Abstract
Jasmonic acid (JA) and methyl jasmonate (MeJA), collectively known as JAs, regulate diverse physiological processes in plants, including the response to wounding. Recent reports suggest that a cyclopentenone precursor of JA, 12-oxo-phytodienoic acid (OPDA), can also induce gene expression. However, little is known about the physiological significance of OPDA-dependent gene expression. We used microarray analysis of approximately 21,500 Arabidopsis (Arabidopsis thaliana) genes to compare responses to JA, MeJA, and OPDA treatment. Although many genes responded identically to both OPDA and JAs, we identified a set of genes (OPDA-specific response genes [ORGs]) that specifically responded to OPDA but not to JAs. ORGs primarily encoded signaling components, transcription factors, and stress response-related genes. One-half of the ORGs were induced by wounding. Analysis using mutants deficient in the biosynthesis of JAs revealed that OPDA functions as a signaling molecule in the wounding response. Unlike signaling via JAs, OPDA signaling was CORONATINE INSENSITIVE 1 independent. These results indicate that an OPDA signaling pathway functions independently of JA/MeJA signaling and is required for the wounding response in Arabidopsis.
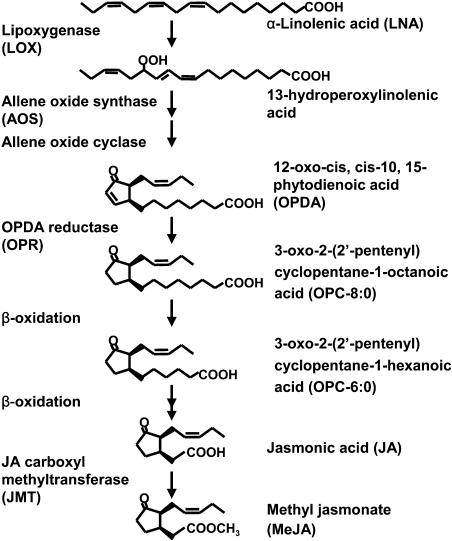
Plants synthesize various fatty acid derivatives having biological activity. Among these, jasmonic acid (JA) and methyl jasmonate (MeJA), collectively known as jasmonates (JAs), are the best characterized. JAs are cyclopentanone compounds derived from linolenic acid via an octadecanoid pathway consisting of several enzymatic steps (Fig. 1). The early steps of this pathway occur in chloroplasts, where linolenic acid is converted to 12-oxo-phytodienoic acid (OPDA) by means of the three enzymes, lipoxygenase, allene oxide synthase (AOS), and allene oxide cyclase (Bell et al., 1995; Laudert et al., 1996; Stenzel et al., 2003). OPDA is subsequently reduced in a cyclopentenone ring by a peroxisome-localized enzyme, 12-oxo-phytodienoic acid reductase 3 (OPR3). The reaction product then undergoes three cycles of β-oxidation in the peroxisome, generating JA (Stintzi and Browse, 2000; Strassner et al., 2002; Reumann et al., 2004).
Figure 1.
Jasmonate biosynthetic pathway.
There have been numerous physiological analyses of the function of JAs. The role of JAs in the response to biotic stresses, such as insect or fungal attack (McConn et al., 1997; Ozawa et al., 2000), and abiotic stresses, such as mechanical wounding (Baldwin et al., 1997; Creelman and Mullet, 1997; Reymond et al., 2000; Richard et al., 2000), has been well documented. JAs also play important roles in anther development (Feys et al., 1994; McConn and Browse, 1996; Xie et al., 1998) and in the regulation of many other plant developmental processes (Creelman and Mullet, 1997). Several components are involved in signaling via JAs. One such component, CORONATINE INSENSITIVE 1 (COI1), contains an F-box motif that has similarity to F-box proteins involved in targeting proteins for removal by ubiquitination (Xie et al., 1998; del Pozo and Estelle, 2000). Thus, COI1 is thought to be involved in protein ubiquitination and degradation regulated by JAs. COI1 is now thought to be a central component of signaling pathways involving JAs.
Recently, OPDA, an intermediate in the biosynthesis of JAs, has been shown to be the biologically active molecule. An opr3 Arabidopsis (Arabidopsis thaliana) mutant that could not produce JA was identified that exhibits delayed anther dehiscence, resulting in male sterility (Sanders et al., 2000; Stintzi and Browse, 2000). Studies using this mutant showed that JA, and not its precursors, is the active signaling molecule that regulates anther development, since the application of JA, but not OPDA, restored fertility (Stintzi and Browse, 2000). However, opr3 plants have shown to be resistant to the dipteran Bradysia impatiens, as are wild-type plants, suggesting that OPDA may be also the active signaling molecule and have a role in the induction of defense response genes (Stintzi et al., 2001). Moreover, using a mini-array system consisting of 150 defense genes, the same investigators showed that OPDA not only up-regulates COI1-dependent genes, which are induced by JA, but also several COI1-independent genes that do not respond to JA (Stintzi et al., 2001). These investigators suggested that OPDA may function cooperatively with JAs to regulate the expression of defense response genes. It was shown that OPDA was more active than JA in eliciting the tendril coiling response in Bryonia dioica (Blechert et al., 1999). More recently, OPDA was shown to induce stomatal opening (Ohashi et al., 2005). However, little is known about OPDA-dependent gene expression or other possible functions of OPDA.
In this study, we analyzed OPDA-dependent gene expression by comparing responses to OPDA and JAs using DNA microarrays covering 80% of the Arabidopsis genome. We identified a group of genes, designated ORGs (OPDA-specific response genes), which responded to OPDA but not to JAs. OPDA treatment of coi1 mutants demonstrated that ORG expression is independent of the COI1-dependent JA signaling pathway. Using the OPDA and JAs biosynthetic mutants aos and opr3 (Fig. 1), we showed that the normal response to wounding was impaired in aos but not in opr3 mutants. These results demonstrate that OPDA is a lipid signal mediator in vivo, regulating ORGs that function during the wounding response.
RESULTS
Identification of Response Genes for OPDA and JAs by Microarray Analysis
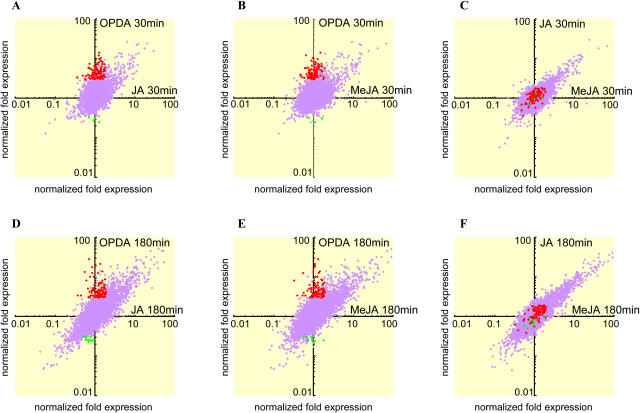
To comprehensively evaluate the effect of OPDA, JA, and MeJA on the transcription of Arabidopsis genes in general, we used an oligonucleotide array covering 21,500 genes, corresponding to 80% of the genome. A time-course experiment was performed to analyze gene expression at 0, 30, and 180 min after treatment with OPDA, JA, or MeJA. After eliminating error spots, as described in “Materials and Methods,” we calculated the normalized fold expression of 20,095 genes for each treatment. Scatter plots comparing expression between the three treatments were drawn on a logarithmic scale using the normalized fold expression of each gene. A higher correlation between JA and MeJA treatment was observed at 30 and 180 min (0.64 and 0.85, respectively) than that observed between OPDA and JAs treatment. Most of the genes that responded to JA treatment responded equivalently to MeJA treatment. The correlation coefficients of OPDA versus JA and OPDA versus MeJA were, respectively, 0.48 and 0.47 at 30 min and 0.77 and 0.76 at 180 min. As shown in Figure 2, A, B, D, and E, spots corresponding to up- or down-regulated genes (red or green spots, respectively) were observed specifically along axial lines. This result demonstrates that a group of genes responded to OPDA but not to JA or MeJA; these genes were termed OPDA-specific response genes, or ORGs. The analysis also revealed a large number of genes that responded to OPDA as well as JA and MeJA. From this comprehensive gene expression analysis, we concluded that OPDA regulates the expression of a distinct set of genes that are not regulated by JAs, in addition to genes that respond to all three compounds.
Figure 2.
Scatter plots of OPDA and JAs response genes. A and D, Normalized fold expression at 30 and 180 min after OPDA treatment plotted against the normalized fold expression at 30 and 180 min after JA treatment. B and E, Normalized fold expression of OPDA versus MeJA treatment at 30 and 180 min. C and F, Normalized fold expression of JA versus MeJA treatment at 30 and 180 min. Red spots denote genes whose expression was induced more than 3-fold by OPDA treatment at 30 min (A, B, and C) or 180 min (D, E, and F), and less than 2-fold by JAs at 30 and 180 min. Green spots denote genes whose expression was repressed more than 3-fold by OPDA treatment after 30 min (A, B, and C) or 180 min (D, E, and F), and responded less than 2-fold by treatment with JAs at 30 and 180 min.
Validation of ORGs by Northern-Blot Analysis
Based on the microarray results, ORGs were defined as follows (for a detailed description, see “Materials and Methods”). Up-regulated ORGs were induced more than 3-fold by OPDA but less than 2-fold by JA and MeJA (Table I; Supplemental Table I). Down-regulated ORGs were repressed more than 3-fold by OPDA but less than 2-fold by JA and MeJA (Table II). Genes corresponding to 172 loci (157 induced and 15 repressed) satisfied these criteria. For identification of ORGs, we also applied hierarchical clustering for 1,078 genes whose expression responded more than 3-fold to any of three treatments. By this clustering, 214 ORG genes were also found (193 induced 21 repressed; see Supplemental Table II and Supplemental Fig. 1). Basically, most of genes identified in former criteria were included in latter criteria (144 induced and 11 repressed). We therefore used the gene list obtained by the former criteria for further analysis.
Table I.
List of up-regulated ORGs
Up-regulated ORGs are shown. Microarray data for normalized fold expression are calculated from two biologically independent experiments. OPDA, JA, and MeJA were treated for plants grown in liquid culture, whereas wounding was performed for plants grown in solid medium. Normalized fold expression more than 3 is in bold. See also Supplemental Table I.
| Normalized Fold Expression
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Annotation | Gene Name | OPDA
|
JA
|
MeJA
|
Wounding
|
||||
| 30 min | 180 min | 30 min | 180 min | 30 min | 180 min | 30 min | 180 min | ||
| Group 1: Signaling Component | |||||||||
| Phophatase | |||||||||
| Protein phosphatase 2C (PP2C), putative | At3g27140 | 3.0 | 1.5 | 1.4 | 1.3 | 0.9 | 0.5 | 1.6 | 0.4 |
| Protein kinase | |||||||||
| Protein kinase family (OXI1) | At3g25250 | 12.1 | 3.1 | 1.3 | 1.8 | 0.6 | 0.9 | 10.0 | 1.1 |
| Protein kinase family | At4g25390 | 1.9 | 3.5 | 1.0 | 1.5 | 1.1 | 1.8 | 2.2 | 2.2 |
| Protein kinase family (MAPKKK18) | At1g05100 | 3.7 | 3.0 | 1.1 | 1.1 | 1.0 | 1.6 | 5.7 | 2.6 |
| Protein kinase, putative | At2g05940 | 4.2 | 1.6 | 1.2 | 1.5 | 1.3 | 1.2 | 1.5 | 0.7 |
| Receptor-related protein kinase like | At5g25930 | 9.6 | 7.1 | 1.3 | 1.4 | 1.3 | 1.3 | 7.4 | 0.9 |
| Calcium-binding protein | |||||||||
| Calcium-binding EF-hand family protein | At4g27280 | 3.5 | 0.8 | 0.6 | 0.3 | 0.7 | 0.4 | 0.4 | 0.4 |
| Calcium-binding EF-hand family protein | At5g39670 | 6.0 | 2.8 | 1.1 | 0.8 | 1.4 | 0.8 | 1.5 | 1.1 |
| Calcium-binding protein, putative (PBP1) | At5g54490 | 3.7 | 0.7 | 0.5 | 0.5 | 0.5 | 0.4 | 1.0 | 0.4 |
| Calcium-transporting ATPase, putative | At3g63380 | 1.2 | 3.4 | 0.7 | 1.1 | 1.1 | 1.0 | 2.4 | 8.1 |
| Calmodulin-binding protein | At5g26920 | 4.4 | 0.9 | 1.0 | 0.6 | 0.8 | 0.7 | 1.1 | 0.3 |
| Calmodulin-related protein, putative | At3g01830 | 8.1 | 0.9 | 1.3 | 0.6 | 1.1 | 0.7 | 12.2 | 1.0 |
| Calmodulin-related protein, putative | At5g42380 | 5.5 | 5.0 | 1.1 | 0.7 | 1.2 | 0.8 | 6.3 | 1.3 |
| Other signaling component | |||||||||
| F-box protein family | At1g61340 | 4.4 | 0.9 | 1.0 | 0.6 | 1.1 | 0.7 | 6.9 | 1.2 |
| Transcription factor inhibitor I κ B like | At5g45110 | 3.4 | 1.4 | 1.4 | 1.0 | 1.1 | 0.9 | 2.2 | 0.7 |
| Response regulator 2 (ATRR2) | At3g48100 | 3.7 | 0.6 | 1.5 | 0.7 | 1.5 | 0.9 | 1.0 | 0.3 |
| SigA-binding protein (SIB1) | At3g56710 | 7.5 | 1.5 | 1.2 | 0.7 | 1.1 | 0.6 | 1.2 | 0.7 |
| Group 2: Transcription Factor | |||||||||
| AP2-EREB-type transcription factor | |||||||||
| AP2 domain transcription factor TINY, putative | At1g22810 | 13.9 | 1.5 | 1.3 | 1.2 | 1.5 | 1.1 | 58.8 | 5.2 |
| AP2 domain transcription factor, putative | At1g19210 | 6.1 | 1.2 | 1.0 | 1.0 | 1.1 | 1.2 | 1.3 | 0.3 |
| C-repeat/DRE-binding protein, putative | At5g52020 | 2.1 | 6.3 | 1.4 | 1.0 | 0.9 | 0.8 | 1.9 | 1.0 |
| DRE-binding protein (DREB2A) | At5g05410 | 7.3 | 4.4 | 0.7 | 0.9 | 1.6 | 1.9 | 7.9 | 2.8 |
| Ethylene responsive element binding factor 5 (AtERF5) | At5g47230 | 4.3 | 0.9 | 0.9 | 0.5 | 0.9 | 0.5 | 0.3 | 0.3 |
| Transcription factor TINY, putative | At1g33760 | 3.6 | 0.6 | 0.9 | 0.7 | 1.0 | 0.5 | 0.6 | 0.1 |
| WRKY-type transcription factor | |||||||||
| WRKY family transcription factor (WRKY30) | At5g24110 | 5.8 | 1.0 | 0.7 | 0.5 | 0.7 | 0.5 | 9.7 | 0.3 |
| WRKY family transcription factor (WRKY46) | At2g46400 | 3.7 | 0.7 | 1.0 | 0.6 | 1.1 | 0.8 | 1.3 | 0.5 |
| WRKY family transcription factor (WRKY53) | At4g23810 | 3.1 | 0.2 | 0.4 | 0.2 | 0.3 | 0.2 | 2.0 | 0.4 |
| MYB-type transcription factor | |||||||||
| Myb family transcription factor (MYB15) | At3g23250 | 5.1 | 1.5 | 1.8 | 1.5 | 1.5 | 1.2 | 34.6 | 4.5 |
| Transcription factor (MYB4) | At4g38620 | 5.6 | 0.9 | 0.7 | 1.1 | 0.7 | 0.8 | 1.0 | 0.9 |
| Zinc-finger-type transcription factor | |||||||||
| C2H2-type zinc-finger protein family | At2g28710 | 2.6 | 4.5 | 0.7 | 0.8 | 0.7 | 0.7 | 0.3 | 0.7 |
| C2H2-type zinc-finger protein related | At2g37430 | 10.2 | 1.1 | 1.4 | 1.5 | 1.0 | 1.0 | 4.5 | 0.7 |
| C2H2-type zinc-finger protein related (FZF) | At2g24500 | 1.2 | 3.1 | 0.9 | 1.1 | 0.9 | 1.4 | 2.0 | 4.7 |
| CONSTANS B-box zinc-finger family protein | At2g47890 | 2.2 | 3.1 | 1.0 | 1.0 | 0.9 | 0.9 | 1.3 | 1.5 |
| CONSTANS B-box zinc-finger family protein | At3g21150 | 3.3 | 1.0 | 0.9 | 0.7 | 0.8 | 1.2 | 1.4 | 0.5 |
| RING-H2 zinc-finger protein related | At5g27420 | 3.4 | 0.5 | 0.4 | 0.3 | 0.5 | 0.3 | 1.3 | 0.2 |
| Salt-tolerance zinc-finger protein (ZAT10) | At1g27730 | 7.0 | 1.7 | 1.2 | 0.7 | 1.3 | 0.7 | 1.6 | 0.7 |
| Zinc-finger protein (PMZ) | At3g28210 | 3.7 | 17.4 | 0.7 | 0.8 | 0.9 | 1.0 | 14.5 | 23.6 |
| Zinc-finger protein Zat12 (RHL41) | At5g59820 | 6.4 | 3.5 | 1.0 | 1.2 | 1.4 | 1.8 | 1.9 | 1.1 |
| Zinc-finger-related protein | At3g46080 | 5.1 | 1.7 | 0.9 | 0.7 | 0.8 | 1.0 | 2.9 | 1.7 |
| Other-type transcription factor | |||||||||
| bHLH protein family | At2g28160 | 4.0 | 3.7 | 1.7 | 1.5 | 1.0 | 1.4 | 1.5 | 1.1 |
| Transcription factor GT-3a | At5g01380 | 1.8 | 4.0 | 1.0 | 1.2 | 0.9 | 0.6 | 15.7 | 12.3 |
| Group 3: Stress Response | |||||||||
| Pathogenesis related | |||||||||
| Disease resistance protein (TIR class), putative | At2g20145 | 3.2 | 1.5 | 1.3 | 0.9 | 1.0 | 0.9 | 1.6 | 2.0 |
| Disease resistance protein (TIR-NBS class), putative | At1g66090 | 5.3 | 0.9 | 0.8 | 0.6 | 0.9 | 0.7 | 2.4 | 0.5 |
| Disease resistance protein family (LRR) | At3g05360 | 1.0 | 3.3 | 0.9 | 1.3 | 1.0 | 1.1 | 1.7 | 3.9 |
| Seven transmembrane MLO protein family (MLO6) | At1g61560 | 3.1 | 3.9 | 1.6 | 1.8 | 1.8 | 1.4 | 11.0 | 0.7 |
| Gly-rich protein | At3g04640 | 4.1 | 3.4 | 1.2 | 0.9 | 1.1 | 0.9 | 1.0 | 0.7 |
| Stress-related protein related | At1g67360 | 3.1 | 2.0 | 1.1 | 0.8 | 1.1 | 0.9 | 3.6 | 1.6 |
| Thaumatin family | At4g36010 | 2.3 | 4.5 | 1.6 | 1.6 | 1.2 | 1.8 | 14.1 | 3.9 |
| Cell wall modification | |||||||||
| BON1-associated protein 1 (BAP1) | At3g61190 | 3.3 | 2.5 | 1.4 | 1.8 | 0.8 | 0.9 | 1.8 | 0.8 |
| Cellulose synthase family (ATCSLE1) | At1g55850 | 1.6 | 3.1 | 1.0 | 1.7 | 0.8 | 1.8 | 1.1 | 1.1 |
| Glucosyltransferase related | At2g15480 | 4.4 | 6.7 | 0.9 | 1.2 | 1.1 | 1.7 | 7.2 | 4.9 |
| Glycosyltransferase family | At1g73880 | 0.9 | 3.6 | 0.9 | 1.8 | 0.8 | 1.3 | 0.9 | 1.0 |
| UDP-glycosyltransferase family | At1g07260 | 1.2 | 3.2 | 0.8 | 1.1 | 1.0 | 1.1 | 0.5 | 0.4 |
| Cinnamyl-alcohol dehydrogenase (CAD) family | At1g09500 | 1.5 | 7.2 | 0.6 | 1.3 | 0.5 | 1.3 | 0.5 | 1.8 |
| Xyloglucan endotransglycosylase (TCH4) | At5g57560 | 3.5 | 0.4 | 0.8 | 0.3 | 0.8 | 0.4 | 0.9 | 0.5 |
| Heat-shock response | |||||||||
| 17.6-kD heat-shock protein (AA 1-156) | At1g53540 | 6.3 | 13.5 | 1.5 | 1.5 | 1.3 | 1.4 | 20.6 | 32.3 |
| Class I heat-shock protein(HSP 17.4) | At3g46230 | 3.3 | 12.4 | 1.3 | 1.2 | 0.8 | 1.3 | 3.5 | 13.8 |
| Class II heat-shock protein | At5g12020 | 3.7 | 12.5 | 0.8 | 0.8 | 0.9 | 0.5 | 2.0 | 10.0 |
| Endomembrane-localized small heat-shock protein | At4g10250 | 5.9 | 5.4 | 1.8 | 1.6 | 1.1 | 1.2 | 1.0 | 4.9 |
| Heat-shock protein 17.6A (AT-HSP17.6A) | At5g12030 | 4.4 | 13.2 | 1.0 | 0.6 | 0.8 | 1.0 | 2.7 | 15.1 |
| Heat-shock protein family | At1g52560 | 8.7 | 28.1 | 1.4 | 0.7 | 0.9 | 1.3 | 1.6 | 18.2 |
| Heat-shock protein family | At5g37670 | 4.6 | 3.0 | 1.3 | 0.8 | 0.8 | 0.9 | 2.6 | 6.0 |
| Heat-shock protein family (HSP18.2) | At5g59720 | 2.4 | 4.7 | 1.3 | 1.8 | 1.1 | 0.9 | 1.7 | 12.1 |
| Heat-shock protein hsp70b | At1g16030 | 4.1 | 10.0 | 1.3 | 0.8 | 1.2 | 1.3 | 5.5 | 10.5 |
| Heat-shock protein hsp70t-2 | At2g32120 | 3.5 | 3.9 | 1.6 | 0.8 | 1.2 | 1.2 | 5.3 | 10.6 |
| Heat-shock protein, putative | At1g59860 | 5.4 | 4.8 | 1.4 | 0.9 | 1.3 | 1.3 | 20.2 | 8.8 |
| Heat-shock protein, putative | At2g20560 | 4.1 | 7.2 | 1.1 | 1.5 | 0.8 | 1.9 | 7.5 | 5.9 |
| Mitochondrion-localized small heat-shock protein | At4g25200 | 5.2 | 2.2 | 1.1 | 0.6 | 1.0 | 1.8 | 1.6 | 7.3 |
| Small heat-shock protein related | At2g19310 | 2.3 | 3.0 | 0.9 | 1.2 | 0.8 | 1.6 | 1.3 | 1.2 |
| Small heat-shock protein, chloroplast precursor (HSP21) | At4g27670 | 5.3 | 3.0 | 1.7 | 1.5 | 1.2 | 1.4 | 1.6 | 14.7 |
| Oxidative burst stress response | |||||||||
| Glutathione peroxidase, putative (AtGPX6) | At4g11600 | 1.5 | 3.2 | 0.8 | 1.1 | 1.0 | 1.4 | 3.0 | 4.5 |
| Glutathione S-conjugate ABC transporter (AtMRP1) | At1g30400 | 1.1 | 3.8 | 0.8 | 1.2 | 0.9 | 1.2 | 1.1 | 1.7 |
| Glutathione transferase, putative (GST21) | At2g29470 | 1.5 | 4.2 | 1.0 | 1.6 | 1.2 | 1.1 | 1.0 | 0.7 |
| Glutathione transferase, putative (GST6) | At2g47730 | 1.7 | 5.1 | 0.8 | 1.0 | 1.0 | 1.8 | 2.3 | 3.4 |
| Iron superoxide dismutase (FSD1) | At4g25100 | 3.1 | 2.5 | 1.0 | 0.9 | 0.9 | 1.0 | 1.0 | 0.6 |
| Group 4: Secondary Metabolism | |||||||||
| FAD-linked oxidoreductase family | At1g30700 | 1.6 | 7.9 | 0.4 | 0.6 | 0.3 | 0.3 | 4.0 | 3.7 |
| FAD-linked oxidoreductase family | At4g20860 | 3.7 | 6.6 | 1.1 | 1.4 | 1.1 | 1.1 | 6.0 | 2.3 |
| FAD-linked oxidoreductase family | At5g44360 | 2.5 | 4.5 | 1.1 | 1.4 | 1.0 | 0.7 | 1.0 | 1.1 |
| Cytochrome P450, putative (CYP72A15) | At3g14690 | 1.8 | 4.0 | 1.0 | 1.3 | 1.1 | 1.7 | 1.4 | 3.8 |
| Cytochrome P450, putative (CYP707A3) | At5g45340 | 3.2 | 0.3 | 0.5 | 0.3 | 1.0 | 0.9 | 0.5 | 0.2 |
| Group 5: Hormonal Response | |||||||||
| Auxin | |||||||||
| Auxin-induced protein family | At3g09870 | 4.5 | 0.7 | 1.0 | 0.8 | 1.1 | 0.9 | 0.6 | 0.4 |
| Auxin-induced protein family | At5g35735 | 4.1 | 3.4 | 1.0 | 1.0 | 0.9 | 1.0 | 6.0 | 1.8 |
| JA | |||||||||
| 12-Oxo-phytodienoate reductase (OPR2) | At1g76690 | 2.7 | 6.2 | 1.1 | 1.7 | 1.2 | 1.8 | 3.1 | 6.7 |
| Abscisic acid | |||||||||
| ABA-responsive protein related | At5g13200 | 2.8 | 3.2 | 1.6 | 1.1 | 1.6 | 1.3 | 8.6 | 4.1 |
| Group 6: Transporter | |||||||||
| ABC transporter family protein | At1g15520 | 1.7 | 18.7 | 0.9 | 1.0 | 0.9 | 1.1 | 3.0 | 16.1 |
| Ammonium transporter, putative (ATAMT1;2) | At1g64780 | 1.6 | 4.2 | 1.5 | 1.9 | 1.3 | 1.3 | 1.6 | 1.2 |
| High-affinity sulfate transporter related (SEL1) | At1g78000 | 0.7 | 10.1 | 0.9 | 0.9 | 1.2 | 1.1 | 0.4 | 1.2 |
| Mitochondrial carrier protein family | At2g22500 | 4.7 | 1.1 | 1.4 | 1.0 | 1.3 | 0.8 | 4.1 | 0.8 |
| Mitochondrial carrier protein family | At4g24570 | 3.9 | 0.4 | 0.7 | 0.3 | 0.5 | 0.3 | 1.1 | 0.4 |
| Nucleoporin 98-related protein | At1g59660 | 4.7 | 0.9 | 1.7 | 0.7 | 1.6 | 1.0 | 0.8 | 0.5 |
| Zinc transporter, putative (MTPa2) | At3g58810 | 1.2 | 3.6 | 0.8 | 1.4 | 1.0 | 1.2 | 2.3 | 2.7 |
| Others | |||||||||
| 60S ribosomal protein L10 (RPL10C) | At1g66580 | 1.2 | 3.8 | 1.0 | 1.3 | 0.9 | 1.6 | 3.5 | 10.2 |
| AAA-type ATPase family | At3g50930 | 6.3 | 1.2 | 1.1 | 0.8 | 1.7 | 1.1 | 1.3 | 0.7 |
| Carbonic anhydrase related | At2g28210 | 3.2 | 4.2 | 1.2 | 0.7 | 1.0 | 0.9 | 1.7 | 2.3 |
| DnaJ protein family | At3g08970 | 4.0 | 5.0 | 0.9 | 1.7 | 1.1 | 1.6 | 2.4 | 4.3 |
| Glycosyl hydrolase family 1 | At2g44460 | 0.9 | 6.1 | 1.3 | 1.7 | 0.9 | 1.3 | 1.3 | 1.6 |
| Glycosyl hydrolase family 1, β-glucosidase (DIN2) | At3g60140 | 1.2 | 3.1 | 1.5 | 1.3 | 1.0 | 1.3 | 0.2 | 0.3 |
| Heavy-metal-associated domain-containing protein | At5g52760 | 3.2 | 0.8 | 1.0 | 0.5 | 1.0 | 0.7 | 1.5 | 0.8 |
| Hydrolase, α/β fold family | At4g24160 | 4.2 | 4.1 | 1.2 | 1.3 | 1.1 | 1.8 | 6.1 | 3.2 |
| Lemir (miraculin) related | At1g17860 | 1.8 | 3.4 | 1.3 | 1.5 | 1.1 | 0.9 | 6.3 | 2.7 |
| Lys decarboxylase-related protein | At5g06300 | 1.3 | 5.3 | 0.8 | 1.7 | 0.8 | 1.1 | 1.0 | 1.4 |
| Nitrilase 4 (sp P46011) (NIT4) | At5g22300 | 1.2 | 3.9 | 1.1 | 1.7 | 1.0 | 1.9 | 1.2 | 1.8 |
| Phospholipid/glycerol acyltransferase family | At4g01950 | 3.2 | 0.7 | 0.7 | 0.7 | 0.6 | 0.5 | 1.9 | 0.6 |
| Pyridine nucleotide-disulfide oxidoreductase family | At3g44190 | 3.6 | 4.3 | 1.1 | 1.1 | 1.0 | 1.2 | 12.7 | 3.2 |
| Transferase family | At5g07870 | 7.0 | 4.7 | 1.7 | 1.6 | 1.1 | 1.5 | 2.0 | 1.8 |
| Pyridine nucleotide-disulphide oxidoreductase family | At5g22140 | 6.6 | 20.9 | 1.0 | 1.4 | 0.9 | 1.1 | 6.9 | 24.2 |
| Aldo/keto reductase family | At1g60730 | 1.1 | 4.6 | 0.9 | 1.4 | 0.9 | 1.8 | 1.8 | 4.1 |
| Aldo/keto reductase family | At1g60750 | 1.4 | 5.8 | 0.9 | 1.2 | 1.4 | 1.2 | 2.9 | 2.1 |
| Ubiquinol-cytochrome-c reductase-related protein | At5g25450 | 2.5 | 5.3 | 1.2 | 1.5 | 0.8 | 1.3 | 5.4 | 8.9 |
| Expressed and putative proteins are 49 | |||||||||
Table II.
List of down-regulated ORGs
Down-regulated ORGs are shown. Microarray data for normalized fold expression are calculated from two biologically independent experiments. OPDA, JA, and MeJA were treated for plants grown in liquid culture, whereas wounding was performed for plants grown in solid medium. Normalized fold expression less than 0.33 is in bold.
| Normalized Fold Expression
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Annotation | Gene Name | OPDA
|
JA
|
MeJA
|
Wounding
|
||||
| 30 min | 180 min | 30 min | 180 min | 30 min | 180 min | 30 min | 180 min | ||
| Group 1: Signaling Component | |||||||||
| Protein kinases | |||||||||
| Ser/Thr kinase-like protein | At4g23190 | 0.9 | 0.3 | 1.1 | 0.9 | 1.1 | 0.7 | 2.0 | 0.2 |
| Phosphatidylinositol 3- and 4-kinase family | At1g64460 | 0.6 | 0.3 | 0.5 | 0.5 | 0.8 | 0.7 | 0.5 | 0.9 |
| Group 2: Transcription Factor | |||||||||
| AP2-EREB-type TF | |||||||||
| C-repeat/DRE binding factor 1 (CBF1) (DREB1B) | At4g25490 | 0.3 | 0.6 | 0.7 | 0.6 | 0.7 | 0.8 | 0.5 | 0.3 |
| Group 3: Stress Response | |||||||||
| Oxidative burst stress response | |||||||||
| Copper/zinc superoxide dismutase (CSD2) | At2g28190 | 0.2 | 0.4 | 1.3 | 1.6 | 1.1 | 1.0 | 0.6 | 0.7 |
| Copper/zinc superoxide dismutase, putative | At1g12520 | 0.3 | 0.4 | 0.9 | 1.1 | 1.0 | 0.9 | 0.3 | 0.7 |
| Peroxidase, putative | At1g34510 | 0.4 | 0.2 | 0.8 | 0.9 | 1.3 | 1.8 | 1.0 | 1.0 |
| Peroxidase, putative | At5g22410 | 0.4 | 0.2 | 0.7 | 0.8 | 1.3 | 1.1 | 1.8 | 1.0 |
| Peroxidase, putative | At1g05240 | 0.5 | 0.3 | 0.7 | 0.6 | 0.7 | 0.6 | 0.9 | 1.2 |
| Cell wall modification | |||||||||
| Expansin, putative | At1g62980 | 0.4 | 0.3 | 0.7 | 0.6 | 1.0 | 1.0 | 0.5 | 0.7 |
| Xyloglucan endotransglycosylase, putative | At4g28850 | 0.4 | 0.3 | 0.7 | 0.6 | 1.2 | 0.9 | 0.6 | 0.6 |
| Others | |||||||||
| Plasma membrane H+-ATPase like | At3g60330 | 0.7 | 0.3 | 1.0 | 0.8 | 1.0 | 0.9 | 1.1 | 1.0 |
| Pro-rich protein | At3g62680 | 0.5 | 0.3 | 0.6 | 0.7 | 1.0 | 1.0 | 1.1 | 1.0 |
| Ribosomal protein L13 homolog | At3g48130 | 0.3 | 1.0 | 1.0 | 0.5 | 1.6 | 0.7 | 1.0 | 1.0 |
| Glycosyl hydrolase family 9 | At1g48930 | 0.5 | 0.2 | 0.7 | 0.7 | 0.9 | 1.0 | 0.7 | 0.4 |
| Invertase/pectin methylesterase inhibitor family | At5g62340 | 0.8 | 0.3 | 0.7 | 0.8 | 1.0 | 0.7 | 1.0 | 0.7 |
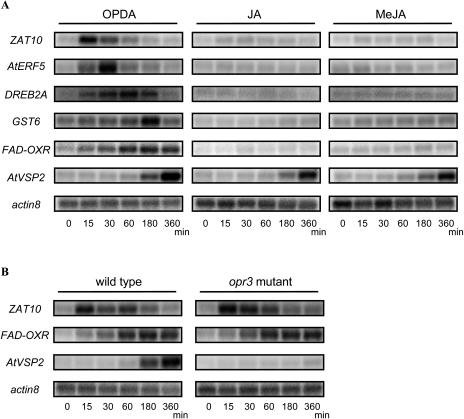
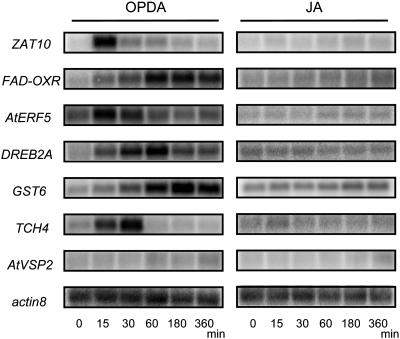
To verify that OPDA modulates ORG expression independently of JAs, we assessed the expression profile of selected putative ORGs by northern blotting. Consistent with the microarray data, northern analysis showed that the expression of ZAT10 (At1g27730), AtERF5 (At5g47230), DREB2A (At5g05410), GST6 (At2g47730), and FAD-OXR (FAD-linked oxidoreductase; At4g20860) increased upon OPDA treatment but did not respond to JAs (Fig. 3A). The expression of AtVSP2 (At5g24770), a known jasmonate-responsive gene (Berger et al., 1995), increased similarly in response to each treatment. ZAT10, AtERF5, and DREB2A are known transcription factors related to the response to various abiotic stresses (Liu et al., 1998; Fujimoto et al., 2000; Ohta et al., 2001; Lee et al., 2002). GST6 and FAD-OXR are also induced in stress responses (Dittrich and Kutchan, 1991; Chen et al., 1996). Expression of the transcription factor genes ZAT10 and AtERF5 was detected within 30 min of OPDA treatment and decreased thereafter. On the other hand, expression of GST6 and FAD-OXR was induced at a later time (around 180 min).
Figure 3.
Northern-blot analysis of OPDA response genes in wild-type and opr3 mutant plants. Five micrograms of total RNA (5 μg) were loaded per lane, and the blot was hybridized with the indicated probes; actin8 served as a loading control. A, Ten-day-old wild-type (Col) plants treated with 30 μm OPDA, JA, or MeJA for 0 (control), 15, 30, 60, 180, or 360 min. B, Ten-day-old wild-type (Ws) and opr3 mutant (Ws background) plants treated with 30 μm OPDA for 0, 15, 30, 60, 180, or 360 min.
To confirm the induction of ORGs, we used an opr3 mutant, which lacks an enzyme catalyzing the reduction of the cyclopentenone ring of OPDA (Fig. 1) and thus is deficient in JAs (Stintzi and Browse, 2000; Stintzi et al., 2001). As shown in Figure 3B, the induction of ZAT10 and FAD-OXR by OPDA was equivalent in the opr3 mutant and wild-type strain (Wassilewskija [Ws] background). Similar results were obtained for AtERF5, DREB2A, and GST6 (data not shown). These results demonstrate that OPDA, but not JAs, regulates the expression of these representative ORGs. In contrast, the induction of AtVSP2 by OPDA was abolished in the opr3 mutant, but was induced by OPDA and JAs in wild-type plants (Fig. 3, A and B), indicating that AtVSP2 responds specifically to JAs. This result also showed that exogenous OPDA could be converted to JA within 180 min under these experimental conditions, given that AtVSP2 expression in OPDA-treated wild-type plants became evident after this time.
Functional Classification of ORGs
Of the 172 ORGs, 123 (70%) were annotated using locus annotation from The Arabidopsis Information Resource (Rhee et al., 2003). Forty-nine (30%) were “hypothetical,” “expressed,” or “putative” genes (Supplemental Table I). We classified the up-regulated ORGs into six groups (Table I) and the down-regulated ORGs into three groups (Table II). Among the up-regulated ORGs, Group 1 comprises genes related to signal transduction, including kinases, calcium-dependent signaling components, and calmodulins. The induction of genes in this group occurred within 30 min of treatment and their expression was transient. Group 2 comprises transcription factors and includes AP2/EREB (Gutterson and Reuber, 2004), WRKY (Eulgem et al., 2000), MYB (Stracke et al., 2001), and zinc-finger types (Tague and Goodman, 1995) of DNA-binding protein families. These genes responded rapidly and transiently to OPDA. Groups 1 and 2 contain 40 genes, accounting for 26% of the up-regulated ORGs we identified. Group 3 contains genes for various proteins involved in stress responses. For example, AtGPX6 is induced under conditions of oxidative stress (Rodriguez Milla et al., 2003), and TCH4 is involved in plant morphogenetic responses to environmental changes (Xu et al., 1995). Group 3 also contains a large number of heat shock protein genes that were expressed at 180 min rather than at 30 min, in contrast to the genes in Groups 1 and 2, which were mainly expressed at 30 min. Groups 4, 5, and 6 contain genes related to secondary metabolism, plant hormones, and transport systems for metals and ions, respectively. We also identified a number of other ORGs that do not fit into the categories of Groups 1 to 6 (see Table I, Others). Compared with the up-regulated ORGs, we identified few that were down-regulated (Table II).
We also identified genes that responded to JAs (JRGs), according to the criteria described in “Materials and Methods.” Briefly, genes that were induced or repressed more than 3-fold by both JA and MeJA were selected as JRGs. A total of 449 loci (371 induced and 78 repressed) satisfied these criteria (Supplemental Table III). This group mainly included genes involved in metabolic pathways, such as ascorbate and glutathione metabolism, JA biosynthesis, and indole glucosinolate synthesis, which are known to be regulated by JAs (Brader et al., 2001; Sasaki et al., 2001; Turner et al., 2002; Sasaki-Sekimoto et al., 2005).
ORG Function in the Wounding Response
Although the exact role of OPDA is unclear since it is a precursor of JA, it is expected to have a related physiological function. JAs have a pivotal role in the wounding response (Reymond and Farmer, 1998). To address the role of OPDA in this response, we performed microarray analysis using RNAs isolated from mechanically wounded plants at 0, 30, and 180 min after wounding. Forty-five percent of ORGs and 46% of JRGs responded to wounding treatment. These results suggest that OPDA is also a signaling molecule involved in the response to wounding.
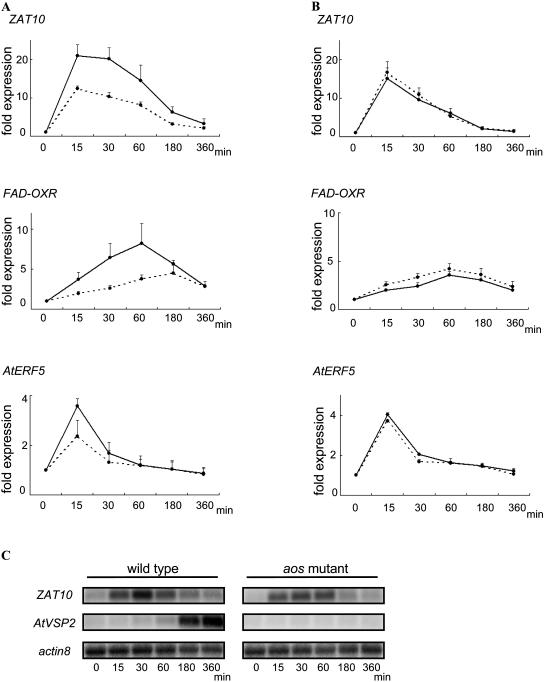
To determine whether OPDA is required to induce expression of ORGs in wounded plants in vivo, the expression of ZAT10, FAD-OXR, and AtERF5 in mechanically wounded plants was quantified for two JA biosynthesis mutants (Fig. 1): opr3, which has a defect in JA biosynthesis, and aos, which lacks both OPDA and JA (Stintzi and Browse, 2000; Park et al., 2002). We used northern blotting to quantify ZAT10, FAD-OXR, AtERF5, and AtVSP2 mRNA expression in wounded aos and opr3 mutants and plants with the corresponding wild-type backgrounds Columbia (Col) and Ws. In aos mutants, expression levels of ZAT10, FAD-OXR, and AtERF5 were reduced by one-half compared with wild-type (Col) plants (Fig. 4A). However, expression levels in opr3 mutants and wild-type plants (Ws) did not differ (Fig. 4B). AtVSP2 expression was detected in wild-type plants but not in either mutant (data not shown). Thus, it is clear that OPDA signaling induces ZAT10, FAD-OXR, and AtERF5 expression in wounded plants.
Figure 4.
Expression of ORGs in wounded wild-type plants and in mutants deficient in jasmonate biosynthesis. A and B, Leaves of 21-d-old wild-type (Col) and aos mutant (Col background) plants (A) or wild-type (Ws) and opr3 mutant (Ws background) plants (B) were wounded several times across the mid-vein with tweezers. RNA was isolated 0 (control), 15, 30, 60, 180, or 360 min after wounding. Five micrograms of total RNA were loaded per lane, and the blots were hybridized with the indicated probes. actin8 served as a loading control. Graphs show fold expression of ZAT10, FAD-OXR, and AtERF5. Fold expression was calculated as the ratio of wounded-to-control band intensity. Solid lines indicate wild-type plants Col (A) and Ws (B), and dotted lines indicate the mutants aos (A) and opr3 (B). Average values ± SEM represent the average of four (ZAT10 and FAD-OXR) or three (AtERF5) independent experiments. C, Ten-day-old wild-type (Col) plants and aos mutants (Col background) were treated with 30 μm linolenic acid for 0 (control), 15, 30, 60, 180, or 360 min. Northern blotting was performed as described above.
Figure 4A also indicates that, even in the aos mutant, about one-half of the wound-induced expression of ZAT10, FAD-OXR, and AtERF5 was not abolished. Recently, various oxylipins were shown to function as signaling molecules that induce gene expression (Gerwick et al., 1991; Howe and Schilmiller, 2002; Weber, 2002). Since most oxylipins are derived from linolenic acid, it would be expected that the residual response to wounding seen in the aos mutant is mediated by oxylipins derived from linolenic acid. Therefore, we treated wild-type (Col) and mutant (aos) plants with linolenic acid and analyzed the expression levels of ZAT10 and AtVSP2 by northern blotting. ZAT10 and AtVSP2 mRNAs accumulated in Col (Fig. 4C). In aos, the ZAT10 mRNA level was reduced compared with wild-type plants (Fig. 4C), and expression of AtVSP2 expression was completely lost.
Expression of ORGs Is Independent of the COI1 Pathway
COI1 is required for the response to JAs (Feys et al., 1994). Because the structure of OPDA is similar to that of JA, responses to OPDA may also be regulated via the COI1 pathway. Therefore, we investigated whether COI1 is required for the induction of ORGs using a coi1-16 mutant (Ellis and Turner, 2002). After application of OPDA or JA to coi1-16, the ORG expression profile was investigated by northern blotting. ZAT10, FAD-OXR, AtERF5, DREB2A, GST6, and TCH4 (At5g57560) expression increased after OPDA treatment (Fig. 5). On the other hand, the induction of AtVSP2 by OPDA or JA was abolished in this mutant. These results indicate that COI1 is not involved in the regulation of ORGs.
Figure 5.
Expression of ORGs in coi1 mutants. Ten-day-old coi1-16 mutants (Col background) were treated with 30 μm OPDA or JA, and total RNA was isolated after 0 (control), 15, 30, 60, 180, or 360 min. For northern blots, 5 μg RNA was loaded per lane, and blots were hybridized with the indicated probes; actin8 served as a loading control.
OPC8:0 and OPC6:0 Also Induce the Expression of ORGs
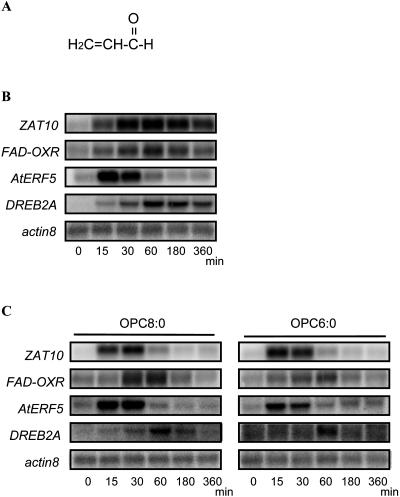
OPDA and JA have a similar structural backbone, a consequence of the fact that OPDA is the precursor of JA before reduction and β-oxidation. However, the above results indicate that OPDA functions as a signaling molecule independently of JAs, suggesting that some structural peculiarity of OPDA is recognized in plants. As shown in Figure 1, there are two major differences in structure between OPDA and JA. First, OPDA has a double bond in the cyclopentenone ring, which forms an α, β-unsaturated carbonyl structure. Various compounds, such as acrolein, having this type of structure can induce gene expression in plants (Alméras et al., 2003). We therefore tested whether ORGs could be induced by acrolein (Fig. 6A). As shown in Figure 6B, acrolein induced the expression of ORGs. Secondly, we noted that the length of the carbon chain with the carboxyl group in OPDA is longer than that of JA. To determine whether this structural difference underlies the transcriptional activation function of OPDA, we analyzed the effect of 3-oxo-2-(2′-pentenyl)cyclopentane-1-octadecanoic acid (OPC8:0) and 3-oxo-2-(2′-pentenyl)cyclopentane-1-hexanoic acid (OPC6:0; downstream intermediates of JA biosynthesis; Fig. 1) on the expression of ORGs. Figure 6C shows that OPC8:0 and OPC6:0, like OPDA, induced the expression of ORGs, although JA with a shorter carbon chain has no effect on the gene expression. These results suggest that the peculiar induction activity of OPDA not only derives from the α, β-unsaturated carbonyl structure but also from the presence of a longer carbon chain than in JA.
Figure 6.
Response of ORGs to structural analogs of OPDA. A, Structure of acrolein, showing the α, β-unsaturated carbonyl group. B, Ten-day-old wild-type (Col) plants were treated with 30 μm acrolein for 0 (control), 15, 30, 60, 180, or 360 min. Five micrograms of total RNA was loaded per lane, and the blots were hybridized with the indicated probes; actin8 served as a loading control. C, Ten-day-old wild-type (Col) plants were treated with 30 μm OPC8:0 or OPC6:0 for 0 (control), 15, 30, 60, 180, or 360 min. Five micrograms of total RNA was loaded per lane, and the blots were hybridized with the indicated probes; actin8 served as a loading control.
DISCUSSION
OPDA Regulation of Gene Expression Is Distinct from JAs
JAs are important regulators of plant responses to environmental stresses, such as wounding, insect attack, and infection (Creelman and Mullet, 1997; Reymond and Farmer, 1998). Under these conditions, the expression of genes regulated by JAs is altered (Farmer and Ryan, 1992; Reymond et al., 2004). OPDA also induces gene expression (Stintzi et al., 2001), but the nature of this activity remains largely unknown.
We used genome-wide expression analysis to identify genes that respond to OPDA, JA, and MeJA, and identified a large number of OPDA response genes, ORGs distinct from those that respond to JAs, or JRGs (Fig. 2; Supplemental Fig. 1). The characteristics of ORGs and JRGs differ greatly (Tables I and II; Supplemental Table III). A significant proportion of ORGs are signaling components and transcriptional factors, whereas JRGs mainly include enzymes involved in metabolic pathways for jasmonates, ascorbate, glutathione, and indole glucosinolate (Brader et al., 2001; Sasaki et al., 2001; Turner et al., 2002; Sasaki-Sekimoto et al., 2005). COI1 is an important signaling component of JA-mediated response (Feys et al., 1994; Benedetti et al., 1995). As shown in Figure 5, expression of JRGs is regulated via a COI1-dependent pathway. In contrast, expression of ORGs was COI1-independent, indicating that the OPDA signaling pathway is independent of the signaling pathway(s) for JAs.
The above results indicate that OPDA and JAs have distinct functions in gene regulation, suggesting that OPDA itself functions as a signaling molecule. The α, β-unsaturated carbonyl structure and the length of carbon chain with the carboxyl group in OPDA appear to be important for its signaling function. Various compounds having an α, β-unsaturated carbonyl group are strong inducers of gene expression in plants (Alméras et al., 2003). Consistent with these findings, acrolein, which has an α, β-unsaturated carbonyl group, induced expression of ORGs (Fig. 6B). Membrane fatty acid-derived prostaglandin plays a signaling role in mammals. Among them described to date, the prostaglandin A and J2 series share structural similarity with OPDA in that they have both an α, β-unsaturated carbonyl group and a long fatty acid chain. In prostaglandin J2, the α, β-unsaturated carbonyl group is critical for its biological activities, which include Michel addition of thiol groups of cellular constituents, namely, glutathione, Cys, and proteins (Atsmon et al., 1990). Therefore, the presence of the α, β-unsaturated carbonyl group may also be important for the specific signaling role of OPDA. Interestingly, our data show that OPC8:0 and OPC6:0 also induce ORG expression (Fig. 6C), indicating that the longer carbon chain length than JA is also important. Although the distinct recognition mechanism of OPDA-mediated signaling remains unclear, the α, β-unsaturated carbonyl group and the length of carbon chain might provide OPDA specificity in plants. Differences in the subcellular localization of OPDA and JA may also influence their signaling functions. OPDA is synthesized in chloroplasts (Bell et al., 1995; Laudert et al., 1996; Stenzel et al., 2003), and it has been reported that OPDA also occurs in an esterified form attached to a plastid-specific galactolipid (Stelmach et al., 2001). OPDA is converted to JA via subsequent action by OPR3 and β-oxidation in peroxisomes (Stintzi and Browse, 2000; Strassner et al., 2002). Therefore, the final subcellular site for emission of each signaling compound is different. In addition, OPDA and JA are differentially distributed in tomato (Lycopersicon esculentum) flowers (Hause et al., 2000), suggesting the different role of OPDA other than the intermediate of JA biosynthesis. Therefore, differences in their synthesis and accumulation sites may explain the significance of the two different signals existing in the same plant cell.
OPDA Function in the Wounding Response
JAs are essential in vivo regulators of defense responses (Reymond and Farmer, 1998), particularly the wounding response. Numerous reports have shown that mechanical wounding of leaves causes JAs to accumulate (Creelman et al., 1992; Peña-Cortés et al., 1995; Parchmann et al., 1997; Reymond et al., 2000), which may be the primary signal that activates a set of wound-inducible genes (León et al., 2001). Consistent with these reports, wounding altered the expression of almost one-half of the JRGs in this study. As described in “Results” section, similar proportions of ORGs and JRGs responded to wounding. Moreover, OPDA also accumulates after mechanical wounding of leaves (Reymond et al., 2000), suggesting its involvement in the wounding response. We showed that wound-induced ORG expression decreases by one-half in the aos mutant (Fig. 4A), whereas this was not the case for the opr3 mutant (Fig. 4B). These results indicate that OPDA functions as a signaling mediator in the wounding response through a mechanism that is distinct from that of JAs. Because AtVSP2 activation requires JA synthesis (shown in Fig. 3B), the AtVSP2 mRNA was not detected in either of the mutants that does not accumulate JA (data not shown).
In the aos mutant, approximately 50% of ORG expression remained. Recently, various oxylipins derived from linolenic acid were shown to accumulate and induce gene expression in plants (Howe and Schilmiller, 2002). Therefore, this residual response to wounding in aos could be mediated by other oxylipins derived from linolenic acid. As shown in Figure 4C, ORG expression was observed in the aos mutant treated with linolenic acid, suggesting the action of an oxylipin(s) derived from the linolenic acid without the AOS reaction. There are several candidate oxylipin compounds in this regard. 13-Hydroperoxylinolenic acid, a precursor of OPDA in the JA biosynthetic pathway (Fig. 1), is not only the substrate of AOS but also of enzymes such as hydroperoxide lyase. Moreover, hydroperoxide lyase- or lipoxygenase-derived types of oxylipins accumulate after wounding or upon infection (Croft et al., 1993; Vollenweider et al., 2000; Vancanneyt et al., 2001; Feussner and Wasternack, 2002). Furthermore, free radical-catalyzed, nonenzymatic reactions can produce phytoprostane, which is similar in structure to OPDA (Krischke et al., 2003). Phytoprostane is also known to induce gene expression in plants, although its induction activity is not as strong as OPDA (Iqbal et al., 2004). These oxylipins may account for the ORG expression observed in the aos mutant.
Wounding induced the expression of 74 ORGs. These included many signal transduction components and transcription factors (Tables I and II), suggesting that OPDA is required upstream of the wounding response. Wounding causes the accumulation of reactive oxygen species, resulting in oxidative stress (Olson and Varner, 1993; Orozco-Cárdenas and Ryan, 1999). Other environmental factors, such as drought, heat, cold, and salinity, also cause oxidative stress (Cheong et al., 2002; Fowler and Thomashow, 2002; Xiong et al., 2002). Remarkably, the expression of many of the ORGs identified in this study is regulated by oxidative stress. The MAPK signaling cascade is activated by oxidative stress (Waskiewicz and Cooper, 1995; Jonak et al., 1996; Bögre et al., 1997; Meskiene et al., 1998; Baudouin et al., 1999). Genes responsible for the MAPK cascade, OXI1 (At3g25250), protein kinase family (At4g25390), MAPKKK18 (At1g05100), and protein phosphatase 2C (At3g27140), were among the ORGs we identified. In particular, OXI1 is an essential component of the signal transduction pathway linked to oxidative burst signals in Arabidopsis (Rentel et al., 2004). Oxidative stress also induces calcium signaling in Arabidopsis (Yang and Poovaiah, 2003; Rentel and Knight, 2004). The up-regulated ORGs (Table I) included a large number of calcium signaling components such as the calcium-binding EF-hand family protein (At4g27280, At5g39670), a putative calcium-binding protein (At5g54490), a calmodulin-binding protein (At5g26920), and a putative calmodulin-related protein (At3g01830, At5g42380). The induction profiles of ORGs involved in these signaling pathways were generally rapid and transient (Table I), suggesting that OPDA plays an important role in the early response to oxidative stress in plants. Moreover, several transcription factors identified as ORGs, ZAT10 (At1g27730), AtERF5 (At5g47230), DREB2A (At5g05410), MYB4 (At4g38620), and ZAT12 (At5g59820), are induced under oxidative stresses such as cold, wounding, dehydration, and salinity (Liu et al., 1998; Fujimoto et al., 2000; Ohta et al., 2001; Lee et al., 2002; Vannini et al., 2004; Davletova et al., 2005). Induction of these genes began within 30 min of OPDA treatment. These results suggest that OPDA signaling and subsequent gene expression occur rapidly under oxidative stress conditions such as those occurring after wounding.
We found that OPDA modulates a distinct set of genes via a COI1-independent signaling pathway. We also propose that OPDA plays a role in the wounding response. The precise function of OPDA in physiological events is still largely unknown. In further analyses of OPDA, isolation of mutants in which OPDA signaling or OPDA level is specifically altered will be a key step to complete our understanding of oxylipin signaling in plant cells.
MATERIALS AND METHODS
Plant Materials, Chemical Treatments, and Wounding
For chemical treatments, Arabidopsis (Arabidopsis thaliana; Col Ws, aos, opr3, and coi1-16) were grown in Murashige and Skoog liquid medium (Murashige and Skoog, 1962) containing 1% (w/v) Suc on an orbital shaker under continuous light at 22°C. The jasmonate signaling mutant coi1-16 was kindly supplied by Dr. John Turner (University of East Anglia). After 10 d, the plants were treated with 30 μm OPDA, JA, MeJA, OPC8:0, OPC6:0, linolenic acid, or acrolein. OPDA, OPC8:0, and OPC6:0 were synthesized and characterized for purity as previously described (Ainai et al., 2003). JA, MeJA, linolenic acid, and acrolein were purchased from Sigma-Aldrich. Plants were harvested at 0 (nontreated), 15, 30, 60, 180, and 360 min after treatment. For wounding studies, Arabidopsis (Col, Ws, aos, and opr3 mutants) were grown on Murashige and Skoog medium plates containing 0.8% agarose under continuous light at 22°C. The OPDA biosynthetic mutant aos was a kind gift from Dr. Joon-Hyun Park and Dr. Kenneth A. Feldmann (Ceres), and the JA biosynthetic mutant opr3 was a kind gift from Dr. John Browse (Washington State University). All rosette leaves of 21-d-old plants were wounded several times across the mid-vein with tweezers. The plants were harvested at 0 (unwounded), 15, 30, 60, 180, and 360 min after the wounding treatment.
RNA Isolation
For northern-blot analyses, total RNA was extracted from Arabidopsis by the phenol/SDS method (Chirgwin et al., 1979). For microarray experiments, the extracted RNA was additionally purified using an RNeasy mini kit (Qiagen). Total RNA was purified from wounded plants using the RNeasy mini kit for both microarray and northern-blot analyses.
DNA Microarray Analysis
The Arabidopsis 2 oligo microarray and the Agilent Linear Amplification/Labeling kit (Agilent Technologies) were used for DNA microarray analyses, all of which were conducted in biological duplicate. The quality of total RNA samples was verified using the RNA 6000 Nano Assay (Agilent Technologies). Sample amplification, labeling, and hybridization essentially followed the protocol recommended by Agilent Technologies. Briefly, 500 ng of each total RNA sample was reverse transcribed into cDNA using the T7 promoter primer. Labeled cRNA was synthesized from the cDNA. The reaction was performed in a solution containing dNTP mix, cyanine 3-dCTP (for treated samples) or cyanine 5-dCTP (for untreated samples; Perkin-Elmer), and T7 RNA Polymerase, and incubated at 40°C for 2 h. To remove unincorporated nucleotides, the labeled cRNA was purified using the RNeasy mini kit (Qiagen). Hybridization was performed in 500 μL of a hybridization mixture containing cRNA probes, the labeled orientation marker (Deposition Control SP300; Operon Technologies), and Cot-1 DNA (Invitrogen) at 60°C for 17 h. The glass slides were then washed and scanned using an Agilent microarray scanner (Agilent Technologies). See also Supplemental File 1, MIAME-compliant description.
Normalization and Analysis of Microarray Data
Spot intensities were quantified, background subtracted, and dye normalized by Agilent Feature Extraction software. Using the spot intensity, a base-10 logarithmic value of relative expression to control (RE) of each gene was determined by the same software (data available in Supplemental Table IV). Then we calculated “normalized fold expression” from RE to unify the duplicate data and to compare between treatments using Microsoft Excel as below.
For treatment tr = {OPDA, JA, MeJA }, replicate r = {1,2}, time t = {30 min, 180 min}, and gene g:
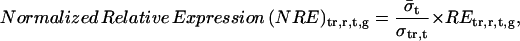 |
where σ is sd. Since OPDA, JA, and MeJA were applied in the same way for liquid cultured plants, for all these data:
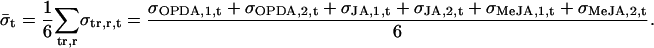 |
Wounding treatment was performed on 21-d-old seedlings grown on MS medium containing agarose. Thus, wounding data were treated independently.
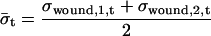 |
We obtained biologically duplicate data for every treatment. To unify the duplicate data, we calculated weighted averages based on a flag (PosAndSignif flag) in Agilent microarray. In the Agilent microarray, if genes showed significantly higher expression than microarray background, the genes were flagged as 1; otherwise 0 for each Green and Red channel. We did not use the data if at least one of the PosAndSignif flags for Green or Red channels was 0. And if the flags for both of the duplicate data were 0, the gene (locus) was removed from all subsequent analyses. For the remaining 20,095 loci, normalized fold expression was determined.
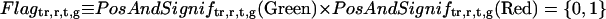 |
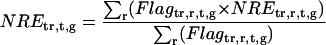 |
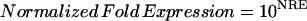 |
Selection of ORGs, JRGs, and WRGs
To screen ORGs, JRGs, and wounding response genes (WRGs), normalized fold expression of each gene was calculated as described above. Then, maximum normalized fold expression (MAF) and minimum normalized fold expression (MIF) were calculated as follows.
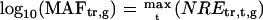 |
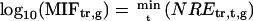 |
The MAF and MIF of OPDA, JA, MeJA, and wound treatment were calculated for each gene (MAFOPDA, MIFOPDA, MAFJA, MIFJA, MAFMeJA, MIFMeJA, MAFwound, and MIFwound, respectively). For ORGs, we selected genes using the following criteria: For up-regulated ORGs, MAFOPDA was more than 3 and both MAFJA and MAFMeJA were less than 2; for down-regulated ORGs, MIFOPDA was less than 0.33 and both MIFJA and MIFMeJA were more than 0.5. A total of 172 genes (157 induced, 15 repressed) fulfilled the criteria. For JRGs, we selected genes having both MAFJA and MAFMeJA of more than 3 or both MIFJA and MIFMeJA of less than 0.33. A total of 449 genes (371 induced, 78 repressed) fulfilled these criteria. For WRGs, we selected genes having MAFwound of more than 3 or MIFwound of less than 0.33; 1,721 genes (1,035 genes induced, 696 repressed, 10 genes overlapped in both categories) fulfilled these criteria.
Northern Blotting
Total RNA (5 μg) was prepared from untreated or treated plants as described. The RNA was electrophoresed on a 1.2% agarose/formaldehyde gel and blotted onto a nylon membrane. Probes were prepared from plasmid DNAs of AV544343, AV521889, AV546998, AV537643, AV544278, and AV532124 (accession nos. reported by Asamizu et al. [2000]) for AtERF5, ZAT10, FAD-OXR, GST6, TCH4, and AtVSP2, respectively. The primer sets DREB2AF (5′-gtgttgttgtattgtagattgtgttg-3′) and DREB2AR (5′-gtcttctctattgtcatatcactgtttcg-3′), and ACTIN8F (5′-cttaggtattgcagaccgtatgagc-3′) and ACTIN8R (5′-gtttttatccgagtttgaagaggct-3′), were used for DREB2A and ACTIN8 genes, respectively. Probes were labeled with [α-32P]dCTP. Hybridization was performed as described (Sasaki et al., 2001). Signals were measured using Image Quant (version 5.1; Molecular Dynamics).
Supplementary Material
Acknowledgments
We thank Dr. J. Browse, Dr. J.H. Park and Dr. K.A. Feldmann, and Dr. J. Turner for kindly providing opr3, aos, and coi1-16 seeds, respectively.
This work was supported in part by the New Energy and Industrial Technology Development Organization, Japan (performed as part of the project Development of Fundamental Technologies for Controlling the Production of Industrial Materials by Plants).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hiroyuki Ohta (hohta@bio.titech.ac.jp).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.067058.
References
- Ainai T, Matsuumi M, Kobayashi Y (2003) Efficient total synthesis of 12-oxo-PDA and OPC-8:0. J Org Chem 68: 7825–7832 [DOI] [PubMed] [Google Scholar]
- Alméras E, Stolz S, Vollenweider S, Reymond P, Mène-Saffrané L, Farmer EE (2003) Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J 34: 205–216 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2000) A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res 7: 175–180 [DOI] [PubMed] [Google Scholar]
- Atsmon J, Sweetman BJ, Baertschi SW, Harris TM, Roberts LJ (1990) Formation of thiol conjugates of 9-deoxy-Δ9, Δ12(E)-prostaglandin D2 and Δ12(E)-prostaglandin D2. Biochemistry 29: 3760–3765 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Zhang ZP, Diab N, Ohnmeiss TE, McCloud ES, Lynds GY, Schmelz EA (1997) Quantification, correlations and manipulations of wound-induced changes in jasmonic acid and nicotine in Nicotiana sylvestris. Planta 201: 397–404 [Google Scholar]
- Baudouin E, Meskiene I, Hirt H (1999) Unsaturated fatty acids inhibit MP2C, a protein phosphatase 2C involved in the wound-induced MAP kinase pathway regulation. Plant J 20: 343–348 [DOI] [PubMed] [Google Scholar]
- Bell E, Creelman RA, Mullet JE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92: 8675–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti CE, Xie D, Turner JG (1995) COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol 109: 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Bell E, Sadka A, Mullet JE (1995) Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol 27: 933–942 [DOI] [PubMed] [Google Scholar]
- Blechert S, Bockelmann C, Füsslein M, Schrader VT, Stelmach B, Niesel U, Weiler EW (1999) Structure-activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta 207: 470–479 [Google Scholar]
- Bögre L, Ligterink W, Meskiene I, Barker PJ, Heberle-Bors E, Huskisson NS, Hirt H (1997) Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell 9: 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G, Tas É, Palva ET (2001) Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol 126: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Chao G, Singh KB (1996) The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J 10: 955–966 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129: 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ (1979) Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18: 5294–5299 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48: 355–381 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Tierney ML, Mullet JE (1992) Jasmonic acid/methyl jasmonate accumulate in wounded soybean hypocotyls and modulate wound gene expression. Proc Natl Acad Sci USA 89: 4938–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft K, Jüttner F, Slusarenko AJ (1993) Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv phaseolicola. Plant Physiol 101: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M (2000) F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol Biol 44: 123–128 [DOI] [PubMed] [Google Scholar]
- Dittrich H, Kutchan TM (1991) Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc Natl Acad Sci USA 88: 9969–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG (2002) A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215: 549–556 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA (1992) Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4: 129–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53: 275–297 [DOI] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwick WH, Moghaddam M, Hamberg M (1991) Oxylipin metabolism in the red alga Gracilariopsis lemaneiformis: mechanism of formation of vicinal dihydroxy fatty acids. Arch Biochem Biophys 290: 436–444 [DOI] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7: 465–471 [DOI] [PubMed] [Google Scholar]
- Hause B, Stenzel I, Miersch O, Maucher H, Kramell R, Ziegler J, Wasternack C (2000) Tissue-specific oxylipin signature of tomato flowers: allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J 24: 113–126 [DOI] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL (2002) Oxylipin metabolism in response to stress. Curr Opin Plant Biol 5: 230–236 [DOI] [PubMed] [Google Scholar]
- Iqbal M, Evans P, Lledó A, Verdaguer X, Pericás MA, Riera A, Loeffler C, Sinha AK, Muller MJ (2004) Total synthesis and biological activity of 13,14-dehydro-12-oxo-phytodienoic acids (deoxy-J1-phytoprostanes). ChemBioChem 5: 1–5 [DOI] [PubMed] [Google Scholar]
- Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H (1996) Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA 93: 11274–11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krischke M, Loeffler C, Mueller MJ (2003) Biosynthesis of 14,15-dehydro-12-oxo-phytodienoic acid and related cyclopentenones via the phytoprostane D1 pathway. Phytochemistry 62: 351–358 [DOI] [PubMed] [Google Scholar]
- Laudert D, Pfannschmidt U, Lottspeich F, Holländer-Czytko H, Weiler EW (1996) Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol Biol 31: 323–335 [DOI] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK (2002) LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J 21: 2692–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León J, Rojo E, Sánchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Bögre L, Glaser W, Balog J, Brandstötter M, Zwerger K, Ammerer G, Hirt H (1998) MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proc Natl Acad Sci USA 95: 1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 493–497 [Google Scholar]
- Ohashi T, Ito Y, Okada M, Sakagami Y (2005) Isolation and stomatal opening activity of two oxylipins from Ipomoea tricolor. Bioorg Med Chem Lett 15: 263–265 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson PD, Varner JE (1993) Hydrogen peroxide and lignification. Plant J 4: 887–892 [Google Scholar]
- Orozco-Cárdenas M, Ryan CA (1999) Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA 96: 6553–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa R, Arimura G, Takabayashi J, Shimoda T, Nishioka T (2000) Involvement of jasmonate- and salicylate-related signaling pathways for the production of specific herbivore-induced volatiles in plants. Plant Cell Physiol 41: 391–398 [DOI] [PubMed] [Google Scholar]
- Parchmann S, Gundlach H, Mueller MJ (1997) Induction of 12-oxo-phytodienoic acid in wounded plants and elicited plant cell cultures. Plant Physiol 115: 1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Peña-Cortés H, Fisahn J, Willmitzer L (1995) Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc Natl Acad Sci USA 92: 4106–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Knight MR (2004) Oxidative stress-induced calcium signaling in Arabidopsis. Plant Physiol 135: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Lecourieux D, Ouaked F, Usher SL, Petersen L, Okamoto H, Knight H, Peck SC, Grierson CS, Hirt H, et al (2004) OXI1 kinase is necessary for oxidative burst-mediated signaling in Arabidopsis. Nature 427: 858–861 [DOI] [PubMed] [Google Scholar]
- Reumann S, Ma C, Lemke S, Babujee L (2004) AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol 136: 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RM, Krishnamurthy V, Dicke M, Farmer EE (2004) A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S, Lapointe G, Rutledge RG, Séguin A (2000) Induction of chalcone synthase expression in white spruce by wounding and jasmonate. Plant Cell Physiol 41: 982–987 [DOI] [PubMed] [Google Scholar]
- Rodriguez Milla MA, Maurer A, Rodriguez Huete A, Gustafson JP (2003) Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J 36: 602–615 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T, et al (2001) Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res 8: 153–161 [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Taki N, Obayashi T, Matsumoto F, Aono M, Sakurai N, Suzuki H, Yokota-Hirai M, Noji M, Saito K, et al (2005) Coordinated activation of metabolic pathways for antioxidants and defense compounds by jasmonates and their roles in stress tolerance in Arabidopsis thaliana. Plant J (in press) [DOI] [PubMed]
- Stelmach BA, Müller A, Hennig P, Gebhardt S, Schubert-Zsilavecz M, Weiler EW (2001) A novel class of oxylipins, sn1-O-(12-oxophytodienoyl)-sn2-O-(hexadecatrienoyl)-monogalactosyl diglyceride, from Arabidopsis thaliana. J Biol Chem 276: 12832–12838 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, Kramell R, Kurz T, Maucher H, Weichert H, Ziegler J, Feussner I, Wasternack C (2003) Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol Biol 51: 895–911 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32: 585–601 [DOI] [PubMed] [Google Scholar]
- Tague BW, Goodman HM (1995) Characterization of a family of Arabidopsis zinc finger protein cDNAs. Plant Mol Biol 28: 267–279 [DOI] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell (Suppl) 14: S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancanneyt G, Sanz C, Farmaki T, Paneque M, Ortego F, Castañera P, Sánchez-Serrano JJ (2001) Hydroperoxide lyase depletion in transgenic potato plants leads to an increase in aphid performance. Proc Natl Acad Sci USA 98: 8139–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37: 115–127 [DOI] [PubMed] [Google Scholar]
- Vollenweider S, Weber H, Stolz S, Chételat A, Famer EE (2000) Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J 24: 467–476 [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Cooper JA (1995) Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol 7: 798–805 [DOI] [PubMed] [Google Scholar]
- Weber H (2002) Fatty acid-derived signals in plants. Trends Plant Sci 7: 217–224 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2003) Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci 8: 505–512 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.