Abstract
The monogenic recessive maize (Zea mays) mutant rootless with undetectable meristems 1 (rum1) is deficient in the initiation of the embryonic seminal roots and the postembryonic lateral roots at the primary root. Lateral root initiation at the shoot-borne roots and development of the aerial parts of the mutant rum1 are not affected. The mutant rum1 displays severely reduced auxin transport in the primary root and a delayed gravitropic response. Exogenously applied auxin does not induce lateral roots in the primary root of rum1. Lateral roots are initiated in a specific cell type, the pericycle. Cell-type-specific transcriptome profiling of the primary root pericycle 64 h after germination, thus before lateral root initiation, via a combination of laser capture microdissection and subsequent microarray analyses of 12k maize microarray chips revealed 90 genes preferentially expressed in the wild-type pericycle and 73 genes preferentially expressed in the rum1 pericycle (fold change >2; P-value <0.01; estimated false discovery rate of 13.8%). Among the 51 annotated genes predominately expressed in the wild-type pericycle, 19 genes are involved in signal transduction, transcription, and the cell cycle. This analysis defines an array of genes that is active before lateral root initiation and will contribute to the identification of checkpoints involved in lateral root formation downstream of rum1.
Maize (Zea mays) displays a complex root stock architecture composed of different root types formed at different stages of plant development (Hochholdinger et al., 2004a, 2004b). The primary root at the basal pole of the seedling develops from the embryonic radicle that is initiated early during embryogenesis while the seminal roots that develop at the scutellar node in variable numbers are initiated from primordia that can be detected in the mature embryo. Later, shoot-borne crown and brace roots are formed from consecutive underground and aboveground stem nodes. A common feature of all root types is the formation of lateral roots from pericycle cells in the differentiation zone some distance from the root apex (Esau, 1965). The initiation of lateral roots, i.e. the first division of founder cells (Lloret and Casero, 2002), is induced by endogenous signals of the maize plant, such as auxin, that stimulate pericycle cell division (Dubrovsky et al., 2000). In contrast to Arabidopsis (Arabidopsis thaliana), the sites of lateral root initiation cannot be predicted in maize roots (Bell and McCully, 1970).
Few mutants specifically affected in root formation have been described in maize (Hochholdinger et al., 2004a, 2004b). Two of these mutants are impaired in root initiation: the mutant rootless concerning the crown and seminal roots (rtcs) is affected in the initiation of all shoot-borne stem-derived postembryonic roots and in the initiation of the embryonically formed seminal roots (Hetz et al., 1996). The mutant lrt1 is impaired in early postembryonic lateral root initiation at the primary and seminal roots (Hochholdinger and Feix, 1998a). In the lateral rootless1 (lrt1) mutant, lateral root development cannot be restored by exogenous application of auxin. A common feature of all maize root mutants described thus far is that they are affected in specific root types during different developmental phases, hence, defining complex genetic networks involved in maize root formation (Hochholdinger et al., 2004b).
In Arabidopsis, two mutants have been identified that are affected in lateral root initiation. The mutants alf4 (Celenza et al., 1995) and Slr (Fukaki et al., 2002) are both blocked in lateral root initiation while primary root elongation is not affected. In both mutants, lateral root formation cannot be induced by exogenous auxin application, and both mutants showed pleiotropic effects in their aerial parts.
Polar auxin transport is necessary for several aspects of root development, including primary root elongation, gravity response, and lateral root formation (Muday and Haworth, 1994). Auxin is transported in two directions in the primary root. In the phloem parenchyma of the central cylinder, polar auxin transport is directed from the base of the root toward the root tip (acropetal). In the region between the root tip and the elongation zone, auxin is transported away from the root tip (basipetal) in the epidermis (Mitchell and Davies, 1975). Experiments with auxin transport inhibitors in Arabidopsis have demonstrated that the auxin movement from the shoot toward the root tip is responsible for lateral root development (Reed et al., 1998), while auxin movement away from the root tip is required for gravitropism and primary root elongation (Rashotte et al., 2000), although involvement of basipetal auxin transport in lateral root formation has also been discussed recently (Casimiro et al., 2001). Several genes involved in auxin transport, including the postulated efflux carriers of the pin family (Gälweiler et al., 1998; Müller et al., 1998; Friml et al., 2002a, 2002b), the auxin importer aux1 (Marchant et al., 1999), and the postulated efflux carriers pgp1, pgp2, and pgp19 (Noh et al., 2001), have been cloned recently in Arabidopsis. The maize ortholog of pgp1 is not expressed in roots (Multani et al., 2003).
Microarray experiments allow for the monitoring of transcriptional activity of thousands of genes in parallel. Until recently, microarray expression profiling in plants was confined to the analysis of whole organs that are composed of various cell types, each displaying a distinct mRNA expression profile. Such microarray experiments provide average gene expression profiles of all cells in an organ and may potentially mask differential gene expression in a particular cell type. Laser capture microdissection (LCM) technology allows for the analysis of cell-type-specific gene expression profiles (Asano et al., 2002; Kerk et al., 2003; Nakazono et al., 2003). In this approach, frozen or fixed cells of interest are physically linked to a thermoplastic film with a low-power laser beam. After isolation and amplification of RNA from these cells, their corresponding labeled cDNAs can be hybridized to microarray chips (Schnable et al., 2004). In a similar approach, a cell-type-specific gene expression map of the Arabidopsis primary root was generated by a combination of protoplasting and subsequent cell sorting of cell-type-specific marker lines (Birnbaum et al., 2003).
This article describes the isolation and characterization of rootless with undetectable meristems 1 (rum1), a novel maize mutant affected in lateral and seminal root initiation, and a comparative transcriptome analysis of the wild type versus rum1 pericycle using a 12k maize cDNA microarray.
RESULTS
Isolation and Genetic Characterization of the rum1 Mutant
The 1,967 F2 families of Mutator-tagged maize stocks were screened for aberrant root phenotypes with the paper roll test, and a novel mutant, rum1, was isolated. The rum1 mutant completely lacks seminal roots and does not develop lateral roots on the primary root within 10 d after germination (DAG; Fig. 1A). The initial screening of the F2 family that led to the isolation of the mutant rum1 revealed a segregation ratio that was not statistically different than the 3:1 expected for a monogenic recessive mutant (Table I). Wild-type and mutant plants of segregating families were selfed over several generations. Selfed heterozygous wild types are expected to generate wild-type and mutant seedlings in a 3:1 ratio. Segregation data of selfed heterozygous wild-type plants (Table I) are most consistent with a monogenic recessive inheritance of the rum1 gene.
Figure 1.
Phenotypic properties of the root mutant rum1. A, Root system of wild-type (left) and rum1 (right) seedlings 10 DAG. The mutant rum1 does not form lateral and seminal roots. P, Primary root, S, seminal root. B, Three of eight additional alleles of rum1 (11 DAG) generated via direct-tagging. C to H, Detection of meristematic tissue by Feulgen staining (magenta precipitate) in the basal part of the primary root. Left, Wild type; right, rum1. Time course of root development: 3 DAG (C), 4 DAG (D), 5 DAG (E), 6 DAG (F), 7 DAG (G and H). C to G, Developing lateral roots; H, primary root tip. J and K, Close up of the coleoptilar node and second node of wild type (J) and rum1 (K) 30 DAG. The mutant rum1 (K) does not initiate crown roots at the coleoptilar node in most instances. C, coleoptilar node. L and M, Crown roots 30 DAG; lateral roots develop normally on wild type (L) and rum1 (M). N, Wild-type and rum1 root system 30 DAG. Left, Wild type; middle and right, rum1. The presence of crown roots at the coleoptilar root determines the formation of lateral roots at the primary root. Middle, Crown roots present, no lateral roots at the primary root; right, no crown roots present, lateral roots formed at primary root late in development. P, primary root. O, Aboveground phenotype of wild-type (left) and rum1 (right) plants 45 DAG does not display any differences. P, Phenotypic properties of the rum1/rtcs double mutant. From left to right: wild-type, rtcs, rum1, and rum1/rtcs seedlings 12 DAG. Double mutant displays an additive phenotype (see text). Q, Phenotypic properties of the double mutant rum1/lrt1. From left to right: wild-type, lrt1, rum1, and lrt1/rum1 seedlings 12 DAG. Double mutant displays a novel phenotype (see text). Bars = 1 cm in A, B, L, M, P, and Q, 5 mm in C to H, 2 mm in J and K, 2 cm in N, and 12 cm in O.
Table I.
Genetic analysis of rum1
| Generation | Wild Type | rum1 | Observed Ratio | Expected Ratio | χ2 |
|---|---|---|---|---|---|
| F2a | 13 | 4 | 3.3:1 | 3:1 | 0.01b |
| F3c | 105 | 31 | 3.4:1 | 3:1 | 0.26b |
| F4c | 376 | 95 | 4.0:1 | 3:1 | 4.48b |
| F5c | 153 | 48 | 3.2:1 | 3:1 | 0.06b |
| Backcrosses in inbred lines (F2 generation)cd | |||||
| B73 | 323 | 53 | 18.3:3 | 13:3e | 5.43f |
| F2 | 211 | 34 | 18.6:3 | 13:3 | 3.85f |
| F7 | 209 | 64 | 9.9:3 | 13:3 | 4.07f |
| Aet | 221 | 36 | 18.3:3 | 13:3 | 3.69f |
| Hi II A | 414 | 64 | 19.5:3 | 13:3 | 9.25f |
Segregation ratio of seeds from the original ear in which the mutation was identified.
Cutoff value for an expected 3:1 ratio: χ1;0.992 < 6.63.
Combined segregation ratios of several segregating plants from that generation.
Segregation data of reciprocal crosses was combined because they yielded similar χ2 values.
Segregation ratios are most consistent with the presence of a negative recessive suppressor of the rum1 gene, which would result in a 13:3 segregation.
Cutoff value for an expected 13:3 ratio: χ1;0.992 < 6.63.
In addition, the mutant rum1 was reciprocally introgressed into the inbred lines B73, F2, F7, Aet, and Hi II A. All backcrosses revealed rum1 phenotypes in their F2 progenies (Table I). For each of the inbred lines, the segregation ratio of wild-type to mutant seedlings was homogenous as determined by the χ2 test irrespective of the pollen donor. Thus, no difference was observed in the transmission rate of the mutant allele via the male or female gametophyte. However, introduction of the rum1 gene into different genetic backgrounds resulted in segregation ratios that were statistically consistent with a 13:3 instead of the expected 3:1 segregation ratio (except for the inbred line F7). This could be explained by a recessive suppressor of the rum1 gene present in these genetic backgrounds.
The 52,800 plants derived from crosses of homozygous rum1-1 mutants to highly active Mutator stocks were screened via the paper roll test for rum1 mutant root phenotypes. Eight new rum1 alleles designated rum1-2 to rum1-9 were identified via this screen (Fig. 1B). All new alleles had the same phenotypic characteristics as the reference allele rum1-1.
The Mutant rum1 Is Deficient in Primordia Formation of Embryonic Seminal and Postembryonic Lateral Roots
Seminal root primordia are initiated in the region above the scutellar node late during embryogenesis (approximately 25 to 30 d after pollination [DAP]; Sprague, 1977; Erdelska and Vidovencova, 1993) in wild-type embryos (Fig. 2A). Transverse sections of wild-type and rum1 embryos 30 DAP revealed that the mutant rum1 does not initiate any seminal root primordia (Fig. 2B). Transplantation of young wild-type and rum1 embryos (15 DAP) on a regeneration medium (Armstrong and Green, 1985) prior to the initiation of seminal root primordia revealed that the competence for seminal root initiation was already lost in the mutant rum1 at this early stage of embryogenesis. Although transplanted rum1 embryos germinated within 3 d after transfer to nutrient medium, they did not develop seminal roots; in contrast, wild-type embryos, germinated under the same conditions, did develop seminal roots (data not shown).
Figure 2.
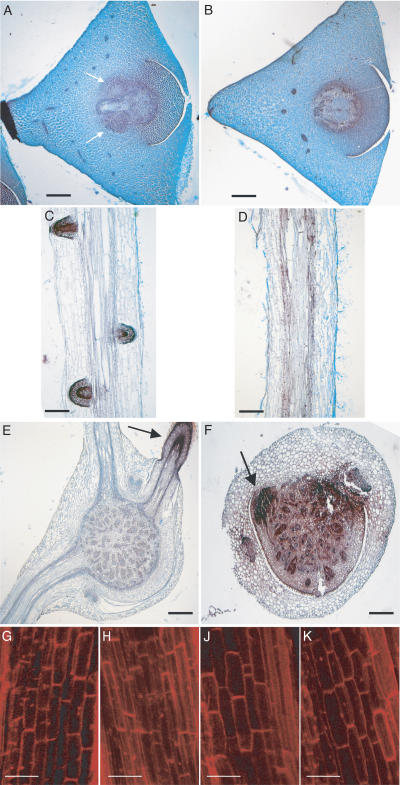
Histological analyses of the mutant rum1. A and B, Cross sections of the scutellar node of wild-type (A) and rum1 (B) embryos 30 DAP. Arrows indicate the seminal root primordia in the wild type. No seminal root primordia were detected in the mutant rum1. C and D, Longitudinal sections of the primary root of wild type (C) and the mutant rum1 (D) 5 DAG. No lateral root primordia were detected in the mutant rum1 (D), while the wild-type (C) displayed well-developed primordia at this developmental stage. E and F, Cross sections through the coleoptilar node of wild type (E) and the mutant rum1 (F) 30 DAG. While the wild-type develops various crown roots, the mutant rum1 displays only a rudimentary crown root primordium (indicated by an arrow). A to F, Stained with Safranin O and Fastgreen. G to K, Confocal laser scanning analysis of the cortical cell structure in the basal part of the primary root after propidium iodide staining of wild-type (G), lrt1 (H), rum1 (J), and the double mutant lrt1/rum1 (K). Despite the significantly reduced primary root length of the double mutant (compare with Fig. 1Q), cortical cell elongation is not affected in the double mutant. Bars = 500 μm in A, B, E, and F, 800 μm in C and D, and 100 μm G to J.
Lateral root primordia are initiated inside the primary root tissue from pericycle cells several days before emerging lateral roots penetrate the epidermis and become visible from outside. Lateral root primordia of the basal and, thus, the most differentiated 2 cm of the primary root were visualized with Schiff's base in a time course between 3 and 7 DAG (Fig. 1, C–G). Lateral root primordia became visible in wild-type primary roots about 4 DAG (Fig. 1D). Lateral root primordia were never detected in wild-type primary roots 3 DAG. While wild-type seedlings displayed fully developed lateral roots on the primary root 7 DAG, no lateral root primordia were detectable at this developmental stage in the mutant rum1 (Fig. 1G). These results were further supported by serial cross sections and longitudinal sections of wild-type (Fig. 2C) and mutant primary roots (Fig. 2D). The mutant rum1 did not display any lateral root primordia at any of these early developmental phases. For wild-type primary roots, the first pericycle cell divisions were detected 4 DAG. In none of the analyzed samples was division of pericycle cells observed at 3 DAG. Feulgen staining pattern of the primary root tip did not show any difference between the wild-type and rum1 primary root region of the apical meristem (Fig. 1H).
Determination of primary root length 10 DAG revealed that the primary root length of the mutant rum1 was reduced by 29.8% in light-grown seedlings and by 23.8% in dark-grown seedlings compared to wild-type primary roots (Table II). Root hair formation on the primary root of the mutant rum1 was not affected (data not shown). Among dark-grown seedlings, the mesocotyl and coleoptile of the mutant rum1 showed a 107% and 77% increase in length, respectively, as compared to the wild type; no difference in mesocotyl length was observed between genotypes when germinated in the light (Table II).
Table II.
Characterization of 5-d-old light- and dark-grown wild-type and rum1 seedlings
| Characteristics
|
Dark
|
Light
|
||
|---|---|---|---|---|
| Wild Type | rum1 | Wild Type | rum1 | |
| Primary root length (cm) | 13.0 ± 2.6 (70)a | 9.9 ± 2.4 (33)**b | 5.7 ± 2.6 (30) | 4.0 ± 2.4 (30)** |
| Mesocotyl length (cm) | 2.7 ± 1.2 (65) | 5.6 ± 1.8 (65)** | 0.7 ± 0.3 (20) | 0.7 ± 0.3 (20) |
| Coleoptile length (cm) | 1.3 ± 0.5 (40) | 2.3 ± 0.5 (40)** | n.d.c | n.d. |
Numbers in parentheses indicate the number of replicates for each measurement.
Two asterisks indicate a significant difference (t test; α = 1%) between wild type and rum1 under a specific light regime.
Light-grown coleoptiles were already penetrated by green leaves and wilted 5 DAG. n.d., Not determined.
Cross sections of coleoptilar nodes (first shoot nodes) of wild-type (Fig. 2E) and rum1 seedlings (Fig. 2F) grown under hydroponics until 30 DAG indicated that crown roots at the coleoptilar node were initiated in rum1 mutants but that these undifferentiated primordia did develop into crown roots in only 17% of the mutant plants. In the remaining 83% of the mutant plants, no crown roots were formed from the undifferentiated crown root primordia at the coleoptilar node of rum1 (Fig. 1, K and wild type in J), although normal shoot-borne roots, including lateral roots, developed on the consecutive stem nodes (Fig. 1, L and wild type in M). The term undifferentiated crown root primordia describes early developmental stages of the primordia where the different cell types that will subsequently form the newly emerging crown roots cannot be distinguished. Two different primary root phenotypes were observed 30 DAG depending on the presence of crown roots at the coleoptilar node. The 17% of the mutant plants that formed crown roots at the coleoptilar node did not initiate lateral roots on the primary root by day 30 after germination. More frequently, mutant plants that failed to initiate crown roots at the coleoptilar node (83% of the mutants) formed lateral roots in the tip region of the primary root approximately 3 d after transfer into hydroponics culture (Fig. 1N) irrespective of the use of nutrient solution or distilled water in the hydroponics culture. This transient phenotype might be indicative of a precise developmental window in which the rum1 gene inhibits lateral root formation or of the activity of another gene that is involved in lateral root initiation on the primary root later in development. No obvious differences were detectable in aboveground development between rum1 and wild-type plants (Fig. 1O).
Interaction of Maize Root Initiation Mutants
Allelism of rum1 was tested with the shoot-borne root initiation mutant rtcs (Hetz et al., 1996) and the lateral root initiation mutant lrt1 (Hochholdinger and Feix, 1998a) by reciprocal crosses of rum1 mutants with rtcs and lrt1 mutants. Since all progenies of these crosses displayed a wild-type phenotype in the F1 generation, we concluded that the mutants were not allelic.
Double mutants were generated to observe possible interactions between the different loci involved in root initiation. The segregation ratios are given in Table III. Crosses between rum1 and rtcs revealed double mutants with an additive phenotype in the F2 generation (Fig. 1P). The double mutants developed only a primary root that completely lacked all lateral roots, thus combining the root phenotypes of rtcs (i.e. a complete lack of crown roots) and rum1 (no lateral roots on the primary root) in an additive way. All double mutant seedlings died shortly after transfer into soil, presumably due to the limited absorbing capacity of their drastically reduced root system. The mutants rum1 and rtcs grew to maturity under the same conditions. Crosses of lrt1 and rum1 revealed seedlings with a novel phenotype in the F2 generation that we concluded to represent the double mutant. The rum1/lrt1 double mutant showed a severely reduced primary root and shoot compared to the wild type and the single mutants. No lateral or seminal roots were observed in the double mutant (Fig. 1Q). The lrt1/rum1 double mutant also died after transfer into soil. Longitudinal scans of the basal end of the primary root showed no difference in cortical cell size and organization between the wild type, single mutants, and lrt1/rum1 double mutants under a confocal laser scanning microscope (Fig. 2, G–K). The reduced size of the double mutant seedling might thus be caused by reduced or arrested meristematic activity of the apical meristems in the double mutant, although longitudinal sections of the root apical meristems did not show any histological difference between the wild type and the double mutant (data not shown).
Table III.
Genetic analysis of double mutants
| Double Mutants (F2 Generation) | Wild Type | rum1 | Mutant | Double Mutant | χ2a |
|---|---|---|---|---|---|
| rtcs × rum1 | 269 | 13 | 67 | 21 | 63.6 |
| rum1 × rtcs | 565 | 26 | 99 | 50 | 157 |
| lrt1 × rum1 | 243 | 28 | 25 | 15 | 61.2 |
| rum1 × lrt1 | 155 | 32 | 18 | 13 | 23.3 |
Cutoff value for an expected 9:3:3:1 ratio: χ3;0.992 < 11.34.
The Mutant rum1 Is Affected in Polar Auxin Transport
Exogenous application of the auxin α-naphthyl acetic acid (α-NAA), which can passively diffuse into the cells (Delbarre et al., 1996), induces lateral root formation and inhibits primary root elongation in wild-type plants. Wild-type and rum1 seeds were germinated in paper rolls in the presence of different concentrations of α-NAA (0, 0.01, 0.1, 1, and 10 μm). At 10 DAG, wild-type and rum1 seedlings showed a similar reduction of primary root length at increasing concentrations of auxin. However, in contrast to wild-type seedlings, the mutant rum1 did not develop any lateral roots on the primary root at any of the applied α-NAA concentrations. Hence, the primary root of the mutant rum1 showed normal sensitivity toward exogenously applied auxin; however, the pericycle cells did not respond by initiating lateral roots.
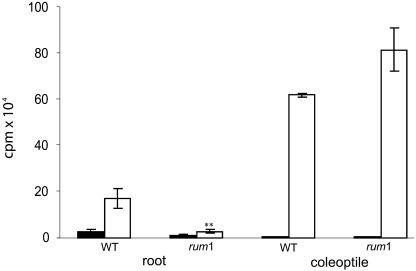
Polar auxin transport was measured in the primary root and the coleoptile of the mutant rum1. The major bioactive auxin in plants, indole-3-acetic acid (IAA), is known to be transported acropetally (toward the root tip) in the basal part of roots (Sachs, 1991) and basipetally (away from the tip) in coleoptiles. 3H-IAA transport in 3-DAG rum1 primary root fragments was reduced by 83% compared to the wild type. Polar auxin transport was enhanced in rum1 compared to wild-type coleoptiles (Fig. 3). This increase was, however, not statistically significant. In contrast to the mutant rum1, polar auxin transport was not affected in the only other known maize lateral root initiation mutant lrt1 when compared to wild-type seedlings (data not shown).
Figure 3.
Polar auxin (3H-IAA) transport in wild-type and rum1 roots and coleoptiles. White columns indicate polar auxin transport (root, transport toward the root tip; coleoptile, transport away from the tip of the coleoptile); black columns represent the control transport experiments. Polar auxin transport in the primary root of the mutant rum1 is reduced by 83% compared to polar auxin transport in wild-type primary roots. In the coleoptile, no statistically significant differences in transport were detected. **, Indicates a significant difference in the t test (unknown and unequal variances of the two populations) at a level α of 1%; error bars indicate standard error of the mean.
The reduced polar auxin transport in the rum1 primary root raised the question if the reduced auxin transport capacity also results in a reduced concentration of biologically active free auxin (IAA) in rum1 primary roots. IAA levels were measured via HPLC. Free IAA levels were similar in wild-type and rum1 roots (wild type, 0.10 ± 0.03 nmol/mg dry weight; rum1, 0.11 ± 0.02 nmol/mg dry weight). Thus, a reduced concentration of free auxin cannot be the cause for reduced auxin transport in the mutant rum1. Interestingly, only a small fraction of the detected IAA was present as free active IAA, while most of it was present in the inactive covalently bound form (wild type, 95.8%; rum1, 73.7%).
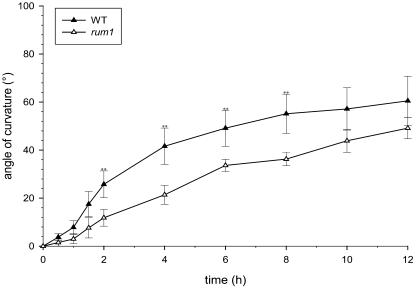
The gravitropic response of the primary root is related to polar auxin transport. The primary root of the mutant rum1 displayed a delayed gravitropic response in comparison to the wild-type primary root. A different gravitropic response of the mutant rum1 was detectable at least between 2 and 8 h after the gravitropic stimulus (Fig. 4). No significant difference in gravitropic response was detectable in coleoptiles of rum1 compared to wild-type seedlings (data not shown).
Figure 4.
Gravitropic response of wild type (black triangles) and the mutant rum1 (white triangles) primary root tips. Seedlings grown in paper rolls for 3 d were turned 90° at time 0. The angle of curvature was measured at the indicated times. Each triangle represents the mean of eight measurements. The mutant rum1 shows a significant reduction in gravitropic response compared to the wild type between 2 and 8 h after induction. **, Indicates significant difference for t test (unknown and unequal variances of the two populations) at a level α of 1%; error bars indicate standard error of the mean.
In an effort to investigate the expression of genes known to be involved in polar auxin transport in the rum1 mutant and to exclude the possibility that the knockout of one of the known auxin transporters is the reason for the rum1 phenotype, the expression of 13 maize orthologs of putative auxin transporters from Arabidopsis, including eight members of the pin and five members of the aux1 gene families, was determined. The 3′ untranslated regions (UTRs) of these genes were analyzed via reverse RNA gel-blot analyses in roots of 5-d-old seedlings. Maize orthologs of the putative auxin transporters from the pin and aux1 gene families were identified via BLAST database searches (Altschul et al., 1997) in the maize genome database (www.maizegdb.org) and in the maize assembled genomic island database (http://maize.ece.iastate.edu/asm.html) using Arabidopsis pin and aux1 sequences as query sequences with a cutoff value of e−10. Specific PCR primers were designed for the 3′ UTR of the eight identified maize orthologs of the pin gene family and the five members of the aux1 gene family. In addition, specific 3′ UTR probes of the maize genes actin1 and glyceraldehyde-3-P dehydrogenase (GAPDH), which are constitutively expressed at similar levels in different tissues, have been generated and used as an internal reference (Coker and Davies, 2003).
None of the eight pin and the five aux1 orthologs showed significantly different expression (>2-fold) in roots of wild-type seedlings versus rum1 (Fig. 5). The maize orthologs of the putative auxin efflux carrier genes pgp2 and pgp19 were not detectable in an RNA gel-blot analysis in the primary root of 5-DAG maize seedlings (data not shown).
Figure 5.
Expression pattern of the maize homologs of putative auxin transporters from the aux1 and pin families in wild-type (WT) and rum1 primary roots. 3′ UTR of the known maize homologs of the pin and the aux1 gene family as well as of actin1 and GAPDH were amplified and slot blotted to a membrane. Radioactively labeled cDNA of wild-type and rum1 primary roots 5 DAG was used as a hybridization probe. Accession numbers of the maize genes are given.
Differential Gene Expression between Wild-Type and rum1 Pericycle Cells
Lateral roots in maize are initiated during postembryonic development via dedifferentiation and subsequent division of pericycle cells. Pericycle cells of the lateral root initiation mutant rum1 and of the corresponding wild-type pericycle were isolated via LCM from 64-h-old primary roots and subsequently hybridized to 12k maize cDNA microarray chips. We chose this very early developmental stage because in our histological analyses of wild-type and mutant primary root development, we did not detect any pericycle cell divisions in 3-DAG primary roots. To account for any variability in lateral root initiation, we took our samples 2.5 DAG (64 h after germination). Statistical analyses using specific variances for each gene revealed 90 genes that displayed significantly higher expression in pericycle cells of the wild type compared to rum1 pericycle cells (Supplemental Table I). For 57% (51/90) of these genes, a functional annotation was possible via BLASTx searches in the NCBI nonredundant database (as of January 3, 2005). Among the remaining 39 genes, for 15 genes, a database hit to proteins with unknown function was retrieved, and for 24 genes, no database hits were found. Differentially expressed genes were annotated according to the Munich Information Center for Protein Sequences database (version 2.0; http://mips.gsf.de/proj/thal/db). Metabolism (15/51), transcription (10/51), and cellular transport (8/51) were the most prominent classes among the genes predominantly expressed in wild-type pericycle cells. Other, however, less frequent categories among the differentially expressed genes were signal transduction/cellular communication (5/51), cell cycle (4/51), protein fate (folding, modification, and destination; 2/51), subcellular localization (2/51), translation (2/51), cell rescue, disease and defense (1/51), cell fate (1/51), and transposable elements and viral proteins (1/51).
In addition, 73 genes were preferentially expressed in the pericycle of the mutant rum1 (Supplemental Table II). In all, 55% (40/73) of the genes predominantly expressed in rum1 were functionally annotated via BLASTx database searches. The remaining genes were unclassified or of unknown function (12/73) or did not yield any database hit (21/73). Metabolism (7/40) and energy generation (7/40) were the most prominent classes, followed by the classes of cell rescue, defense, and virulence (5/40) and protein fate (5/40), transcription (4/40), signal transduction/cellular communication (3/40), subcellular localization (3/40), unclassified proteins (3/40), translation (1/40), cellular transport (1/40), and transposable elements and viral proteins (1/40). The estimated false discovery rate of this pericycle-specific gene expression data set was calculated as 13.8%.
DISCUSSION
The mutant rum1 defines a novel locus involved in lateral and seminal root initiation in maize and is the first root mutant identified so far that displays a phenotype restricted to the late embryonic and early postembryonic developmental phases. This phenotype extends the concept of complex overlapping developmental programs active during root formation in maize (Hochholdinger et al., 2004b). Other known root initiation mutants of maize are either exclusively affected in early postembryonic development (lrt1; Hochholdinger and Feix, 1998a) or during late embryonic and late postembryonic development (rtcs; Hetz et al., 1996).
Analyses of double mutants between rum1 and rtcs showed an additive phenotype, thus demonstrating that shoot-borne root initiation and lateral root initiation at the primary root are independent processes. Moreover, the initiation of seminal root formation that is affected in rum1 and rtcs mutants is regulated at least via two independent developmental pathways. Although the number of seminal roots is inherited in a complex polygenic manner (Hochholdinger et al., 2004b), initiation of seminal roots can be blocked by a mutation in a single gene. The double mutant rum1/lrt1 displayed a novel phenotype with a considerable reduction of root and shoot size in addition to the lack of lateral roots. Since cortical cell elongation and root organization was not affected in the primary root of the rum1/lrt1 double mutant, the rum1 and lrt1 genes could have a cooperative function in the control of meristematic activity in the root and shoot apex and in lateral root initiation. A similar phenomenon was observed for the maize double mutant slr1/slr2. The lateral root elongation genes slr1 and slr2 cooperate, however, in pattern formation of the primary root in addition to lateral root elongation (Hochholdinger et al., 2001).
The plant hormone auxin plays a crucial role during embryogenesis (Liu et al., 1993; Hadfi et al., 1998) and is involved in various aspects of root development (Friml, 2003). Auxin transport experiments demonstrated severely reduced acropetal auxin transport toward the root tip in the primary root of the mutant rum1. In Arabidopsis, inhibition of polar auxin transport in primary roots was correlated with a block of lateral root formation (Reed et al., 1998). Therefore, the reduced transport of auxin in the primary root of the mutant rum1 might explain the lack of lateral roots in rum1. The observed delayed gravitropic response of the mutant rum1 during the first hours after gravistimulus could also be attributed to the reduced polar auxin transport (Chen et al., 1998) by not allowing asymmetric auxin distribution in the root tip, which is necessary for asymmetric inhibition of root elongation during gravitropic growth (Schurzmann and Hild, 1980). However, reduced auxin transport might not be the single reason for the complete lack of lateral roots in the mutant rum1. Exogenously applied auxin inhibits primary root elongation at increasing auxin concentrations and induces lateral roots (Chadwick and Burg, 1967; Hetz, 1996; Inukai et al., 2005; Liu et al., 2005). While the primary root of the mutant rum1 exhibited a normal response to exogenously applied auxin, i.e. growth inhibition, this treatment did not induce lateral roots in the primary root of the mutant rum1. This implies that the auxin sensitivity of the cells in the apical region of the rum1 primary root, which are responsible for the elongation of the primary root, is not impaired in rum1, while the pericycle cells that initiate lateral roots in the differentiation zone might be impaired in auxin sensitivity or auxin signal transduction. Hence, the rum1 gene might have a pleiotropic regulatory function with respect to auxin action in roots, which is manifested by reduced polar auxin transport in the primary root but also by a reduced sensitivity of the pericycle cells toward auxin. The maize developmental mutant semaphore1 (sem1; Scanlon et al., 2002) is also affected in the regulation of polar auxin transport. However, in contrast to rum1, the mutant sem1 displays a pleiotropic phenotype affecting specific domains of the shoot, the embryo, the endosperm, pollen, and lateral root formation. Therefore, the sem1 gene appears to be a general regulator of polar auxin transport in maize, while our data suggest that the rum1 gene is a specific regulator of auxin transport and auxin perception in the primary root. The rum1 gene also has a defined function during embryonic seminal root initiation. Disruption of polar auxin transport in Brassica embryos resulted in severe defects in bilateral symmetry (Hadfi et al., 1998). The embryo of rum1 does not display such aberrations, indicating normal polar auxin transport in the embryo. Since embryonic development of the rum1 mutant is only affected by the absence of seminal root primordia, the function of the rum1 gene during embryogenesis seems to be restricted to the region of the scutellar node.
In Arabidopsis, two mutants, Slr (Fukaki et al., 2002) and alf4 (Celenza et al., 1995), were identified that are affected in lateral root initiation but do not significantly affect primary root formation. The slr gene encodes for IAA14, a member of the Aux/IAA protein family (Fukaki et al., 2002). The alf4 gene encodes for a plant-specific protein with no structural similarity to proteins of known function (DiDonato et al., 2004). Since IAA14 and ALF4 are believed to be required downstream of auxin synthesis and transport, they are most likely no candidates for the rum1 gene.
Reverse RNA gel-blot hybridization experiments indicated only subtle changes in the expression levels of all 13 available maize homologs of the auxin transporters from the pin and aux1 gene families between wild-type and rum1 primary roots. Therefore, it is unlikely that one of the analyzed putative auxin transporters is responsible for the reduced auxin transport detected in the primary root of the mutant rum1.
Global gene expression profiling of pericycle cells using a 12k cDNA microarray was conducted to identify genes that are differentially expressed between wild-type and rum1 primary root pericycle cells and thus gain a better understanding of the transcriptional networks active prior to lateral root initiation. None of the previously published cell-type-specific transcriptome analyses in plants (Asano et al., 2002; Birnbaum et al., 2003; Nakazono et al., 2003) dissected the root pericycle. Our analyses identified 90 genes that were preferentially expressed in the wild-type pericycle and 73 genes that were preferentially expressed in the rum1 pericycle. Remarkably, 19 of the 51 functionally annotated proteins that were preferentially expressed in wild-type pericycle cells were regulatory genes involved in signal transduction, transcription, and cell cycle. Preferential expression of these gene classes in the wild-type pericycle might indicate that those genes are involved in the regulatory network that finally leads to the dedifferentiation of the pericycle cells (Dubrovsky et al., 2000) and consequentially to cell division and lateral root primordia formation, a process that is blocked in the rum1 mutant. Several developmental processes in plants, e.g. root hair formation or radial organization of the primary root in Arabidopsis, are controlled by a hierarchy of transcription factors (Benfey and Weigel, 2001). Transcriptional regulators represented a subset of 10 genes preferentially expressed in wild-type pericycle cells. The exact function of these transcriptional regulators is yet unknown. However, the function of the two differentially expressed helix-loop-helix transcription factors (NP_194827.1 and AAO72577.1), which control cell proliferation and development of specific cell lineages (Heim et al., 2003), might support their role in lateral root initiation. The biological significance of the differentially expressed genes in the class of transcriptional regulators is supported by the coordinated up-regulation of the genes HMGB1 (T03640) and CK2 (AAG36872.1) in the wild-type pericycle. These genes are known to interact with each other via phosphorylation of HMGB1 by CK2 (Stemmer et al., 2002). Cyclin T2 (BAD17160.1) was up-regulated in the wild-type pericycle. Cyclins are primary regulators of the activity of cyclin-dependent kinases, which play a crucial role in controlling eukaryotic cell cycle progression (Wang et al., 1994) and are activated when quiescent cells become ready to reenter the cell cycle (Martinez et al., 1992). Lateral root initiation is such a process in which quiescent pericycle cells are activated and reenter the cell cycle. The maize mutant rtcs, which is completely blocked in shoot-borne root initiation, is comparable to rum1, where the initiation of lateral roots is blocked. None of the maize cyclins is expressed in rtcs coleoptilar nodes, while all four cyclins are expressed in wild-type coleoptilar nodes that proliferate crown roots (Hochholdinger and Feix, 1998b).
This pericycle-specific gene expression data set defines genes related to signal transduction, cell cycle, and transcription active before lateral root initiation, which might mediate the developmental cues for the initiation of lateral root formation. Preferential expression of genes from other functional classes in the wild-type pericycle also appears to be linked to lateral root initiation. For example, argonaute proteins (NP_175274.1) are required for stem cell function and organ polarity (Kidner and Martienssen, 2005). In addition to the genes preferentially expressed in the wild-type pericycle, 73 genes were preferentially expressed in rum1 pericycle cells. Among the 40 annotated genes, at least 10 can be assigned to the processes of cell wall formation (expansins, methyltransferases, and mannan endohydrolases) and defense. Cell walls give shape and structure to plant cells and organs, while at the same time maintaining strength, flexibility, and plasticity to accommodate growth and respond to biotic and abiotic environmental changes. Thus, cell walls play a pivotal role particularly with respect to their development and differentiation (McCabe et al., 1997; Fleming et al., 1999; Lally et al., 2001; O'Neill et al., 2001). As a consequence of the lack of lateral roots, the mutant rum1 is considerably challenged in water and nutrient acquisition during its early developmental phases. This might explain why stress- and cell-wall-related genes that facilitate water and nutrient uptake by modifying the chemical composition of the cell wall are preferentially expressed in the mutant rum1. Expansins are, for example, cell wall proteins that induce pH-dependent wall extension and stress relaxation to loosen cell walls during cell enlargement and root hair growth (Cosgrove et al., 2002). A similar up-regulation of genes involved in lignin biosynthesis, e.g. methyltransferases, has also been observed in a proteome study of the primary root of the maize lateral root initiation mutant lrt1 (Hochholdinger et al., 2004c).
The results of this global gene expression profiling of pericycle cells are an initial step toward a better understanding of the regulatory networks active in the pericycle prior to lateral root initiation. Functional analyses of the up- and down-regulated genes of this data set will reveal further aspects of the mechanisms regulating lateral root initiation.
In summary, the rum1 gene is an important novel checkpoint in auxin-mediated lateral and seminal root initiation in maize, which might act via the regulation of auxin transport in the primary root and auxin perception in the primary root pericycle. Global expression profiles of pericycle cells have identified regulatory genes that may be involved in lateral root initiation. Cloning of the rum1 gene will help to elucidate the function of the rum1 gene that considerably affects the molecular networks involved in maize root system formation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The mutant rum1 was isolated from mutagenized F2 families generated from selfed F1 crosses between the inbred line B73 and active Mutator stocks. The screening of the F2 generation for aberrant root phenotypes was conducted in the paper roll test. Seeds were surface sterilized with 6% hypochlorite for 6 min, rinsed in distilled water, and germinated on moistened filter paper (20×70 cm Grade 603 N; Sartorius). Seedling root phenotypes were analyzed 10 DAG. To study the older root system, 10-DAG seedlings were transferred from paper rolls into a hydroponic culture box containing 36 L of gramineae nutrient solution (Marschner, 1995). Oxygen was supplied with an aquarium pump (Schego). Plant growth chamber conditions for the paper roll test and hydroponics were 60% humidity, 26°C, 16-h-light, 8-h-dark regime.
Assessment of Phenotypic Seedling Parameters
Determinations of phenotypic seedling parameters, including root, mesocotyl, and coleoptile lengths, were performed by digitalizing seedling pictures via a scanner (HP scanjet 7400C; Hewlett-Packard Company) and subsequently analyzing the scans with Image Pro Express software (Media Cybernetics).
The gravitropic curvature of the primary root was measured in seedlings grown in paper rolls at 28°C in the dark. Three days after germination, seedlings were glued in their initial position to germination paper with Fixo Gum (Marabu) and turned 90° relative to their former growth direction. The gravitropic response was documented by digital photographs every 10 min for 12 h (PC-Cam 600; Creative).
Genetic Analyses
Propagation and backcrossing of the rum1 mutant was performed according to standard genetic procedures (Larson and Hanway, 1977). The parents into which the mutants were backcrossed were B73, F2, F7, Aet, and Hi II A. B73 is a commonly used inbred line developed from Stiff Stalk Synthetic (BSSS). Aet is a highly inbred genetic stock primarily composed of New England Flint germplasm, and Hi II A was generated by inbreeding an A188×B73 hybrid. F2 and F7 are two very early and cold-resistant European (French) inbred lines.
Novel mutant alleles of the rum1 locus were obtained from a directly tagged population generated by crosses of homozygous rum1-1/rum1-1 reference mutants as females with pollen from highly active Mutator stocks (rum1-1/rum1-1×Rum1/Rum1 Mu). The 52,800 progenies of these crosses were screened in the paper roll test for rum1 root phenotypes.
Histological Analyses
Safranin/Fast Green Staining
Maize (Zea mays) primary roots, coleoptilar nodes (first shoot node), second shoot nodes, and mature embryos were fixed in 4% (w/v) paraformaldehyde for 12 h at 4°C as described by Lim et al. (2000). The 10- to 12-μm sections were prepared using a Leica RM2145-microtome. Staining of deparaffinized specimens with Safranin O (Applichem) and Fast Green (Serva) was performed according to standard histological procedures (Johansen, 1940). Stained specimens were analyzed under a Zeiss-Axioskop HBO 100W/2 microscope and documented with an Olympus SC35, Type12 digital camera.
Feulgen Staining
The basal 2 cm of the primary root of 3- to 7-d-old wild-type and rum1 seedlings that represent the most differentiated part of the root were fixed. After 10 min of hydrolysis in a 1 n solution of HCl at 60°C, the samples were stained according to the Feulgen protocol (De Tomasi, 1936) for 30 min. The staining pattern of the roots was documented with an Olympus SC35, Type12 digital camera under a binocular (Stemi SV8; Zeiss).
Propidium Iodide Staining and Confocal Laser Scanning Microscopy
Cortical cell elongation of the primary root was analyzed with a confocal laser scanning microscope (DM IRBE; Leica) with a TRITC wide filter. Hand sections of the proximal end of the primary root 10 DAG were stained for 10 min in 5 μg/mL of propidium iodide and then washed for 2 min in 50 mL of water (Oparka and Read, 1994). Samples were mounted in glycerol prior to analysis with the confocal laser scanning microscope.
Measurement of Auxin Transport
Polar auxin transport was determined in 2-cm-long proximal fragments of the primary root (3 DAG) excluding at least 0.4 cm of the root tip, which contains the meristematic and distal elongation zone (Ishikawa and Evans, 1995), and in 1-cm fragments of the basal part of the coleoptile (5 DAG) after removing at least 0.3 cm of the coleoptile tip in paper rolls at 28°C in the dark. Only seedlings with a primary root length of about 2.5 cm were used for the experiments. Samples were incubated in both orientations in 30 μL of 10 mm potassium phosphate buffer (pH 5.5) containing 12.5 μCi 3-[5(n)-3H]-indolylacetic acid (3H-IAA; Amersham Biosciences). Primary roots were incubated for 16 h and coleoptiles for 2.5 h at room temperature. The upper 0.3 cm of the samples were removed after incubation and analyzed in an Ultima Gold multipurpose scintillation counter (Packard Instruments).
Measurement of Auxin Content
Homozygous wild-type (B73) and rum1 plants were grown at 28°C for 64 h in the dark. The apical 0.5 cm of the root apex, including the meristematic zone, were discarded. Extraction and determination of free and bound IAA were conducted according to a modified method described by Sweeter and Vatvars (1976). IAA and its conjugates were extracted in dim light for 30 min with 30 mL of 80% (v/v) methanol at 3°C. The extract was filtered, methanol was evaporated, and the aqueous fraction was adjusted to pH 8.0 and extracted twice with diethyl ether. The ether fractions were discarded, and the remaining aqueous fraction was acidified to pH 2.8 and extracted as described before. The combined ether fractions were concentrated, and the residue was dissolved in 50% (v/v) acetonitrile in HPLC-grade water and used for determinations of free IAA. For determination of the bound IAA content, the remaining aqueous fraction was adjusted to pH 11.0 and heated for 1 h at 60°C. The hydrolyzed fraction was acidified to pH 2.8 and treated as described before. Estimation and quantification of free and bound IAA levels were performed using a modular HPLC system (Pumps 420, Autosampler 360, KromaSystem 2000 software; all from Kontron Instruments) equipped with a Kontron Techsphere ODS column (5-μm particles, 280-Å pore diameter, length 150 mm) with the following parameters: a gradient of mobile phase from 15% to 30% (v/v) acetonitrile in HPLC-grade water containing 0.1% (w/v) trifluoroacetic acid and a flow rate of 0.8 mL/min. IAA was identified and quantified with a diode array detector (Kontron DAD 440) and a fluorescence detector (Shimadzu RF 535, excitation 280 nm, emission 370 nm) and cochromatography with authentic IAA standards. Additionally, 1H-IAA (9CI) (87–51-4) was identified using gas chromatography-mass spectrometry as 1H-IAA, 1-(trimethylsilyl)-, trimethylsilyl ester (9CI) (56114–66-0).
Reverse RNA Gel-Blot Analysis
PCR probes of the 3′ UTR of the analyzed genes were generated using the following oligonucleotide primers: ZmPina, ZMtuc03-08-11.220 forward (Fw), 5′-ACTCTCCCACCGCACCTC-3′, reverse (Rev), 5′-GACGCACCAAGAAACACTTG-3′; ZmPinb, ZMtuc03-08-11.14930 Fw, 5′-TCTCTCGCTCGCTTCTTCAG-3′, Rev, 5′-TTCAGACAGCATGAAGCAAGAT-3′; ZmPinc, ZMtuc03-08-11.76 Fw, 5′-CCTGAGCCCTACAACCACTC-3′, Rev, 5′-ACGAGCAGTGCCAATTTGTT-3′; ZmPind, ZMtuc03-08-11.19547 Fw, 5′-GGAGGCAAGTGAGGAGACTG-3′, Rev, 5′-CGTGACGTGCTTAAACTGGA-3′; ZmPine, ZMtuc03-08-11.805 Fw, 5′-TACGCGGGGCAGACTCATA-3′, Rev, 5′-GGGAATTGGAGGGACCTGA-3′; ZmPinf, ZMtuc03-08-11.19548 Fw, 5′-GGAGGCAAGTGAGGAGACTG-3′, Rev, 5′-CCGGAGGTGAGCTGTTTATC-3′; ZmPing, ZMtuc03-08-11.13048 Fw, 5′-GATGTGGAGCAGGAGGAGAC-3′, Rev, 5′-GCGCATGAAAAGTCCCTAAC-3′; ZmPinh, ZMtuc03-08-11.14931 Fw, 5′-GCGCCCTGCTACTACTGAAG-3′, Rev, 5′-TTCAGACAGCATGAAGCAAGA-3′; ZmAux1a, ZMtuc03-08-11.52 Fw, 5′-GTGGCACACTGCTGCTTTAG-3′, Rev, 5′-GAAATGGATTCCCCTTCTTTG-3′; ZmAux1b, AJ011794 Fw, 5′-GTCACGTACGGCCGATTACT-3′, Rev, 5′-TCTCGAAAGCAGCTACACACA-3′; ZmAux1c, ZMtuc03-08-11.24489 Fw, 5′-ATTGCAGCCGGTGTTAATTG-3′, Rev, 5′-AGCCAAACGAAAACAAAACG-3′; ZmAux1d, ZMtuc03-08-11.24490 Fw, 5′-CGCTCTCTCTCTCACCGAAT-3′, Rev, 5′-AGATTTGAAGATCGCGGATG-3′; ZmAux1e, MAGI _83218 Fw, 5′-ATCACTGAAGCAGCGAGCGA-3′, Rev, 5′-TCGGTTTACGGTCGTTAGCC-3′; Actin1, AY104722 Fw, 5′-ATGTGACAATGGCACTGGAA-3′, Rev, 5′-GACCTGACCATCAGGCATCT-3′; GAPDH, ×75326 Fw, 5′-GTCTCCCTGGTAATGAACGA-3′, Rev, 5′-TGCTCCTTTCTCCTTTGCAT-3′.
The PCR products were heat denatured, transferred on a Hybond NX nylon membrane (Amersham Biosciences) via slot blotting, and cross-linked to the membrane at 0.12 J/cm2 (BioLink BLX). The membrane was then prehybridized with 10 mL of a solution containing 50% (v/v) Formamid, 5× Denhardt's reagent, 6× SSC, 0.5% (w/v) SDS, 0.01 m EDTA, and 100 μg herring sperm DNA for 2 h at 42°C followed by a hybridization with a radioactively labeled cDNA probe that was generated from RNA of the primary root of 5-DAG wild-type and rum1 seedlings. The probes were synthesized by incubating 1.5 μg RNA with 2.5 μL 10 mm dithiothreitol, 20 units of RNase-inhibitor (MBI Fermentas), 625 ng random hexamer primers, 1× RT-Puffer, 20 mm each dATP, dTTP, and dGTP, 8 μL 10 mCi/mL [α-32P]dCTP, and 200 units of reverse transcriptase for 1 h at 42°C. Prior to hybridization at 42°C overnight, the probe was denatured at 95°C for 5 min. Washing of the membrane was conducted at 55°C for 30 min with washing buffer I (2× SSC, 0.1% [w/v] SDS) and washing buffer II (0.1× SSC, 0.1% [w/v] SDS), respectively. Slot-blot signal intensities where determined with Bio-1D software (Vilber Lourmat).
Pericycle-Specific Transcriptome Profiling
Plant Material, Growth Conditions, and Fixation of Primary Root Samples for LCM
Primary roots grown at 28°C for 64 h in the dark with lengths of between 1.5 and 2 cm were utilized. The apical 0.3 cm of the root apex, including the meristematic zone and the distal elongation zone, were discarded. The remaining differentiation and elongation zones of the roots were collected in 0.5-cm samples, fixed, and embedded in TissueTek OCT medium according to the protocol described by Nakazono et al. (2003). The corresponding parts of three roots were embedded in one cryomold representing one biological replicate. Circadian effects on gene expression were taken into account by always germinating and harvesting roots of the same length at the same time of the day (germination, 6 pm; harvest, 10 am). This sampling strategy was chosen because in maize, lateral roots are initiated in the differentiation zone (Ishikawa and Evans, 1995), although the site of lateral root formation in this zone cannot be predicted (Bell and McCully, 1970). Since elongation and differentiation zones in maize are partly overlapping, only pericycle cells from these two zones were collected. Primary roots in maize emerge around 2 DAG. Thus, 2.5-d-old (64-h-old) primary roots are the earliest developmental stage that is allowing for the isolation of pericycle cells from the differentiation zone. At this early developmental stage, no lateral root primordia can be detected in the primary root in wild-type seedlings. Consequently, this developmental stage represents an early stage before lateral root initiation.
Cryosectioning and LCM
Preparation of 10-μm primary root cross sections was performed at −20°C using a cryostat (Leica CM1850) and mounted on adhesive slides using the CryoJane tape-transfer system (Instrumedics). At most, only every fifth section of a series was collected on an adhesive tape window that was brought into contact with the specimen before sectioning. The tape-window containing the cross section was transferred and firmly cross-linked to an adhesive-coated slide with a flash of 360-nm UV light. After removal of the tape window, cross sections were dehydrated by a series of 70% (−20°C), 95%, 100% (v/v) ethanol each for 1 min followed by three xylene steps for 2 min on ice. Sections were kept in fresh xylene until they were used for LCM.
In the PixCell II LCM system (Arcturus Bioscience), air-dried samples were placed on the microscope and brought into focus. CapSure caps (Arcturus) were then placed over the cross section. After focusing the laser, a circle of pericycle cells was captured using the following parameters: laser spot size of 7.5 μm, laser power of 60 mW, and laser pulse duration of 550 to 650 μs. Each circle of pericycle cells contained approximately 200 cells.
RNA Extraction and Amplification
RNA of approximately 13,000 captured and pooled pericycle cells was extracted using the Absolutely RNA Microprep kit (Stratagene) and treated with 30 units of RNase-free DNase I (Stratagene). Extracted RNA was amplified according to the protocol of Nakazono et al. (2003), first synthesizing double-stranded cDNA of the mRNA followed by RNA synthesis via in vitro transcription with T7-polymerase. The amplification procedure was repeated twice to increase the amplified RNA yield. The efficiency of amplification was quantified using the RiboGreen RNA-quantitation reagent (Molecular Probes) by measuring RNA yield after the first and second rounds of amplification.
Microarray Hybridization, Scanning, and Spot Quantification
Microarray probe synthesis and hybridization of four spotted 12k maize cDNA microarray slides (Generation II, Version A) from the Iowa State MicroArray Facility (http://www.plantgenomics.iastate.edu/maizechip/) were conducted as described by Nakazono et al. (2003). Three independent biological replications from each genotype were profiled on the four arrays. Samples from differing cell types were paired on each array. Dyes were assigned to samples in such a way that each genotype was measured an equal number of times with both dyes.
Dried slides were scanned three times at different scan settings with a ScanArray 5000-scanner (Packard) for each channel (Cy3 and Cy5) with laser power and PMT gain settings adjusted so that the signal intensity for both channels was equal for one slide. ImaGene software (Biodiscovery) was used to quantify the spot intensities on the slides. Spots were removed from the data set before analysis if they did not represent a single-band PCR product or if they were empty spots.
Data Normalization
The lowess normalization method originally described by Dudoit et al. (2002) was applied to the log of background-corrected raw signal intensities to remove signal-intensity-dependent dye effects from each sector on each slide. Following lowess normalization, the normalized data for each slide/dye combination were median centered so that expression measures would be comparable across slides. Median centering simply involves subtracting the median value for a particular slide/dye combination from each individual value associated with the particular slide/dye combination. Thus, negative (positive) values indicated that a particular transcript was expressed below (above) the median for a particular slide/dye combination.
Data Analysis
For each of 10,767 sequences, a mixed linear model analysis of the normalized log-scale signal intensities was conducted to identify transcripts whose expression differed significantly between rum1 and wild-type pericycle cells. The mixed linear model included genotype and dye terms as fixed effects as well as slide terms, sample terms, and a general error term as random effects. A t test for genotype differences was conducted as part of a mixed linear model analysis for each gene (Wolfinger et al., 2001) yielding 10,767 P-values. As described by Allison et al. (2002), a mixture of uniform and a β distribution was fit to the observed distribution of the 10,767 P-values obtained from the mixed linear model analysis. The estimated parameters from the fit of the mixture model were used to estimate the posterior probability of differential expression for each gene and to estimate the proportion of false positive results among all genes with P-values ≤0.01 and estimated fold change > 2.
Sequence data from this article can be found in the GenBank/EMBL data libraries. Accession numbers are given in the supplemental tables.
Supplementary Material
Acknowledgments
We thank Dr. An-Ping Hsia (Iowa State University) for assistance with the direct tagging of rum1, Dr. Peter Peterson (Iowa State University) for Aet seeds, Drs. Rolf Reuter and Reinhard Schröder (University of Tübingen, Germany) for support with confocal laser scanning microscopy, Drs. Günter Feix and Rainer Hertel (University of Freiburg, Germany) for helpful comments on the manuscript, and Ingrid Blumberg, Caroline Marcon, Katharina Maurer, Dirk Middendorf, and Bettina Stadelhofer (University of Tuebingen, Germany) for excellent technical assistance in the field and laboratory.
This work was supported by the Sonderforschungsbereich 446 “Mechanisms of cell behaviour.” Additional support was provided by Hatch Act and State of Iowa funds. K.W. was supported in part by the “Wilhelm-Schuler” and the “Reinhold-und-Maria-Teufel” Foundations.
The author responsible for distribution of materials integral to the findings presented in the article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Frank Hochholdinger (frank.hochholdinger@zmbp.uni-tuebingen.de).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.067330.
References
- Allison DB, Gadbury GL, Heo M, Fernández JR, Lee CK, Prolla TA, Weindruch R (2002) A mixture model approach for the analysis of microarray gene expression data. Comput Stat Data An 39: 1–20 [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CL, Green CE (1985) Establishment and maintenance of friable, embryogenic maize callus and the involvement of L-proline. Planta 164: 207–214 [DOI] [PubMed] [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 32: 401–408 [DOI] [PubMed] [Google Scholar]
- Bell JK, McCully ME (1970) A histological study of lateral root initiation and development in Zea mays. Protoplasma 70: 179–205 [Google Scholar]
- Benfey PN, Weigel D (2001) Transcriptional networks controlling plant development. Plant Physiol 125: 109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, Bennett M (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Grisafi PL, Fink GR (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Chadwick AV, Burg SP (1967) An explanation of the inhibition of root growth caused by indole-3-acetic acid. Plant Physiol 42: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson P (1998) The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci USA 95: 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker JS, Davies E (2003) Selection of candidate housekeeping controls in tomato plants using EST data. Biotechniques 35: 740–742 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D (2002) The growing world of expansins. Plant Cell Physiol 43: 1436–1444 [DOI] [PubMed] [Google Scholar]
- Delbarre A, Müller P, Imhoff V, Guern J (1996) Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541 [DOI] [PubMed] [Google Scholar]
- De Tomasi JA (1936) Improving the technique of the Feulgen stain. Stain Technol 11: 137–144 [Google Scholar]
- DiDonato RJ, Arbuckle E, Buker S, Sheets J, Tobar J, Totong R, Grisafi P, Fink GR, Celenza JL (2004) Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J 37: 340–353 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colon-Carmona A, Doerner P (2000) Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol 124: 1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit S, Fridlyand J, Speed TP (2002) Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc 97: 77–87 [Google Scholar]
- Erdelska O, Vidovencova Z (1993) Development of adventitious seminal root primordia during embryogenesis. Biologia (Bratisl) 48: 85–88 [Google Scholar]
- Esau K (1965) Plant Anatomy, Ed 2. John Wiley & Sons, New York
- Fleming AJ, Caderas D, Wehrli E, McQueen-Mason S, Kuhlemeier C (1999) Analysis of expansin-induced morphogenesis on the apical meristem of tomato. Planta 208: 166–174 [Google Scholar]
- Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, Palme K (2002. a) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K (2002. b) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Friml J (2003) Auxin transport: shaping the plant. Curr Opin Plant Biol 6: 7–12 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Hadfi K, Speth V, Neuhaus G (1998) Auxin-induced developmental patterns in Brassica juncea embryos. Development 125: 879–887 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hetz W (1996) Charakterisierung einer Wurzelbildungsmutante von Mais und Versuche zur Isolierung der mutierten Genstruktur. PhD thesis. University of Freiburg, Germany
- Hetz W, Hochholdinger F, Schwall M, Feix G (1996) Isolation and characterisation of rtcs a mutant deficient in the formation of nodal roots. Plant J 10: 845–857 [Google Scholar]
- Hochholdinger F, Feix G (1998. a) Early post-embryonic root formation is specifically affected in the maize mutant lrt1. Plant J 16: 247–255 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Feix G (1998. b) Cyclin expression is completely suppressed at the site of crown root formation in the nodal region of the maize root mutant rtcs. Plant Physiol 153: 425–429 [Google Scholar]
- Hochholdinger F, Guo L, Schnable PS (2004. c) Lateral roots affect the proteome of the primary root of maize (Zea mays L.). Plant Mol Biol 56: 397–412 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Feix G (2001) Cooperative action of SLR1 and SLR2 is required for lateral root specific cell-elongation in maize. Plant Physiol 125: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K (2004. a) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9: 42–48 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Woll K, Sauer M, Dembinsky D (2004. b) Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programmes. Ann Bot (Lond) 93: 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML (1995) Specialized zones of development in roots. Plant Physiol 109: 725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen DA (1940) Plant Microtechnique. McGraw-Hill, New York
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM (2003) Laser capture microdissection of cells from plant tissues. Plant Physiol 132: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2005) The role of ARGONAUTE (AGO1) in meristem formation and identity. Dev Biol 280: 504–517 [DOI] [PubMed] [Google Scholar]
- Lally D, Ingmire P, Tong H-Y, He Z-H (2001) Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 13: 1317–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson WE, Hanway JJ (1977) Corn production. In GF Sprague, ed, Corn and Corn Improvement. American Society of Agronomy, Madison, WI, pp 626–669
- Lim J, Helariutta Y, Specht CD, Jung J, Sims L, Bruce WB, Diehn S, Benfey PN (2000) Molecular analysis of the SCARECROW gene in maize reveals a common basis for radial patterning in diverse meristems. Plant Cell 12: 1307–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Xu ZH, Chua NH (1993) Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 5: 621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Jie Yu, He X, Zhang S, Shou H, Wu P (2005) ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J 43: 47–56 [DOI] [PubMed] [Google Scholar]
- Lloret F, Casero PJ (2002) Lateral root initiation in plant roots. In Y Waisel, A Eshel, U Kafkafi, eds, Plant Roots—The Hidden Half. Marcel Dekker, New York, pp 127–155
- McCabe PF, Valentine TA, Forsberg LS, Pennell RI (1997) Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. Plant Cell 9: 2225–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perot-Rechenmann C, Bennett MJ (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within the root apical tissues. EMBO J 8: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London
- Martinez MC, Jorgensen JE, Lawton MA, Lamb CJ, Doerner PW (1992) Spatial pattern of cdc2 expression in relation to meristem activity and cell proliferation during plant development. Proc Natl Acad Sci USA 89: 7360–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell EK, Davies PJ (1975) Evidence for three different systems of movement of indoleacetic acid in intact roots of Phaseolus coccineus. Plant Physiol 33: 290–294 [Google Scholar]
- Muday GK, Haworth P (1994) Tomato root growth, gravitropism, and lateral development: correlation with auxin transport. Plant Physiol Biochem 32: 193–203 [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS (2003) Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302: 81–84 [DOI] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS (2003) Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 3: 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP (2001) Multi drug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MA, Eberhard S, Albersheim P, Darvill AG (2001) Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846–849 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Read ND (1994) The use of fluorescent probes for studies of living plant cells. In N Harris, KJ Oparka, eds, Plant Cell Biology: A Practical Approach. Oxford University Press, New York, pp 27–50
- Rashotte AM, Brady SR, Reed RC, Ante SJ, Muday GK (2000) Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol 122: 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK (1998) Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T (1991) Cell polarity and tissue patterning in plants. Development (Suppl) 1: 83–93 [Google Scholar]
- Scanlon MJ, Henderson DC, Bernstein B (2002) SEMAPHORE1 functions during the regulation of ancestrally duplicated knox genes and polar auxin transport in maize. Development 129: 2663–2673 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Hochholdinger F, Nakazono M (2004) Global expression profiling applied to plant development. Curr Opin Plant Biol 7: 50–56 [DOI] [PubMed] [Google Scholar]
- Schurzmann M, Hild V (1980) Effect of indoleacetic acid, abscisic acid, root tips and coleoptile tips on growth and curvature of maize roots. Planta 150: 32–36 [DOI] [PubMed] [Google Scholar]
- Sprague GF (1977) Corn and Corn Improvement. American Society of Agronomy Publishers, Madison, WI
- Stemmer C, Schwander A, Bauw G, Fojan P, Grasser KD (2002) Protein kinase CK2 differentially phosphorylates maize chromosomal high mobility group B (HMGB) proteins modulating their stability and DNA interactions. J Biol Chem 277: 1092–1098 [DOI] [PubMed] [Google Scholar]
- Sweeter PB, Vatvars A (1976) High-performance liquid chromatographic analyses of abscisic acid in plant extracts. Anal Biochem 71: 68–78 [DOI] [PubMed] [Google Scholar]
- Wang XL, McCully ME, Canny MJ (1994) The branch roots of Zea. IV. The maturation and openness of xylem conduits in first-order branches of soil-grown roots. New Phytol 126: 21–29 [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8: 625–637 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.