|
| OsWAK18 | AK103831 | Tourist | 3′ end of FL-cDNA in the element |
| OsWAK23 | AK108554 | LINE | 3′ end of FL-cDNA in the element |
| OsWAK35c | AK100568 | SoloLTR | 5′ end of FL-cDNA in the element |
| OsWAK35c | AK100568 | Stowaway | 38 bp from 3′ end of FL-cDNA |
| OsWAK52 | AK111909 | MULE | 73 bp from 3′ end of FL-cDNA |
| OsWAK101 | AK111067 | MULE | 15 bp from 5′ end of FL-cDNA |
Further analyses indicated that the inserted TEs likely did not interrupt the OsWAKs; however, they either provided novel splicing sites or initialized antisense transcriptions, which resulted in short ORFs for the OsWAKs. For example, the 3′ end of the OsWAK23 FL-cDNA is located inside a long interspersed nuclear element (LINE) that appears to provide a novel splicing acceptor site. As a result, both the EGF-like and kinase domains were spliced out and thus do not present in the FL-cDNA sequence of OsWAK23 (Fig. 4). OsWAK35 has one sense and three antisense transcripts, and one of the antisense transcripts (AK100568) appears to be initialized from a solo long-terminal repeat (LTR; Fig. 4).
OsWAK Pseudogenes
Twelve putative OsWAKs were initially annotated as pseudogenes in GenBank because of the apparent lack of the 5′ exon, when compared to other OsWAK-RLKs, or the presence of stop codons or frame shifts in their coding regions (Supplemental Table II). However, four (OsWAK21, OsWAK76, OsWAK82, OsWAK84) had the corresponding FL-cDNA sequences available (Fig. 5), indicating they were transcribed in rice. Since there are stop codons in the coding regions, it is possible that they are nonfunctional. However, the ORFs derived from the FL-cDNAs contained either a kinase domain (OsWAK21, OsWAK76, OsWAK82) or the EGF-like domains (OsWAK84), suggesting they could also be functional in rice. Even though further analysis is needed to verify their status, we have reannotated them as OsWAKs. Three other OsWAKs (OsWAK3, OsWAK6, OsWAK80) were initially annotated as pseudogenes because the 5′ exon was missing; however, truncation does not always lead to loss of function (see “Discussion”). As a result, OsWAK3 is reannotated as an OsWAK-RLCK because it contains a kinase domain. OsWAK6 (524 amino acids) is shorter than other OsWAK-RLKs (>700 amino acids); however, it encodes an EGF-like domain and a kinase domain and was reannotated as an OsWAK-RLK member. For OsWAK80, the missing 5′ exon was recovered by extending the 5′ side from the bacterial artificial chromosome (BAC) sequence. These three putative pseudogenes, plus the four putative pseudogenes described above, were reannotated as true OsWAKs, leaving five annotated as OsWAK pseudogenes (OsWAK66, OsWAK68, OsWAK83, OsWAK86, OsWAK88).
Figure 5.
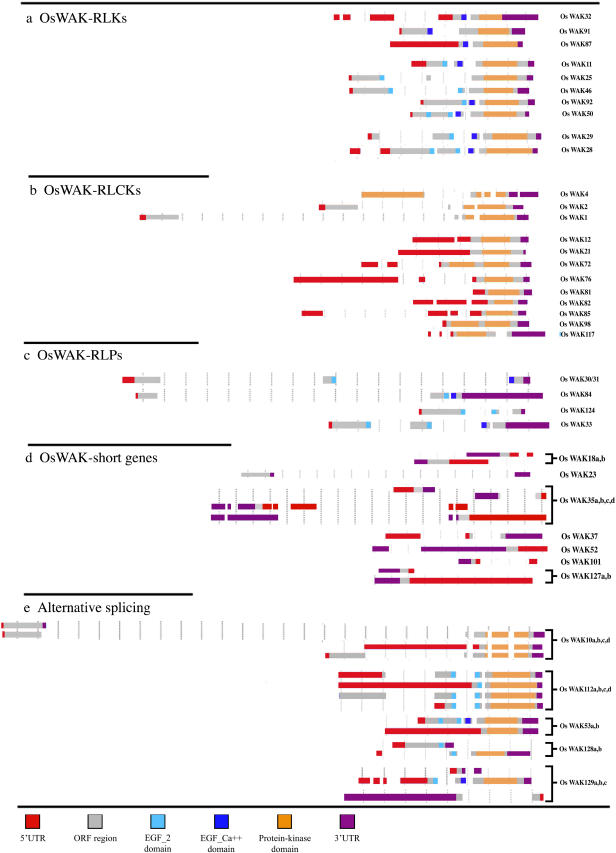
Gene structure and domain composition of 38 OsWAKs having FL-cDNA sequences. Intron number and position are shown along with domains for each OsWAK; OsWAKs having alternative splicing forms are indicated with brackets; distance between each vertical bar is 500 bp.
Intron Number and Position of OsWAKs
Arabidopsis RLK-type WAK/WAKLs have a two-intron/three-exon gene structure pattern within their coding regions (Verica and He, 2002). Both introns are located within the extracellular regions; the first intron is between the two EGF-like domains and the second is between an EGF-like domain and the kinase domain. Based on predicted coding sequences or FL-cDNA sequences, intron numbers in the coding regions for each of the 125 OsWAKs were determined (Supplemental Table II). For the 38 OsWAKs having a FL-cDNA sequence, intron numbers and positions, along with domain positions, are shown in Figure 5. One to three introns were observed in the 5′ UTR regions in 14 of the 38 OsWAKs, and one intron was found in the 3′ UTR regions in three OsWAKs. Of the 10 OsWAK-RLKs (Fig. 5A), five (OsWAK11, OsWAK25, OsWAK46, OsWAK50, OsWAK92) have the two-intron/three-exon pattern as observed in Arabidopsis WAK/WAKLs. However, three others (OsWAK32, OsWAK87, OsWAK91) have only one intron, likely due to a missing EGF-like domain in their extracellular regions. Another two (OsWAK28, OsWAK29) have three introns; both acquired an extra EGF-like domain in their extracellular regions. Of the 12 OsWAK-RLCKs (Fig. 5B), nine do not contain introns and three have one to two extra introns in their kinase domains. Of the four OsWAK-RLPs (Fig. 5C), three have the conserved two-intron pattern in their presumed extracellular regions, and one has an extra intron because of an additional EGF domain. Two of the seven OsWAK short genes have one to two introns in their coding regions (Fig. 5D). Various numbers of introns were observed in the different splicing forms of the five OsWAKs that have more than one FL-cDNA (Fig. 5E).
Domain Composition and Organization of OsWAKs
Excluding the five OsWAK pseudogenes and the 38 OsWAKs with FL-cDNAs (Fig. 5), domain composition and organization were analyzed for the other 82 OsWAKs, using their predicted protein sequences to search the Simple Modular Architecture Research Tool (SMART) database (http://smart.embl-heidelberg.de) for EGF-like domains, the kinase domain, and the transmembrane domain. Of the 82 OsWAKs, 53 OsWAK-RLKs contain both extracellular EGF-like domains (EGF-2 and/or EGF-Ca2+) and a cytoplasmic protein kinase domain; 15 OsWAK-RLCKs contain only one to two kinase domains; nine OsWAK-RLPs contain only the extracellular EGF-like domains; and five OsWAK short genes contain no domain (Fig. 6).
Figure 6.
Diagrams of domain composition and organization of 82 OsWAKs without FL-cDNA sequences. Number to the left of each OsWAK is chromosome number on which OsWAK is encoded. Superscripts indicate gene type: 1, OsWAK-RLP; 2, OsWAK-RCLK; and 3, OsWAK short genes.
Expression Analysis of OsWAKs
Thirty-eight OsWAKs have corresponding FL-cDNA sequences (Fig. 5), indicating that they are expressed in rice. Rice EST database searches in Sputnik (http://mips.gsf.de/proj/sputnik/oryza) were also performed for each of the 125 OsWAKs. Eleven OsWAKs that previously had no FL-cDNA matched to one to five different EST sequences (details in Supplemental Table III), indicating they are also expressed. The matched EST sequences were derived from various rice tissues and also from libraries made from plants following biotic and abiotic stress treatments. Of the 38 OsWAKs with FL-cDNAs, four (OsWAK18, OsWAK52, OsWAK101, OsWAK127) appear to be expressed as antisense genes because of the antisense orientation of their FL-cDNA sequences (Fig. 5D). Six OsWAKs (OsWAK10, OsWAK53, OsWAK112, OsWAK118, OsWAK18, OsWAK127) appear to be alternatively spliced, since they have more than one FL-cDNA sequence; and two (OsWAK35, OsWAK129) are transcribed in both directions (Fig. 5E).
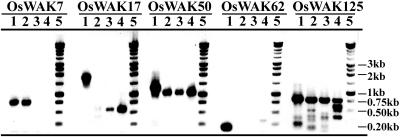
In addition, five different OsWAKs were chosen for expression analysis using reverse transcription (RT)-PCR, OsWAK7, OsWAK50, OsWAK125, plus two short genes, OsWAK17 (48% amino acid identity to the C terminus of OsWAK53) and OsWAK62 (92% amino acid identity to the extracellular region of OsWAK63). DNA samples from leaf tissues were used in PCR analyses; three RNA samples from the root tip, root base, and shoot tissues were used in RT-PCR analyses. OsWAK50, which has a FL-cDNA sequence available, had RT-PCR products of an expected size (1,084 bp) from all three tissues; because two introns were present, its PCR product was larger (1,308 bp). This result confirmed OsWAK50 is an expressed gene and showed there was no DNA contamination in RNA samples used for RT-PCR reactions. The same DNA and RNA samples were used for the other four OsWAKs. OsWAK7 and OsWAK125 that are without FL-cDNA sequences also had RT-PCR products from one or more rice tissues; because they had no intron, their RT-PCR products (OsWAK7, 758 bp; OsWAK125, 800 bp) were the same sizes as their PCR products (Fig. 7). The short gene, OsWAK17, which had an RT-PCR product from the root base and shoot, indicating the short gene, resembling the C-terminal region of an RLK, is also expressed. Because of the presence of two introns, the PCR product of OsWAK17 was much larger (2,178 bp) than its RT-PCR product (731 bp). Only a PCR product (207 bp) was obtained from OsWAK62, indicating it may not be expressed or may be expressed in other tissues.
Figure 7.
RT-PCR gene expression analyses of five OsWAK genes in rice. One DNA sample isolated from leaf tissue (lane 1) and three RNA samples isolated from root tip (lane 2), root base (lane 3), and shoot (lane 4) were used for all OsWAKs. Lane 5 contains Mr markers for all OsWAKs, as indicated.
For functional studies of all OsWAKs, further experimental confirmation of gene expression patterns is needed. To perform this type of analysis, rice microarrays containing all 125 OsWAKs would be ideal; however, this type of custom microarray is not yet available. We therefore took advantage of available rice tiling-path microarrays representing the entire rice chromosome 10 that have been successfully used to detect gene transcription activity (Li et al., 2005). Two independent sets of 36-mer probes, with 10-nucleotide intervals, tiled throughout chromosome 10 of japonica and indica, were designed for the microarrays; rice subsp. japonica cv Nipponbare and rice subsp. indica cv 93-11, respectively, were used for the experiments. The mRNA samples used for the microarray hybridization were from four normally growing tissues: 7-d-old seedling shoot and root, panicle (heading and filling stages), and suspension culture cells. Determination of the signal-to-noise ratio and expression level of a given gene model was previously described (Li et al., 2005). Based on the results of that analysis, we examined transcription activity for the 20 OsWAKs on chromosome 10. Fifteen out of the 20 OsWAKs were shown to be expressed in japonica tissues, and 10 in both japonica and indica (Table III). Surprisingly, of the three OsWAKs (OsWAK98, OsWAK101, OsWAK112) already having FL-cDNA sequences available, only OsWAK101 was expressed based on the microarray experiment, suggesting the other two might be expressed under specific stress or developmental conditions.
Table III.
Tiling-microarray analysis of transcriptional activity in japonica and indica rice of 20 OsWAKs from chromosome 10
Two independent sets of tiling microarrays were used for japonica cv Nipponbare and indica cv 93-11, respectively. Analysis was performed using mRNA extracted from four tissues: 7-d-old seedling shoots and roots, panicles (heading and filling stages), and suspension culture cells from japonica cv Nipponbare and indica cv 93-11.
| OsWAK Gene
|
Stranda
|
Expression
|
|
|---|---|---|---|
| japonica | indica | ||
| OsWAK93 | − | Pb | P |
| OsWAK94c | − | P | P |
| OsWAK95 | − | P | P |
| OsWAK97 | + | P | |
| OsWAK98de | − | ||
| OsWAK99 | − | P | |
| OsWAK100d | + | ||
| OsWAK101ef | + | P | P |
| OsWAK102d | + | P | P |
| OsWAK103 | + | P | |
| OsWAK104 | + | P | |
| OsWAK105 | − | P | P |
| OsWAK106 | − | P | P |
| OsWAK107c | − | P | |
| OsWAK108 | − | P | P |
| OsWAK110 | + | P | P |
| OsWAK111 | − | ||
| OsWAK112e | − | ||
| OsWAK113c | + | P | P |
| OsWAK114 | + | ||
Strand, Gene model orientation relative to the sequence; +, from the same orientation; −, from the complement.
P, Positively expressed.
OsWAK-RLP.
OsWAK-RLCK.
Genes in bold have FL-cDNA sequences.
Short genes.
Phylogenetic Analyses of Rice OsWAKs, Arabidopsis WAK/WAKLs, and Barley HvWAKs
Intracellular protein kinase domains are typically the conserved regions of RLKs and are used in phylogenetic analyses (Shiu and Bleecker, 2001). Ninety-five OsWAKs (67 OsWAK-RLKs plus 28 OsWAK-RCLKs) containing one or two kinase domains were initially analyzed phylogenetically together with the 21 Arabidopsis WAKs/WAKLs (Fig. 8).
Figure 8.
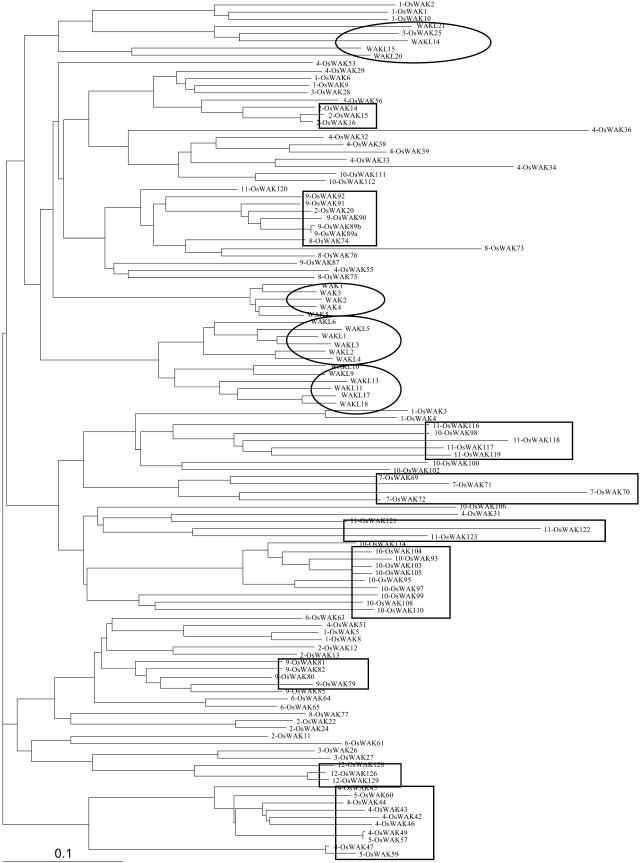
Phylogenetic analysis using conserved protein kinase domain regions of 95 OsWAKs and 21 Arabidopsis WAKs/WAKLs. Distinct species-specific clades of Arabidopsis WAK/WAKLs are circled. Rectangles highlight several OsWAK gene groups closely located on the same chromosomes and clustered in the same subclades.
This analysis revealed that most OsWAKs and Arabidopsis WAK/WAKLs cluster in species-specific distinct clades, except for four OsWAKs (OsWAK1, OsWAK2, OsWAK10, OsWAK25) and four WAKL-IV members (WAK14, WAK15, WAK20, WAK21) that cluster in the same clade. This result indicates that most OsWAKs and Arabidopsis WAK/WAKLs expanded in a species-specific manner; only a few members likely originated from the common ancestral genes that existed before divergence of monocots and dicots.
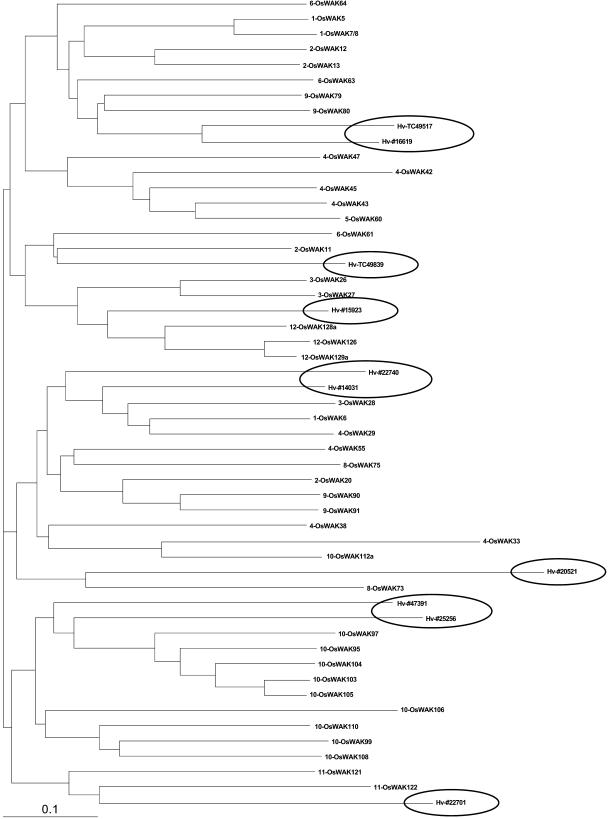
To further investigate whether expansion of OsWAKs is rice specific or due to lineage-specific expansion of this family in monocots, we identified 10 barley WAKs (HvWAKs) from the barley EST database (HarvEST; http://harvest.ucr.edu). Because only partial predicted protein sequences (of either extracellular regions or kinase domains) of HvWAKs were obtained (Supplemental Data 3), we used predicted, FL protein sequences of 43 OsWAK-RLKs containing both extracellular regions and kinase domains in the phylogenetic analyses. Ten HvWAKs cluster with individual OsWAKs, rather than diverging in a group unique to barley (Fig. 9), suggesting that OsWAK expansion in rice likely resulted from lineage-specific expansion of the family in monocots.
Figure 9.
Phylogenetic analysis using predicted FL protein sequences of 43 OsWAK-RLK members and 10 partial protein sequences of HvWAKs. Circled HvWAK members aligned with individual OsWAK members.
Localized Duplications Resulted in Expansion of OsWAKs
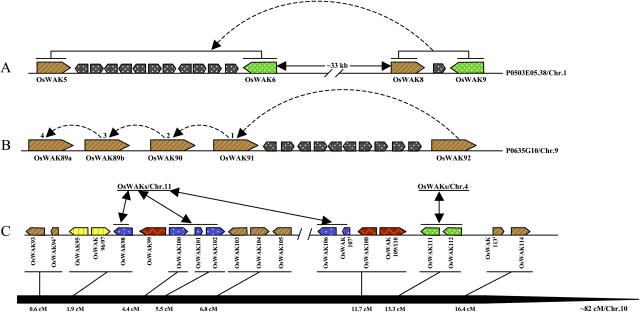
Examination of individual OsWAKs in different subclades of the rice-specific clades (Fig. 8) revealed that many small OsWAK groups (two to seven genes) located close together on the same chromosomes are clustered in the same subclades, i.e., OsWAKs 14 to 16 from chromosome 2; OsWAKs 42 to 47 and 49 from chromosome 4; OsWAKs 70 to 72 from chromosome 7; OsWAKs 79 to 82, 85, and 89 to 92 from chromosome 9; OsWAKs 116 to 119 and 121 to 123 from chromosome 11; and OsWAKs 126, 128, and 129 from chromosome 12. The high sequence similarity of these physically, closely located OsWAKs indicates that likely they arose from localized gene duplications. For example, based on sequence similarity and physical location, the four OsWAKs (OsWAK5, OsWAK6, OsWAK8, OsWAK9) on chromosome 1 likely derived from duplication of a gene pair and, after duplication, 11 genes inserted between OsWAK5 and OsWAK6 (Fig. 10A). The five OsWAKs (OsWAK89a, OsWAK89b, OsWAK90, OsWAK91, OsWAK92) on chromosome 9 likely derived from four gene duplications and, after the duplication, 10 genes inserted between OsWAK91 and OsWAK92 (Fig. 10B). The 20 OsWAKs on chromosome 10 are physically located as two clusters within 16 cM at the end of the short arm. Twelve likely derived from several localized gene duplications; six were duplicated to or from the OsWAKs on chromosome 11, and two were duplicated to or from the OsWAKs on chromosome 4 (Fig. 10C).
Figure 10.
Examples of localized gene duplications resulting in OsWAK gene clusters in rice chromosomes. Genes of the same color indicate duplications, and throughout the figure all symbols like those between OsWAK5 and OsWAK6 in A indicate inserted genes. A, Pair of OsWAKs duplicated on chromosome 1 with 11 genes inserted between OsWAK5 and OsWAK6. BAC clone designation is at the right. B, Four localized duplications, resulting in a five-OsWAK gene cluster on chromosome 5; 10 genes were inserted between OsWAK91 and OsWAK92. BAC clone designation is at the right. C, Twenty OsWAKs, clustered within approximately 16 cM at the end of the short arm of chromosome 10, arose from several different gene duplication events (different colors represent different gene duplications). Six OsWAKs (depicted in blue) were duplicated to or from OsWAKs on chromosome 11; two OsWAKs (in green) duplicated to or from OsWAKs on chromosome 4. The remaining 12 OsWAKs derived from four gene duplications, which are highly similar to each other, as indicated by their being clustered in the same subclade (Fig. 8). Due to space limitation, genes between OsWAKs were not indicated in C.
Ratios of Nonsynonymous versus Synonymous Substitution Rates between OsWAK EGF-Ca2+ Domain Regions
The EGF-Ca2+ domains, the typical extracellular domains in Arabidopsis WAK/WAKLs (Verica and He, 2002), are more conserved than the other extracellular regions and were shown to be under purifying selection (Verica et al., 2003). The EGF-Ca2+ domain regions of OsWAKs, which were identified by searching for EGF-Ca2+ domains ([DEQN]-x-[DEQN](2)-C-x(3,14)-C-x(3,7)-C-x-[DN]-x(4)-[FY]-x-C) using the SMART database (http://smart.embl-heidelberg.de), are also more conserved than other extracellular regions (data not shown). To determine whether OsWAK EGF-Ca2+ domains are also under purifying selection, ratios of nonsynonymous versus synonymous substitution rates among EGF-Ca2+ domain regions from 15 randomly selected OsWAKs were analyzed using an improved analytical method (Yang and Nielsen, 2000). Analysis of results shows that most ratios were significantly less than 1 (P < 0.05; Supplemental Table IV), indicating OsWAK EGF-Ca2+ domain regions are also under purifying selection.
Physical Locations of OsWAKs on Chromosomes
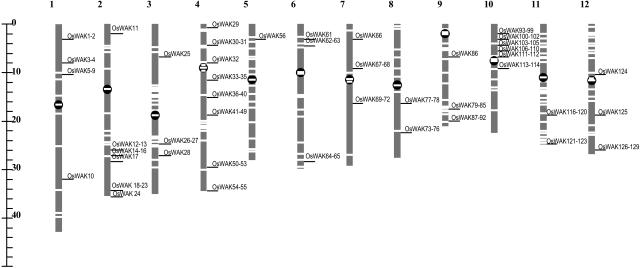
Since all rice clones used for genomic sequencing in the International Rice Genome Sequencing Project can be physically anchored on the 12 rice chromosomes, as are the pseudomolecules assembled in TIGR (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml), the 125 OsWAKs encoded on individual clones can be correspondingly mapped on japonica chromosomes (Fig. 11). This analysis showed that OsWAKs are clustered on chromosomes in most cases; however, in a few cases only a single OsWAK is present at a given chromosomal location, e.g., OsWAK10 on chromosome 1. The majority of OsWAKs (56.8%; 71 of 125) are located on chromosomes 2, 4, 9, and 10.
Figure 11.
Physical locations of 125 OsWAKs on the 12 japonica rice chromosomes, based on TIGR rice pseudomolecules (Osa1, release 2). Horizontal white lines within each chromosome indicate sequence gaps remaining; horizontal black lines indicate locations of OsWAKs on each chromosome. Three OsWAKs (OsWAK57/58, OsWAK59, OsWAK60) remain to be located on the map because, to date, the contig (OJ1480_H01) encoding these three OsWAKs has not been mapped to the chromosome 5 pseudomolecule in TIGR.
DISCUSSION
Identification of OsWAK Gene Family Members
Through a reiterative database search and manual reannotation, we identified 125 OsWAK gene family members from rice japonica cv Nipponbare. Thirty-seven (approximately 30%) of the identified OsWAKs were corrected/reannotated from their earlier automated annotations, such as five merged genes from 10 split genes, five split genes from four fused genes, seven corrected as short genes, and six reannotated from putative pseudogenes. A few of the reannotations were based on sequence similarities to the other OsWAK family members; a few other reannotations/corrections were found by sequence extension of the previously annotated genes to recover the missing exons from the 5′ or 3′ ends.
Most reannotations, however, were based on the corresponding FL-cDNA sequences available in rice (Kikuchi et al., 2003). This also occurred in the analysis of the Arabidopsis genome, where approximately 35% of automated annotations were reannotated using FL-cDNA sequences (Haas et al., 2003). Misannotations from the current automated annotation programs could occur for several reasons, including insufficient numbers of experimentally confirmed genes being available to train the gene-prediction programs, unexpectedly large introns, presence of TE insertions that interrupt normal gene sequences and transcription, and possible sequencing errors (Haas et al., 2003; Wang et al., 2003; Castelli et al., 2004). In addition, few of the annotation errors in rice were due to incomplete sequencing information; of the 63 BACs that encode the 125 OsWAKs, six BACs were not yet completed at the time the annotation was generated (see Supplemental Table II, column D). None of the five OsWAK pseudogenes came from incomplete BACs; however, the merged OsWAK57/58 was from an incomplete BAC (OJ1480_H01). Also, two OsWAKs (OsWAK10, OsWAK50) were truncated because they were located at the end of the BAC sequences; the full length of these two OsWAKs was recovered from their FL-cDNA sequences. Therefore, further experimental validations of the reannotated OsWAKs are warranted.
Classification of the OsWAK Gene Family
Of the 125 OsWAK gene family members, 67 are OsWAK-RLKs containing both extracellular EGF-like domains and an intracellular kinase domain; 28 are OsWAK-RLCKs containing only the kinase domain; 13 are OsWAK-RLPs with only the extracellular EGF-like domains; 12 are OsWAK short genes; and five are pseudogenes. Functions of these various OsWAKs are yet to be determined.
The unique character of OsWAK-RLPs and OsWAK-RLKs is the EGF-like domain at the N-terminal extracellular region. The function of EGF-like domains in OsWAKs and Arabidopsis WAK/WAKLs is yet to be determined. Analysis of other EGF-containing proteins from plants and animals suggests several possible roles for the EGF-like domains in the OsWAK-RLKs and OsWAK-RLPs. In the plant vacuolar sorting protein, BP-80, EGF domains were shown to alter the structural conformation of the ligand-binding domains, thereby increasing their affinity for ligand binding (Cao et al., 2000). EGF-like domains in certain proteins from animals directly participate in protein-protein interactions (Appella et al., 1987; Rebay et al., 1991; Kuroda and Tanizawa, 1999; Stenberg et al., 1999). For example, in the fruit fly (Drosophila melanogaster), the transmembrane protein, Notch, which contains 36 tandem EGF-like repeats, interacts with other EGF-containing transmembrane proteins, including Delta and Serrate. Two of the Notch EGF repeats were shown to be both necessary and sufficient for mediating these interactions (Rebay et al., 1991). The formation of both homodimeric and heterodimeric receptor complexes has been proposed for a large number of receptor kinases (Heldin, 1995), including members of the EGF family of receptor kinases in animals (Yarden and Schlessinger, 1987; Sliwkowski et al., 1994) and the CLV1 receptor complex in plants (Clark et al., 1997; Trotochaud et al., 1999). It remains to be seen whether the EGF-like domains in OsWAK-RLKs and OsWAK-RLPs are involved in protein-protein interactions.
In plants, several different RLK members were characterized and found to function in a diverse array of signaling processes, including phytohormone responses (Chang et al., 1993 [ETR1]; Li and Chory, 1997 [BRI1]; Clark et al., 1998 [CTR1]), reproduction (Stein et al., 1991 [SRK]; Mu et al., 1994 [PRK1]), developmental regulation (Becraft et al., 1996 [CRINKLY4]; Clark et al., 1997 [CLV1]; Yokoyama et al., 1998 [ERECTA]; Jinn et al., 2000 [HAESA]), and plant disease resistance responses (Song et al., 1995 [Xa21]). These RLKs are divided into different subclasses based on sequence relationships between their predicted extracellular regions (Shiu and Bleecker, 2001).
Several RLP members in plants were also shown to have important functions. For example, CLV2 functions in shoot meristem development in Arabidopsis (Kayes and Clark, 1998; Jeong et al., 1999); Fasciated Ear 2, an ortholog of CLV2, in the regulation of ear inflorescence meristem proliferation in maize (Taguchi-Shiobara et al., 2001); Too Many Mouths (TMM) in stomatal patterning in Arabidopsis (Nadeau and Sack, 2002); Cf-9 in disease resistance in tomato (Jones et al., 1994); and Xa21D in pathogen resistance in rice (Wang et al., 1998). Arabidopsis WAKL7 is also an RLP gene that is wound inducible (Verica et al., 2003). A few plant RLCKs were found to be functional in disease resistance signaling processes, including Pto in pathogen resistance (Martin et al., 1993) and EDR1 in negative regulation of defense responses (Frye et al., 2001).
Especially interesting are recent studies showing that plant RLK, RLP, and RLCK members could function together in the same signaling pathways. For example, in the self-incompatibility signaling pathway in Brassica, the S-receptor kinase (an RLK member), the S-locus glycoprotein (an RLP member resembling the extracellular part of the S-receptor kinase), and the M-locus protein kinase (an RLCK member) function as a single complex in signaling (Cui et al., 2000; Takasaki et al., 2000; Murase et al., 2004). Another example is the complex interaction of TMM (Leu-rich repeat [LRR]-RLP) and three ERECTA family members (LRR-RLKs) in stomatal patterning and differentiation (Shpak et al., 2005); TMM negatively regulates specific ERECTA family members at critical steps in stomatal differentiation. The mechanism for regulation of the ERECTA family members by TMM is still unknown. It was suggested that TMM forms a receptor heterodimer with ERECTA family RLKs, preventing signaling; the lack of a signal transducer domain in TMM supports the idea of this inhibitory function. Alternatively, TMM may have the same ligands as the ERECTA family, hence repressing the ERECTA family signaling pathway. These examples suggest that different OsWAKs could also function by heterodimerization in multimeric assemblies or alone in transduction cascades.
Twelve OsWAK family members are short genes with less than 300 amino acids and no known domain. This type of short gene was also identified in the Arabidopsis FL-cDNA sequence analyses (Haas et al., 2003). In some cases, the short genes appear to be functional in Arabidopsis, e.g. the CLE gene family members (Hobe et al., 2003; Sharma et al., 2003). A possible clue into the genesis of OsWAK short genes comes from the observation that recognizable TEs are found within or near the 5′ or 3′ end of five of the short genes (OsWAK18, OsWAK23, OsWAK35, OsWAK52, OsWAK101). They either provide a novel splicing site or initialize antisense transcription, which resulted in the short ORFs. Further studies of the function, if any, of the OsWAK short genes, especially of the seven OsWAK short genes with corresponding FL-cDNA sequences, should provide interesting insights into the role of OsWAK short genes in rice.
Structure and Expression of OsWAKs
The two-intron gene structure pattern of Arabidopsis WAK/WAKLs (Verica and He, 2002) was generally conserved in OsWAKs. However, in OsWAKs with an extra/missing EGF-like domain, an extra/missing intron was observed in their extracellular regions. Extra introns were also observed in the kinase domains of a few OsWAK-RLCKs. Intron variation in OsWAKs demonstrates that OsWAK gene structure is not as conserved as it is in the Arabidopsis WAK/WAKLs and is a reflection of its expansion in rice.
Antisense expression, bidirectional transcription, and alternative splicing were observed in a few of the 38 OsWAKs with FL-cDNA sequences. This type of antisense and bidirectional transcription was also observed from FL-cDNA sequence analysis of many other rice genes (Osato et al., 2003). However, biological implications of the different directions of transcription and the nature of the resulting products remain to be determined. Expression analyses based on the tiling-microarray experiment of the 20 OsWAKs on chromosome 10 and RT-PCR for five additional OsWAKs suggest that the majority of OsWAKs are expressed in rice.
Evolutionary Expansion of OsWAKs
Compared to the 26 Arabidopsis WAKs/WAKLs (Verica and He, 2002), the japonica rice genome has nearly a 5 times greater number of OsWAKs. Previous comparative analyses between the two plant species of other gene families, e.g. the CONSTANS-like gene family (Griffiths et al., 2003), the Dof family (Lijavetzky et al., 2003), and the LRR extensin family (Baumberger et al., 2003), did not reveal the magnitude of difference in family sizes that is seen with the OsWAKs. Therefore, the evolutionary expansion of the OsWAK gene family does not appear to be due simply to the larger genome size of rice.
Phylogenetic analyses of OsWAKs and Arabidopsis WAK/WAKLs show that most OsWAKs and Arabidopsis WAK/WAKLs are clustered in distinct species-specific clades, suggesting species-specific expansion in both plants. Further phylogenetic analyses, comparing OsWAKs with barley HvWAKs, indicate that OsWAK expansion was mainly due to its lineage-specific expansion in monocot species. This type of divergence between monocot (rice) and dicot (Arabidopsis) species was also observed for a large gene family involved in pathogen resistance, the nucleotide-binding site (NBS)-LRR gene family (Bai et al., 2002). In rice, greater than 600 members of the OsNBS-LRR gene family were identified, 3 to 4 times greater than the number in Arabidopsis. Also, most OsNBS-LRRs do not encode Toll and mammalian interleukin-1 receptor domains; however, most Arabidopsis NBS-LRRs do contain an interleukin-1 receptor domain. This lineage-specific divergence of domains and expansion of certain gene families involved in pathogen resistance in both monocot and dicot species likely occurs to enable reaction of plants to pathogens unique to each species.
It has been suggested that the expansion of Arabidopsis WAK/WAKLs is due to both local tandem duplications and large-scale genomic duplications (Verica and He, 2002). However, it appears from our analyses in rice that expansion of OsWAKs is probably due in large part to localized gene duplications, since many small OsWAK groups, located in close proximity on the same chromosome, have high sequence similarity. Localized duplication was also suggested as the main reason for WAK gene family expansion in indica rice (Shiu et al., 2004). A recent study that looked at the history of rice genome duplications showed that massive individual gene duplications are ongoing in the rice genome, providing a robust source of raw material for the genesis of new genes and gene functions (Yu et al., 2005). The results of our study provide the necessary genomic information for further in-depth study of the functions of OsWAKs and of their evolutionary expansion in the rice genome.
MATERIALS AND METHODS
Sequence Retrieval and Analysis
The DNA and predicted protein sequences, annotated as putative OsWAKs from the genome sequence of rice (Oryza sativa) japonica cv Nipponbare, were retrieved from three public databases: (1) TIGR Rice Genome Annotation Database (Osa1; http://www.tigr.org/tdb/e2k1/osa1; Yuan et al., 2005); (2) Rice Protein Database in Gramene (http://www.gramene.org; Ware et al., 2002); and (3) GenBank in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The same sequences from the same or overlapping contigs, but annotated by different putative OsWAK names in the various databases, were checked using the multiple sequence alignment tool of ClustalW from the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/index.html). Genes from rice varieties other than japonica cv Nipponbare were manually removed. BLAST search tools, BLASTp and tBLASTn (Altschul et al., 1997), were used to identify additional OsWAK sequences from Osa1 and GenBank, using 10 putative OsWAK protein sequences as queries.
Domain Searches of OsWAK Protein Sequences
The SMART database (http://smart.embl-heidelberg.de) was used to search for EGF-like domains, protein kinase domains, and transmembrane domains. BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) was utilized as a tool for diagramming domain positions for OsWAKs.
RT-PCR Analysis of OsWAKs
Total DNA and RNA, used in PCR and RT-PCR respectively, were isolated from 2-week-old seedlings of rice japonica cv Nipponbare. DNA extraction was performed on young leaf tissues (Cone, 1989); shoot and root tissues were used for total RNA isolation with TRIzol reagent (Gibco-BRL, Life Technologies), according to the manufacturer's instructions. RNA purification, PCR, and RT-PCR were conducted as described (Meng et al., 2003). PCR or RT-PCR products were purified using QIAquick PCR purification kit (Qiagen) and sequenced (Elim Biopharmaceuticals). Gene-specific RT-PCR primers were designed for each OsWAK as follows: OsWAK7 (F-AGTGTTGCACTAGTCATGCTGCA, R-CTATGGCATGCATATGAAGTCATG); OsWAK17 (F-GTTGATTGGTTCCTCTTGATGCAG, R-GAAGAGTGGAGAGTGGAGGATGA); OsWAK50 (F-CAACTCAAGCTTAACGTCAACTC, R-GAGCTCACTGGTGGTGAATATC); OsWAK62 (F-ACTCATGGACATTATAGGTCATC, R-GAATGTTGCACCATCTCCTCC); and OsWAK125 (F-AACCTCACCTGCAGCAGCAAC, R-TCATGGCATCCACCAGCAACG).
Multiple Sequence Alignment and Phylogenetic Tree Analysis
Predicted protein sequences of OsWAKs were retrieved from public databases as described in “Results.” Arabidopsis (Arabidopsis thaliana) WAK/WAKL protein sequences were obtained from GenBank as described in Verica and He (2002). Barley (Hordeum vulgare) HvWAKs were retrieved from the barley EST database, HarvEST (http://harvest.ucr.edu), and their predicted protein sequences were translated by frame matching to known WAK protein sequences. Multiple sequence alignment analysis was performed using ClustalW, with default parameters set as in the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/index.html) and BioEdit. The phylogenetic tree was produced using TreeView software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Physical Mapping of OsWAKs on Rice Chromosomes
All sequenced contigs from japonica cv Nipponbare were physically constructed as pseudomolecules (release 2) in TIGR (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml), representing the 12 rice chromosomes. OsWAKs were then mapped on individual chromosomes, based on corresponding contigs on rice chromosomes.
Supplementary Material
Acknowledgments
The authors are grateful to Barbara Alonso for providing expert assistance with the figures, and to the anonymous reviewers for their helpful comments.
This work was supported by the National Science Foundation (NSF) Plant Genome Research Program (grant no. 0110512) and NSF Research Experience for Undergraduate (REU; to P.G.L.), and by the National Institutes of Health (NIH grant no. S06 GM52588 to Z.-H.H.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Peggy G. Lemaux (lemauxpg@nature.berkeley.edu) and Shibo Zhang (shibo@nature.berkeley.edu).
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CM, Wagner TA, Perret M, He ZH, He D, Kohorn BD (2001) WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol Biol 47: 197–206 [PubMed] [Google Scholar]
- Appella E, Robinson EA, Ullrich SJ, Stoppelli MP, Corti A, Cassani G, Blasi F (1987) The receptor-binding sequence of urokinase: a biological function for the growth-factor module of proteases. J Biol Chem 262: 4437–4440 [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, et al (2002) Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 12: 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Doesseger B, Guyot R, Diet A, Parsons RL, Clark MA, Simmons MP, Bedinger P, Goff SA, Ringli C, et al (2003) Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol 131: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR (1996) CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273: 1406–1409 [DOI] [PubMed] [Google Scholar]
- Brownlee C (2002) Role of the extracellular matrix in cell-cell signalling: paracrine paradigms. Curr Opin Plant Biol 5: 396–401 [DOI] [PubMed] [Google Scholar]
- Cao X, Rogers SW, Butler J, Beevers L, Rogers JC (2000) Structural requirements for ligand binding by a probable plant vacuolar sorting receptor. Plant Cell 12: 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli V, Aury JM, Jaillon O, Wincker P, Clepet C, Menard M, Cruaud C, Quetier F, Scarpelli C, Schachter V, et al (2004) Whole genome sequence comparisons and “full-length” cDNA sequences: a combined approach to evaluate and improve Arabidopsis genome annotation. Genome Res 14: 406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Cone K (1989) Yet another rapid plant DNA prep. Maize Genet Coop Newsl 63: 68 [Google Scholar]
- Cui Y, Bi YM, Brugiere N, Arnoldo M, Rothstein SJ (2000) The S locus glycoprotein and the S receptor kinase are sufficient for self-pollen rejection in Brassica. Proc Natl Acad Sci USA 97: 3713–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, et al (2002) Sequence and analysis of rice chromosome 4. Nature 420: 316–320 [DOI] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131: 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL (2003) Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol 3: RESEARCH0029 [DOI] [PMC free article] [PubMed]
- He ZH, Fujiki M, Kohorn BD (1996) A cell wall-associated, receptor-like protein kinase. J Biol Chem 271: 19789–19793 [DOI] [PubMed] [Google Scholar]
- He ZH, He D, Kohorn BD (1998) Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J 14: 55–63 [DOI] [PubMed] [Google Scholar]
- Heldin CH (1995) Dimerization of cell surface receptors in signal transduction. Cell 80: 213–223 [DOI] [PubMed] [Google Scholar]
- Hobe M, Muller R, Grunewald M, Brand U, Simon R (2003) Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol 213: 371–381 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequence Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573 [DOI] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793 [DOI] [PubMed] [Google Scholar]
- Kayes JM, Clark SE (1998) CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125: 3843–3851 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Kohorn BD (2000) Plasma membrane-cell wall contacts. Plant Physiol 124: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD (2001) WAKs; cell wall associated kinases. Curr Opin Cell Biol 13: 529–533 [DOI] [PubMed] [Google Scholar]
- Kuroda S, Tanizawa K (1999) Involvement of epidermal growth factor-like domain of NELL proteins in the novel protein-protein interaction with protein kinase C. Biochem Biophys Res Commun 265: 752–757 [DOI] [PubMed] [Google Scholar]
- Lally D, Ingmire P, Tong HY, He ZH (2001) Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 13: 1317–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li L, Wang X, Xia M, Stolc V, Su N, Peng Z, Li S, Wang J, Wang X, Deng X-W (2005) Tiling microarray analysis of rice chromosome 10 to identify the transcriptome and relate its expression to chromosomal architecture. Genome Biol 6: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijavetzky D, Carbonero P, Vicente-Carbajosa J (2003) Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262: 1432–1436 [DOI] [PubMed] [Google Scholar]
- Meng L, Bregitzer P, Zhang S, Lemaux PG (2003) Methylation of 5′ untranslated exon and intron in Ubi1 promoter complex is correlated with transcriptional transgene silencing in barley. Plant Mol Biol 53: 327–340 [DOI] [PubMed] [Google Scholar]
- Mu JH, Lee HS, Kao TH (1994) Characterization of a pollen-expressed receptor-like kinase gene of Petunia inflata and the activity of its encoded kinase. Plant Cell 6: 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Shiba H, Iwano M, Che FS, Watanabe M, Isogai A, Takayama S (2004) A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303: 1516–1519 [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Osato N, Yamada H, Satoh K, Ooka H, Yamamoto M, Suzuki K, Kawai J, Carninci P, Ohtomo Y, Murakami K, et al (2003) Antisense transcripts with rice full-length cDNAs. Genome Biol 5: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park AR, Cho SK, Yun UJ, Jin MY, Lee SH, Sachetto-Martins G, Park OK (2001) Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J Biol Chem 276: 26688–26693 [DOI] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S (1991) Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67: 687–699 [DOI] [PubMed] [Google Scholar]
- Rice Chromosome 10 Sequencing Consortium (2003) In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300: 1566–1569 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, Wu J, Niimura Y, Cheng Z, Nagamura Y, et al (2002) The genome sequence and structure of rice chromosome 1. Nature 420: 312–316 [DOI] [PubMed] [Google Scholar]
- Sharma VK, Ramirez J, Fletcher JC (2003) The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol 51: 415–425 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Sivaguru M, Ezaki B, He Z-H, Tong H, Osawa H, Baluska F, Volkmann D, Matsumoto H (2003) Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol 132: 2256–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwkowski MX, Schaefer G, Akita RW, Lofgren JA, Fitzpatrick VD, Nuijens A, Fendly BM, Cerione RA, Vandlen RL, Carraway KL (1994) Coexpression of erbB2 and erbB3 proteins reconstitutes a high affinity receptor for heregulin. J Biol Chem 269: 14661–14665 [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhe LH, et al (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB (1991) Molecular cloning of a putative receptor protein kinase encoded at the self incompatibility locus of Brassica oleracea. Proc Natl Acad Sci USA 88: 8816–8820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg Y, Muranyi A, Steen C, Thulin E, Drakenberg T, Stenflo J (1999) EGF-like module pair 3–4 in vitamin K-dependent protein S: modulation of calcium affinity of module 4 by module 3, and interaction with factor X. J Mol Biol 293: 653–665 [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K (2000) The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403: 913–916 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE (1999) The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11: 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica JA, Chae L, Tong H, Ingmire P, He ZH (2003) Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol 133: 1732–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica JA, He ZH (2002) The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol 129: 455–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, Kohorn BD (2001) Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13: 303–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Ruan DL, Song WY, Sideris S, Chen L, Pi LY, Zhang S, Zhang Z, Fauquet C, Gaut BS, et al (1998) Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10: 765–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li S, Zhang Y, Zheng H, Xu Z, Ye J, Yu J, Wong GK (2003) Vertebrate gene predictions and the problem of large genes. Nat Rev Genet 4: 741–749 [DOI] [PubMed] [Google Scholar]
- Ware DH, Jaiswal P, Ni J, Yap IV, Pan X, Clark KY, Teytelman L, Schmidt SC, Zhao W, Chang K, et al (2002) Gramene, a tool for grass genomics. Plant Physiol 130: 1606–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17: 32–43 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Schlessinger J (1987) Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry 26: 1443–1451 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Takahashi T, Katom A, Toriim KU, Komeda Y (1998) The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J 15: 301–310 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, et al (2005) The institute for genomic research Osa1 rice genome annotation database. Plant Physiol 138: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.