Abstract
Phytochrome A (phyA) is the primary photoreceptor mediating responses to far-red light. Among the phyA downstream signaling components, Far-red Elongated Hypocotyl 1 (FHY1) is a genetically defined positive regulator of photomorphogenesis in far-red light. Both physiological and genomic characterization of the fhy1 mutants indicated a close functional relationship of FHY1 with phyA. Here, we showed that FHY1 is most abundant in young seedlings grown in darkness and is quickly down-regulated during further seedling development and by light exposure. By using light-insensitive 35S promoter-driven functional β-glucuronidase-FHY1 and green fluorescent protein-FHY1 fusion proteins, we showed that this down-regulation of FHY1 protein abundance by light is largely at posttranscriptional level and most evident in the nuclei. The light-triggered FHY1 protein reduction is primarily mediated through the 26S proteasome-dependent protein degradation. Further, phyA is directly involved in mediating the light-triggered down-regulation of FHY1, and the dark accumulation of FHY1 requires functional pleiotropic Constitutive Photomorphogenic/De-Etiolated/Fusca proteins. Our data indicate that phyA, the 26S proteasome, and the Constitutive Photomorphogenic/De-Etiolated/Fusca proteins are all involved in the light regulation of FHY1 protein abundance during Arabidopsis (Arabidopsis thaliana) seedling development.
Light is one of the most important environmental signals that affect plant growth and development. Plants have evolved a series of photoreceptors to sense all facets of light, such as direction, duration, quantity, and wavelength. Three major classes of photoreceptors have been characterized by the different wavelengths of the light that they perceive: the red (R)/far-red (FR) light (600–750 nm)-absorbing phytochromes, the blue (B)/UV-A light (320–500 nm)-absorbing cryptochromes and phototropins, and the unidentified UV-B light (282–320 nm)-absorbing UV-B receptors (Kendrick and Kronenberg, 1994; Briggs and Olney, 2001; Briggs et al., 2001). In Arabidopsis (Arabidopsis thaliana), there are five phytochromes (phyA to phyE) that are encoded by five distinct gene family members, and they are classified into two groups according to their stability in light (Sharrock and Quail, 1989). PhyA is type I (light-labile) phytochrome, which is most abundant in dark-grown seedlings, and its level drops rapidly upon exposure to R or white light. PhyB to phyE are all type II (light-stable) phytochromes. In light-grown plants, phyB is the most abundant phytochrome and plays a major role (Hirschfeld et al., 1998). In vivo, phytochrome exists in two distinct but reversible forms: the R-light-absorbing form (Pr) and the FR-light-absorbing form (Pfr). The Pfr form is generally considered to be the biological active form, but there is evidence suggesting that the Pr form also has some biological activity (Reed, 1999; Shinomura et al., 2000).
PhyA is the primary photoreceptor responsible for the very-low-fluence response and the high-irradiance response to continuous FR (cFR) light. Mutants for several signaling components, which are affected in the high-irradiance response branch of phyA pathways, have been identified, including fhy1, fhy3, spa1, fin2, far1, pat1, laf1, fin219, eid1, and hfr1 (Whitelam et al., 1993; Hoecker et al., 1998, 1999; Soh et al., 1998; Hudson et al., 1999; Bolle et al., 2000; Büche et al., 2000; Fairchild et al., 2000; Hsieh et al., 2000; Ballesteros et al., 2001). Although many of them have been cloned, their specific functional roles in phyA pathway and relationship with phyA are largely unknown. Among these intermediates, SPA1 and EID1 are negative regulators, while the others are all positive regulators. The fhy1 mutants, together with fhy3 and far1 mutants, exhibit the strongest elongated hypocotyl in FR light (Whitelam et al., 1993; Barnes et al., 1996; Hudson et al., 1999; Wang and Deng, 2002). Thus, those three loci may define components that play major roles in phyA-mediated FR-light control of seedling development. An examination of effect of known phyA signaling mutants on FR-light control of genome expression profiles in Arabidopsis indicated that mutation in Far-red Elongated Hypocotyl 1 (FHY1) resulted in genome expression deficiency that was most similar to that of phyA mutants (Wang et al., 2002). Additionally, it has been shown that FHY1, but not FAR1 and FHY3, is also involved in mediating the very-low-fluence response signaling (Yanovsky et al., 1997). Therefore, FHY1 might act closest to phyA in phyA-mediated signal transduction among all the available FR-light-signaling components examined (Wang et al., 2002).
At the cellular level, phytochromes (including phyA) are synthesized and exist in the cytosol in the Pr form in the absence of light, and migrate to the nucleus upon irradiation with R and FR light (Kircher et al., 1999, 2002; Yamaguchi et al., 1999). Accordingly, possible interactive partners of phyA have been reported to have both nuclear (PIF3 [Ni et al., 1998], NDPK2 [Choi et al., 1999], PIF1 [Huq et al., 2004]) and cytoplasmic (PKS1 [Fankhauser et al., 1999], NDPK2 [Choi et al., 1999]) localizations. Interestingly, it appears that loss-of-function mutants of these biochemically defined phyA partners all exhibited only subsets of phyA pathway-deficient phenotypes in photomorphogenic development. Thus, it is of great interest to find out what is the molecular role of those genetically defined phyA-signaling components such as FHY1, whose mutations result in pleiotropic defects in phyA-mediated responses.
It has become increasingly clear that regulated proteolysis, especially ubiquitin/proteasome-mediated protein degradation, plays important roles in regulating phytochrome-mediated signaling pathways. Early on, it was clear that phyA abundance accumulated to high levels in darkness and was rapidly reduced upon exposure to light (Quail et al., 1995). This light-triggered rapid decline of phyA was attributed largely to the 26S proteasome-mediated protein degradation (Jabben et al., 1989; Somers and Quail, 1995; Clough and Vierstra, 1997; Cantón and Quail, 1999; Clough et al., 1999; Nagy and Schäfer, 2002; Sharrock and Clack, 2002). EID1, a negative regulator of phyA pathway, encodes an F-box protein, which is a subunit of SCF-type E3 ubiquitin ligase (Dieterle et al., 2001). HY5, LAF1, and HFR1, transcription factors that promote phyA signaling, are ubiquitinated and down-regulated by Constitutive Photomorphogenic 1 (COP1), a RING family E3 ubiquitin ligase, whose activity is enhanced by SPA1, another negative regulator of phyA pathway (Osterlund et al., 2000; Saijo et al., 2003; Seo et al., 2003; Duek et al., 2004; Jang et al., 2005; Yang et al., 2005). Further, phyA itself has been reported to be a target of the COP1 E3 ubiquitin ligase activity (Seo et al., 2004). More recently, the R-light-signaling repressor PIF3 was also shown to be regulated by protein degradation (Bauer et al., 2004).
FHY1 has been genetically characterized as a positive signal transducer specific for the phyA pathway (Desnos et al., 2001; Zeidler et al., 2004). FHY1 gene encodes a predicted polypeptide of approximately 23 kD (202 amino acids; Desnos et al., 2001). However, little is known as to how FHY1 protein acts in phyA signaling at the protein level. In this study, we carried out the cell biological and biochemical characterization of the FHY1 protein in Arabidopsis. We showed that FHY1 exists in both cytosol and nucleus, with clear enrichment in the nucleus. We further demonstrated that FHY1 protein level is regulated by light and phyA through the 26S proteasome-mediated degradation, and Constitutive Photomorphogenic/De-Etiolated/Fusca (COP/DET/FUS) proteins are required for the dark accumulation of FHY1 protein.
RESULTS
FHY1 Protein Level Is Regulated by Light and Developmental Cues
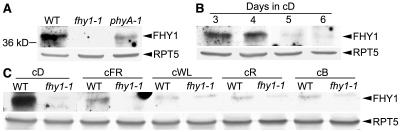
To examine the endogenous FHY1 protein, we raised rabbit polyclonal antibody (anti-FHY1) against a His-tagged recombinant FHY1 protein. Protein gel-blot analysis of total protein extracts from dark-grown seedlings revealed that anti-FHY1 antibody is able to detect a protein band migrating at the size of around 40 kD (Fig. 1A). This apparent size is larger than FHY1's predicted size of 23 kD. Nevertheless, the presence of this band in wild type and the phyA-1 mutant seedlings but not in the fhy1-1 mutant seedlings supports that it is indeed the endogenous Arabidopsis FHY1 protein. It is interesting to note that FHY1 protein has a lower abundance in the dark-grown phyA-1 mutant than in the wild type (Fig. 1A), implying a dependence of optimal FHY1 accumulation on functional phyA photoreceptor in darkness.
Figure 1.
Monitoring FHY1 protein at various developmental time points and in different light conditions by anti-FHY1 antibodies. A, Protein extracts prepared from 3-d-old dark-grown wild-type (WT), fhy1-1, and phyA-1 seedlings were subjected to protein-blot analysis with anti-FHY1 and anti-RPT5 antibodies. B, Protein samples were prepared from dark-grown (cD) wild-type seedlings of 3 to 6 d old, and blotted with anti-FHY1 and anti-RPT5 antibodies. C, Protein samples were extracted from 3-d-old wild-type (WT) and fhy1-1 seedlings grown in continuous dark (cD), FR light (cFR), white light (cWL), R light (cR), and B light (cB), and subjected to protein-blot analysis with anti-FHY1 and anti-RPT5 antibodies. The faint protein band in the anti-FHY1 blot of both wild type and fhy1-1 mutant in the three visible light conditions, whose size is slightly larger than FHY1, is presumably an unspecific band that is detected by anti-FHY1 antibody. In A to C, RPT5 was used as a loading control.
With our FHY1 antibodies, we examined FHY1 protein levels at different time points during Arabidopsis seedling development. The FHY1 protein abundance reaches its highest level in 3- to 4-d-old dark-grown seedlings and then decreases rapidly (Fig. 1B). In extracts from older tissues, the FHY1 protein is hardly detectable (data not shown). The high abundance of FHY1 during early seedling development is consistent temporally with an important role of FHY1 in seedling photomorphogenesis and the absence of fhy1 mutant phenotype in later developmental stages. Interestingly, as shown in Figure 1C, the FHY1 protein level is significantly reduced in light conditions. The FHY1 protein is most abundant in dark-grown seedlings, and its level drops about 10-fold in FR light. In B, R, and white light, FHY1 protein is hardly detectable. The reported FHY1 mRNA level decrease in light conditions (Desnos et al., 2001) coincides with these FHY1 protein level changes.
Generation of Arabidopsis Stable Transgenic Lines with Functional FHY1 Reporter Fusion Proteins
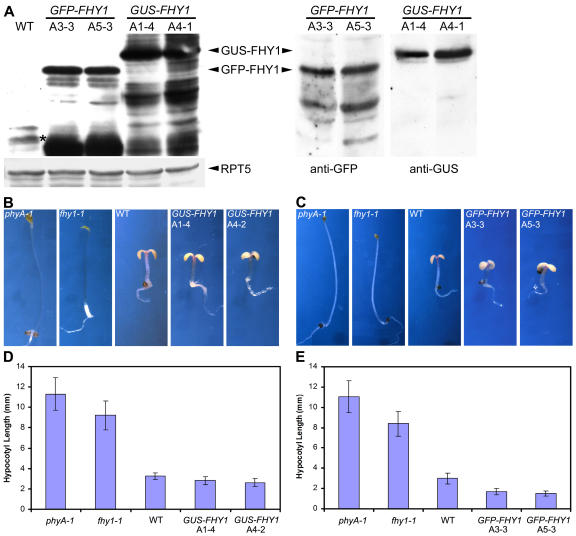
To examine the contribution of transcription regulation of FHY1 toward observed FHY1 abundance regulation by light and the subcellular location of this regulation, we introduced cauliflower mosaic virus 35S promoter-driven full-length FHY1 cDNA fused with either green fluorescent protein (GFP) or β-glucuronidase (GUS) into the fhy1-1 mutant background. Homozygous progeny carrying a single-locus transgene from at least three independent lines were selected for further studies. The expression of the fusion proteins in the transgenic lines was confirmed by protein gel-blot analyses with FHY1 and GFP or GUS antibodies, with two representative lines each shown in Figure 2A. Evidently, both 35S:GFP-FHY1 and 35S:GUS-FHY1 transgenes can rescue the fhy1-1 mutant phenotype in FR condition (Fig. 2, B–E), suggesting that the two FHY1 fusion proteins are functional in vivo. Moreover, FR-grown 35S:GFP-FHY1 transgenic seedlings (in fhy1-1 mutant background) exhibited significantly shorter hypocotyls than wild type (Fig. 2, C and E), indicating that a possible higher level of GFP-FHY1 conferred by the strong 35S promoter enhanced the FR responses in these transgenic plants. Protein-blot analysis of the GFP-FHY1 transgenic lines shown above indicated a higher level of the fusion protein than that of the endogenous FHY1 (Fig. 2A). It is interesting to note that both GFP-FHY1 and GUS-FHY1 protein levels are much higher than endogenous FHY1 protein level (Fig. 2A). However, only 35S:GFP-FHY1 transgenic plants showed increased response to FR light (Fig. 2, B–E). Thus, the GUS fusion might interfere with the function of FHY1 so that the GUS-FHY1 protein is less active than the GFP-FHY1 protein.
Figure 2.
Characterization of Arabidopsis FHY1 fusion protein overexpression lines. A, Protein extracts were prepared from dark-grown wild-type, 35S:GFP-FHY1, and 35S:GUS-FHY1 transgenic seedlings, and blotted with anti-FHY1, anti-RPT5, anti-GFP, and anti-GUS antibodies. The asterisk marks the endogenous FHY1 protein band on the left. RPT5 was used as a loading control. B and C, Morphological comparison of fhy1-1 mutant seedlings expressing GUS-FHY1 (B) or GFP-FHY1 (C) with wild type, phyA-1, and fhy1-1 mutants. The fhy1-1 mutant phenotypes in FR light are rescued by 35S:GUS-FHY1 and 35S:GFP-FHY1 transgenes, respectively. All pictures were taken under the same magnification. D and E, Quantitative analysis of hypocotyl length of transgenic lines, 35S:GUS-FHY1 (D) or 35S:GFP-FHY1 (E), compared with phyA-1 and fhy1-1 mutants and wild-type controls.
FHY1 Is Localized in Both Cytoplasm and Nucleus, with Nuclear Enrichment in Darkness
The intracellular localization of FHY1 was investigated using at least three independent transgenic lines each for both GFP and GUS fusion proteins. The subcellular distribution of GUS-FHY1 and GFP-FHY1 fusion proteins in all lines examined exhibited essentially the same localization pattern. As shown in Figure 3, A and B, GUS staining or GFP fluorescence was detected intensely in the nucleus and also weakly in the cytoplasm. The nuclear signals were most strong in darkness and evidently reduced in all light conditions examined, especially in the case of GFP-FHY1 (Fig. 3, A, B, and D). These results are consistent with the previous reports (Desnos et al., 2001; Zeidler et al. 2004). Although light exposure causes a decrease in the ratio of nuclear FHY1 versus cytoplasmic FHY1, this is likely due to a selective degradation of the nuclear FHY1 triggered by light, which will be further addressed in later sections.
Figure 3.
Subcellular localization and abundance of GUS-FHY1 and GFP-FHY1 fusion proteins in Arabidopsis seedlings under different light conditions. A and B, 4-d-old wild type and 35S:GUS-FHY1 (A) or 35S:GFP-FHY1 (B) transgenic Arabidopsis seedlings grown in different light conditions were monitored for GUS staining or viewed directly by fluorescence microscope. The left portions of each section are the GUS staining of GUS-FHY1 or GFP fluorescence of GFP-FHY1, while the right portions are DAPI staining of the corresponding images on the left. The positions of the nuclei are indicated by arrows. Bars = 10 μm. C, Protein samples were extracted from 3-d-old 35S:GFP-FHY1 transgenic Arabidopsis seedlings grown in continuous dark (cD), FR light (cFR), white light (cWL), R light (cR), and B light (cB), and subjected to protein-blot analysis with anti-FHY1 and anti-RPT5 antibodies. RPT5 was used as a loading control. D, Subcellular localization of GFP-FHY1 protein in hypocotyl cells of 4-d-old 35S:GFP-FHY1 transgenic Arabidopsis seedlings grown under different light conditions as indicated on the top. Wild-type Arabidopsis seedlings were used as negative control. Photographs of seedlings were taken at the same magnification. Bar = 100 μm.
FHY1 Protein Level Regulation by Light Is Largely at Posttranscriptional Level
The reported FHY1 mRNA level decrease in light conditions (Desnos et al., 2001) coincides with these FHY1 protein level changes. To further define the specific contribution of transcriptional and posttranscriptional regulation toward FHY1 abundance by light, GFP-FHY1 fusion protein driven by the light-insensitive 35S promoter (35S:GFP-FHY1) was analyzed. As shown in Figure 3C, GFP-FHY1 fusion protein driven by the light-insensitive 35S promoter (35S:GFP-FHY1) also exhibited similar light-regulated abundance changes in transgenic seedlings. Fluorescence microscopic examination revealed that the reduction of GFP-FHY1 level is most drastic in its nuclear abundance (Fig. 3D). On the other hand, due to unknown reasons, the stability of GUS-FHY1 protein is less sensitive to light than endogenous FHY1 or GFP-FHY1 (data not shown), which resulted in better-retained nuclear signals of GUS-FHY1 than GFP-FHY1 in light conditions (Fig. 3A). Together with previous studies, our results suggest that the FHY1 protein accumulation level is the result of coordinated transcriptional and posttranscriptional regulations, which is similar to the phyA protein (Quail et al., 1995).
The Proper Light Regulation of FHY1 Abundance Requires PhyA
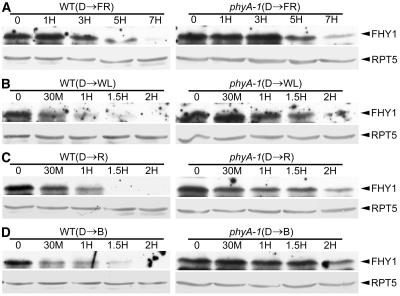
To assess the role of phyA in the regulation of FHY1 protein level, we examined FHY1 protein levels during dark-to-light transitions. Wild-type and phyA-1 mutant seedlings were grown in complete darkness for 3 d and then treated with cFR light for 1, 3, 5, and 7 h (Fig. 4A), or continuous white, B, and R light for 30 min, and 1, 1.5, and 2 h (Fig. 4, B–D). In wild-type seedlings, endogenous FHY1 decreased rapidly after exposure to white, B, and R light. After just 1 h, most of the FHY1 protein had disappeared. When exposed to FR light, FHY1 protein level also showed a decrease, although less dramatic and slower than those in other light conditions. On the other hand, in phyA-1 mutant seedlings, this reduction of the FHY1 protein was slowed down significantly in all the light conditions tested. However, FHY1 was eventually degraded with extended time under all those light conditions. This result indicates that phyA plays a role in this light-mediated FHY1 protein reduction, but other photoreceptors are likely involved as well.
Figure 4.
FHY1 protein level is decreased during dark-light transitions. A, Wild type (WT) and phyA-1 seedlings were grown in dark (D) for 3 d first, and then transferred to FR light. Protein samples were collected at different time points starting from the transfer (0, 1, 3, 5, and 7 h), and blotted with anti-FHY1 and anti-RPT5 antibodies. B to D, Wild-type (WT) and phyA-1 seedlings were grown in dark (D) for 3 d first, and then transferred to white light (WL), R, and B light. Protein samples were collected at different time points starting from the transfer (0, 30 min, 1, 1.5, and 2 h), and blotted with anti-FHY1 and anti-RPT5 antibodies. In A to D, RPT5 was used as a loading control.
FHY1 Protein Is Degraded through the 26S Proteasome
To examine the mechanism responsible for the decrease of the FHY1 protein upon light treatment, we first tested the effect of 26S proteasome-specific inhibitors, MG132, PSI, and ALLN, on FHY1 protein abundance changes during the transition of 3-d-old dark-grown seedlings to white light. As shown in Figure 5, each of these inhibitors was able to efficiently inhibit the decrease of the FHY1 protein. This result indicates that the FHY1 protein is likely degraded through the 26S proteasome pathway upon light exposure.
Figure 5.
Degradation of endogenous FHY1 protein is largely mediated by the 26S proteasome pathway. A to D, The same degradation assay as described in Figure 4B was carried out in wild-type seedlings treated by DMSO (solvent for the inhibitors) only, or by DMSO supplemented with 40 μm 26S proteasome-specific inhibitor MG132, ALLN, or PSI, respectively. In A to D, RPT5 was used as a loading control.
To further assess the involvement of 26S proteasome-mediated protein degradation toward FHY1 protein abundance regulation, we checked the GFP-FHY1 protein level changes and the effect of proteasome inhibitors in the similar dark-to-light transitions. As the expression of GFP-FHY1 fusion protein was driven by a light-insensitive 35S promoter, the contribution of possible transcriptional regulation by light toward the fusion protein should be minimal. As shown in Figure 6, the GFP-FHY1 fusion protein exhibited a similar degradation pattern to endogenous FHY1 during the dark-to-light transitions. Further, the degradation of GFP-FHY1 upon light exposure can be prevented by each of the three distinct 26S proteasome-specific inhibitors (Fig. 7, A–D). Fluorescence microscopic examination confirmed that MG132, but not dimethyl sulfoxide (DMSO), ethanol, or cycloheximide (CHX, an inhibitor of de novo protein biosynthesis), effectively blocked GFP-FHY1 degradation in the nucleus (Fig. 7E). These results further confirm that the light-triggered reduction of the FHY1 protein in planta is largely due to the 26S proteasome-mediated protein degradation.
Figure 6.
GFP-FHY1 fusion protein is degraded similarly during dark-light transitions. A, 35S:GFP-FHY1 transgenic Arabidopsis seedlings were grown in dark (D) for 3 d first, and then transferred to FR light. Protein samples were collected at different time points starting from the transfer (0, 1, 3, and 5 h), and blotted with anti-FHY1 and anti-RPT5 antibodies. B to D, 35S:GFP-FHY1 transgenic Arabidopsis seedlings were grown in dark (D) for 3 d first, and then transferred to white light (WL), R, and B light. Protein samples were collected at different time points starting from the transfer (0, 30 min, 1, and 2 h), and blotted with anti-FHY1 and anti-RPT5 antibodies. In A to D, RPT5 was used as a loading control.
Figure 7.
Degradation of GFP-FHY1 fusion protein is mediated by the 26S proteasome pathway. A to D, The same degradation assay as described in Figure 6B was performed in 35S:GFP-FHY1 transgenic seedlings treated by DMSO only, or by DMSO supplemented with 40 μm 26S proteasome-specific inhibitor MG132, ALLN, or PSI, respectively. In A to D, RPT5 was used as a loading control. E, Fluorescence images of hypocotyl cells of 35S:GFP-FHY1 transgenic Arabidopsis seedlings. Seedlings were grown in darkness for 4 d and then either treated with 100 μm MG132 or mock-treated with DMSO, or treated with 100 μm CHX or mock-treated with ethanol (ethanol), or treated with a mixture of MG132 and CHX. The seedlings were then kept in white light for 3 h before fluorescence observation. Representative photographs were taken at the same magnification. Bar = 100 μm.
PhyA Is the Primary Photoreceptor Mediating FR-Light Regulation of FHY1 Abundance
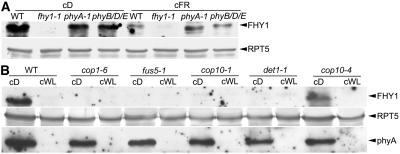
We further examined the role of phyA as well as other phytochromes in mediating light regulation of FHY1 protein abundance. As shown in Figure 8A, phyA-1 and phyB/D/E triple mutants had lower FHY1 protein levels in their dark-grown seedlings compared to wild-type seedlings, which suggests that optimal FHY1 accumulation in darkness requires phyA and other phytochromes (see also Fig. 3A). However, the FR-light-grown phyA-1 mutants were more similar to their dark-grown seedlings and exhibited higher abundance of FHY1 than both wild type and phyB/D/E (Fig. 8A). This result not only supports the early observation that phyA negatively regulates FHY1 in light, but also indicates that phyA is the primary photoreceptor that mediates FR-light-dependent regulation of FHY1 protein abundance. Other phytochromes, such as phyB, phyD, and phyE, do not contribute significantly to the regulation of FHY1 abundance in FR light.
Figure 8.
FHY1 protein levels in different photomorphogenic mutants. A, Protein extracts of 3-d-old wild-type (WT), fhy1-1, phyA-1, and phyB/D/E seedlings grown in continuous dark (cD) and FR light (cFR) were subjected to protein-blot analysis with anti-FHY1 and anti-RPT5 antibodies. B, Comparison of FHY1 and phyA protein levels in wild type (WT) and various cop/det/fus mutants in continuous white light (cWL) and dark (cD) conditions. Protein extracts from 3-d-old seedlings of wild type (WT), cop1-6, fus5-1, cop10-1, det1-1, and cop10-4 grown in cD and cWL were prepared and blotted with anti-FHY1, anti-phyA, and anti-RPT5 antibodies. In A and B, RPT5 was used as a loading control.
The Pleiotropic COP/DET/FUS Proteins Play Positive Roles in the Dark Accumulation of FHY1 Protein
The COP/DET/FUS proteins play important roles in mediating the repression of photomorphogenic development in darkness (Deng et al., 1991; Kwok et al., 1996). They accomplish this by targeting photomorphogenesis-promoting transcription factors (such as HY5) for degradation via the 26S proteasome (Osterlund et al., 2000; Saijo et al., 2003; Seo et al., 2003; Yanagawa et al., 2004). To examine a possible involvement of those proteins in light regulation of FHY1 protein abundance, we analyzed FHY1 protein levels in different cop/det/fus mutants. Protein extracts from 3-d-old cop1-6, fus5-1, cop10-1, det1-1, cop10-4 mutants, and wild type seedlings grown in both white-light and dark conditions were prepared and blotted with FHY1 and phyA antibodies. Except cop10-4, which is a very weak allele of COP10 (Suzuki et al., 2002), all the cop/det/fus mutants exhibited similarly reduced FHY1 protein levels in both light and dark conditions, while phyA protein levels were not significantly affected by the cop/det/fus mutations (Fig. 8B). This lack of effect on phyA protein level in the cop/det/fus mutants is similar to that of a previous study (Seo et al., 2004). It suggests that the accumulation of FHY1 protein in darkness requires the presence of functional COP/DET/FUS proteins. This is consistent with the cop/det/fus mutant phenotype, which essentially mimics light-grown seedlings when grown in darkness (Kwok et al., 1996). By contrast, the cop10-4 mutant behaves like wild type in 3- to 4-d-old seedlings, and only displays very weak cop phenotype after about 8 d growth in darkness (Suzuki et al., 2002). Thus, it is understandable why FHY1 protein level in the 3-d-old cop10-4 mutant seedlings is similar to wild type. Therefore, the COP/DET/FUS proteins are essential for the dark accumulation of FHY1 but not phyA protein in planta.
DISCUSSION
Previous studies have revealed that expression of both PHYA and FHY1 genes is negatively regulated at transcriptional level by light exposure (Somers and Quail, 1995; Cantón and Quail, 1999; Zeidler et al., 2001; Desnos et al., 2001). It was shown for the regulation of phyA that both transcriptional and posttranscriptional mechanisms are involved (Quail et al., 1995). The phyA protein is most abundant in dark-grown seedlings, and its level drops up to 100-fold after exposure to light. One major control is the phyA protein degradation through ubiquitination pathway triggered by light (Jabben et al., 1989; Clough et al., 1999). Our results (Figs. 1, 3, 4, and 6), together with prior studies of FHY1 transcriptional regulation (Desnos et al., 2001; Zeidler et al., 2001), suggested that the FHY1 protein level is also negatively regulated by light, through both mRNA abundance and protein stability regulations.
We further showed that the decrease of endogenous FHY1 as well as GFP-FHY1 fusion protein could be efficiently prevented by 26S proteasome-specific inhibitors (Figs. 5 and 7). This analysis suggests that the protein stability regulation is likely through the 26S proteasome system, which may primarily be responsible for FHY1 protein abundance regulation. Therefore, the regulation of both phyA and FHY1 protein levels is the result of coordinated transcriptional and posttranscriptional regulations, and in both cases the protein degradation may play the dominant role in Arabidopsis. The similarity in the light regulation of phyA and FHY1 abundance would be consistent with a close functional relationship of the two in the FR-light signaling as suggested by both phenotypic and genomic analyses (Fig. 2; Wang et al., 2002).
Degradation of phyA itself (Jabben et al., 1989; Clough et al., 1999), together with the isolation of EID1 (a gene encoding an F-box protein; Dieterle et al., 2001) and the identification of SPA1 as a cofactor in COP1-mediated degradation of LAF1 and HY5 (Saijo et al., 2003; Seo et al., 2003), has suggested a critical role for the proteasome-mediated protein degradation in the phyA-dependent FR-light signal transduction. It is interesting to point out that a phyB signaling component, PIF3, is also sensitively regulated by 26S proteasome-mediated protein degradation (Bauer et al., 2004). Similar to our observation for FHY1, it is demonstrated that COP1 is required for the high-level accumulation of PIF3 in the dark (Bauer et al., 2004). Here, we showed a similar role of COP1 as well as other related COP/DET/FUS proteins in the regulation of FHY1 protein levels (Fig. 8). However, PIF3 acts negatively in R-light signaling and is not involved in FR-light signaling regulating hypocotyl elongation (Bauer et al., 2004), while FHY1 is a positive component in phyA-mediated FR-light pathway. Therefore, it is likely that light-promoted degradation of FHY1 might serve as a desensitizing mechanism of phyA signaling. In this regard, it is interesting to note that COP1 has been suggested to act as an E3 ubiquitin ligase for targeted degradation of phyA (Seo et al., 2004). The requirement of functional COP1 and other COP/DET/FUS proteins for dark accumulation of FHY1 suggests that these COP proteins might be involved in repressing the function of a negative regulator(s) of FHY1 accumulation in darkness. Nevertheless, since FHY1 seems to highly accumulate only in etiolated seedlings, the effect of the COP proteins on FHY1 could also be indirectly mediated through their repression of de-etiolation in darkness. Although COP1 has been demonstrated as a ubiquitin E3 ligase involved in the proteasome-mediated degradation of several proteins such as HY5, LAF1, phyA, and HFR1 (Saijo et al., 2003; Seo et al., 2003, 2004; Duek et al., 2004; Jang et al., 2005; Yang et al., 2005), whether or not COP1 can act as a ubiquitin E3 ligase of FHY1 protein under light conditions still needs further investigation. Meanwhile, COP1's role of promoting FHY1 accumulation in darkness does not rule out a possible involvement of COP1 in the light-triggered degradation of FHY1 protein, because different mechanisms might be involved in the control of FHY1 protein level in darkness and in light conditions.
MATERIALS AND METHODS
Plant Materials
The wild type Arabidopsis (Arabidopsis thaliana) used in this study were of the Landsberg erecta ecotype, unless otherwise indicated. The phyA-1 (Whitelam et al., 1993), fhy1-1 (Desnos et al., 2001), phyB/D/E (Franklin et al., 2003), cop1-6 (McNellis et al., 1994), fus5-1 (Karniol et al., 1999), det1-1 (Chory et al., 1989), cop10-1 (Suzuki et al., 2002), and cop10-4 (Suzuki et al., 2002) mutants have been described previously.
Growth Conditions
To grow Arabidopsis seedlings, seeds were surface sterilized, plated on Murashige and Skoog (Gibco) medium containing 0.3% or 1% Suc, and incubated at 4°C for 3 to 5 d before being placed in a standard continuous-white-light growth chamber at 22°C. After 12 h of incubation, plates were transferred to corresponding light conditions or complete darkness for 3 to 5 d. The fluence rates of the light growth chambers (Percival Scientific) were 111.0 μmol m−2 s−1 for FR light, 150.6 μmol m−2 s−1 for white light, 172.6 μmol m−2 s−1 for R light, and 8.1 μmol m−2 s−1 for B light. To obtain adult plants, 7- to 9-d-old seedlings were transferred to soil and grown in a standard long-day (16 h light/8 h darkness) growth room.
For the experiments testing 26S proteasome-specific inhibitors and protein synthesis inhibitors, MG132, ALLN, and PSI were dissolved in DMSO, and CHX was dissolved in ethanol, and Arabidopsis seedlings were vacuum-infiltrated with these inhibitors for 10 min before being transferred from darkness to white light. The incubation temperature is 22°C.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
The full-length cDNA of FHY1 were amplified by reverse transcription-PCR with forward primer (5′-CTGAATTCGGGATCCCTATGCCTGAAGTGGAAGTGGATAACAACAACGAGAAGCC-3′) and reverse primer (5′-GACTCGAGGTTACAGCATTAGCGTTGAG-3′), and cloned into the pCR2.1-TOPO vector (Invitrogen). This construct served as a PCR template for subsequent cloning of FHY1 into other vectors. A BamHI-SpeI fragment containing FHY1 cDNA was cloned into the BglII-XbaI site of the pRTL2-mGFP (S65T) and pRTL2-GUS/NIa vectors (Torii et al., 1998). For generating 35S:GUS-FHY1 and 35S:GFP-FHY1 binary constructs, a HindIII fragment (containing the 35S promoter, transgene, and 3′ end terminator) was subcloned from the above-mentioned pRTL2 constructs into pPZP221 (Hajdukiewicz et al., 1994).
The 35S:GUS-FHY1 and 35S:GFP-FHY1 constructs were introduced into fhy1-1 mutant, via Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). Transgenic plants were selected with 200 μg/mL Gentamycin (Sigma), and more than 20 independent lines for each construct were generated. At least three independent T3 lines with single T-DNA locus were used for detailed molecular and biochemical analyses.
Cellular Localization Studies
Four-day-old 35S:GUS-FHY1 transgenic seedlings were selected and stained as previously described (von Arnim et al., 1997). The staining time varied depending on the light condition that the seedlings were grown in. All stained seedlings were fixed for 30 min in 3.7% formaldehyde. After bleaching overnight with 70% ethanol, the seedlings were rehydrated with a graded ethanol series before being mounted on slides in 1 mg/L 4′,6-diamino-phenylindole (DAPI), and viewed under a microscope (Zeiss).
For subcellular localization of GFP-FHY1 protein in the 35S:GFP-FHY1 transgenic Arabidopsis, 4-d-old whole seedlings were mounted on slides in 1 mg/L DAPI and viewed under a fluorescence microscope with GFP filter sets (Zeiss).
Representative photographs were taken using a digital camera (Zeiss), and the figures were assembled using Adobe Photoshop software (Adobe Systems).
Protein-Blot Analysis
Arabidopsis tissues were homogenized in extraction buffer A containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10 mm MgCl2, 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor (Roche), or extraction buffer B containing 50 mm Tris-HCl (pH 7.5), 200 mm NaCl, 1 mm EDTA, 10 mm NaF, 2 mm Na3VO4, 25 mm β-glycerolphosphate, 10% glycerol, 0.1% Tween 20, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor (Roche). The extracts were centrifuged twice at 4°C for 15 min each, and the protein concentration in the supernatant was determined by Bradford assay (Bio-Rad). Protein samples were boiled in sample buffer, run on SDS-PAGE gels, and blotted onto polyvinylidene difluoride membranes (Millipore). The protein-blot analysis with indicated primary and secondary antibodies is essentially following previously described protocol (Feng et al., 2003). All protein-blot analyses were repeated at least three times to ensure the reproducibility of the results presented in this article.
FHY1 Antibodies Production and Other Antibodies Used
An EcoRI/NotI fragment containing the full-length FHY1 open reading frame was cloned into pET-28a (Novagen). This construct encodes a fusion protein with 6× His tags and FHY1. The fusion protein was expressed in Escherichia coli and purified with nickel-nitrilotriacetic acid beads (Qiagen). Polyclonal antibodies were raised by immunizing rabbits using purified fusion protein as antigen. The EcoRI/NotI fragment containing FHY1 was also cloned into pGEX-4T1 (Amersham Biosciences), generating a construct that encodes GST-FHY1 fusion protein. The GST-FHY1 fusion protein was expressed in E. coli and purified with Glutathione beads (Amersham Biosciences). Polyclonal FHY1 antibodies were then purified using HITRAP N-hydroxysuccinimide-activated column (Amersham Biosciences) coupled with GST-FHY1.
Other primary antibodies used in this study include anti-RPT5 (Kwok et al., 1999), anti-phyA (Xu et al., 1995), anti-GFP (Clontech), and anti-GUS (Sigma).
Acknowledgments
We are grateful to Peter Quail for anti-phyA antibody and to Jessica Habashi for critical commenting on the manuscript.
This work was supported by the National Institutes of Health (grant no. GM47850 to X.W.D.), the National Science Foundation of China (strategic international cooperation project grant no. 30221120261), the National Institute of Biological Sciences at Beijing, and a Boyce Thompson Institute start-up fund (to H.W.). Y.S. was a Peking-Yale Center Monsanto fellow, and L.M. was a long-term fellow of the Human Frontier Science Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.067645.
References
- Ballesteros M, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Quaggio RB, Whitelam GC, Chua NH (1996) fhy1 defines a branch point in phytochrome A signal transduction pathways for gene expression. Plant J 10: 1155–1161 [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KCS, Ádám É, Fejes E, Schäfer E, et al (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua NH (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14: 1269–1278 [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Beck CF, Cashmore AR, Christie JM, Hughes J, Jarillo JA, Kagawa T, Kanegae H, Liscum E, Nagatani A, et al (2001) The phototropin family of photoreceptors. Plant Cell 13: 993–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Olney MA (2001) Photoreceptors in plant photomorphogenesis to date: five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol 125: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büche C, Poppe C, Schäfer E, Kretsch T (2000) eid1: a new Arabidopsis mutant hypersensitive in Phytochrome A-dependent high-irradiance responses. Plant Cell 12: 547–558 [PMC free article] [PubMed] [Google Scholar]
- Cantón FR, Quail PH (1999) Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol 121: 1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka S, Hahn TR, Song PS (1999) Phytochrome signaling is mediated through nucleoside diphosphate kinase 2. Nature 401: 610–613 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F (1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991–999 [DOI] [PubMed] [Google Scholar]
- Clough RC, Jordan-Beebe ET, Lohman KN, Marita JM, Walker JM, Gatz C, Vierstra RD (1999) Sequences within the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J 17: 155–167 [DOI] [PubMed] [Google Scholar]
- Clough RC, Vierstra RD (1997) Phytochrome degradation. Plant Cell Environ 20: 713–721 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deng XW, Caspar T, Quail PH (1991) cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev 5: 1172–1182 [DOI] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15: 2980–299011711433 [Google Scholar]
- Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541 [DOI] [PubMed] [Google Scholar]
- Feng S, Ma L, Wang X, Xie D, Dinesh-Kumar SP, Wei N, Deng XW (2003) The COP9 signalosome interacts physically with SCFCOI1 and modulates jasmonate responses. Plant Cell 15: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC (2003) Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131: 1340–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA (1998) Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH (1999) SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496–499 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH (1998) SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10: 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh SL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14: 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH (1999) The FAR1 locus encodes a novel nuclear protein specific to Phytochrome A signaling. Genes Dev 13: 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Jabben M, Shanklin J, Vierstra RD (1989) Ubiquitin-phytochrome conjugates: pool dynamics during in vivo phytochrome degradation. J Biol Chem 264: 4998–5005 [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol B, Malec P, Chamovitz DA (1999) Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of the COP9 complex. Plant Cell 11: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM (1994) Photomorphogenesis in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Ádám E, Schäfer E, Nagy F (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bogner L, Kim L, Adam E, Harter K, Schäfer E, Nagy F (1999) Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Piekos B, Miséra S, Deng XW (1996) A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol 110: 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Staub JM, Deng XW (1999) Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J Mol Biol 285: 85–95 [DOI] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng XW (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Schäfer E (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol 53: 329–355 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D (1995) Phytochromes: photosensory perception and signal transduction. Science 268: 675–680 [DOI] [PubMed] [Google Scholar]
- Reed JW (1999) Phytochromes are Pr-ipatetic kinases. Curr Opin Plant Biol 2: 393–397 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang J, Ishikawa M, Bolle B, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Sharrock RA, Clack T (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol 130: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3: 1745–1757 [DOI] [PubMed] [Google Scholar]
- Shinomura T, Uchida K, Furuya M (2000) Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol 122: 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG (1998) Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J 16: 411–419 [DOI] [PubMed] [Google Scholar]
- Somers DE, Quail PH (1995) Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J 7: 413–427 [DOI] [PubMed] [Google Scholar]
- Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng XW (2002) Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev 16: 554–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, McNellis TW, Deng XW (1998) Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J 17: 5577–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Osterlund MT, Kwok SF, Deng XW (1997) Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol 114: 779–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ma L, Habashi J, Zhao H, Deng XW (2002) Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J 32: 723–733 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Parks BM, Short TW, Quail PH (1995) Missense mutations define a restricted segment in the C-terminal domain of phytochrome A critical to its regulatory activity. Plant Cell 7: 1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagtani A (1999) Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol 3: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa Y, Sullivan JA, Komatsu S, Gusmaroli G, Suzuki G, Yin J, Ishibashi T, Saijo Y, Rubio V, Kimura S, et al (2004) Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Casal JJ, Luppi JP (1997) The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J 12: 659–667 [DOI] [PubMed] [Google Scholar]
- Zeidler M, Bolle C, Chua NH (2001) The phytochrome A specific component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol 42: 1193–1200 [DOI] [PubMed] [Google Scholar]
- Zeidler M, Zhou Q, Sarda X, Yau CP, Chua NH (2004) The nuclear localization signal and the C-terminal region of FHY1 are required for transmission of phytochrome A signals. Plant J 40: 355–365 [DOI] [PubMed] [Google Scholar]